Abstract
Following estrogenic activation, the estrogen receptor-α (ERα) directly regulates the transcription of target genes via DNA binding. MicroRNAs (miRNAs) modulated by ERα have the potential to fine tune these regulatory systems and also provide an alternate mechanism that could impact on estrogen-dependent developmental and pathological systems. Through a microarray approach, we identify the subset of microRNAs (miRNAs) modulated by ERα, which include upregulation of miRNAs derived from the processing of the paralogous primary transcripts (pri-) mir-17–92 and mir-106a-363. Characterization of the mir-17–92 locus confirms that the ERα target protein c-MYC binds its promoter in an estrogen-dependent manner. We observe that levels of pri-mir-17–92 increase earlier than the mature miRNAs derived from it, implicating precursor cleavage modulation after transcription. Pri-mir-17–92 is immediately cleaved by DROSHA to pre-miR-18a, indicating that its regulation occurs during the formation of the mature molecule from the precursor. The clinical implications of this novel regulatory system were confirmed by demonstrating that pre-miR-18a was significantly upregulated in ERα-positive compared to ERα-negative breast cancers. Mechanistically, miRNAs derived from these paralogous pri-miRNAs (miR-18a, miR-19b, and miR-20b) target and downregulate ERα, while a subset of pri-miRNA-derived miRNAs inhibit protein translation of the ERα transcriptional p160 coactivator, AIB1. Therefore, different subsets of miRNAs identified act as part of a negative autoregulatory feedback loop. We propose that ERα, c-MYC, and miRNA transcriptional programs invoke a sophisticated network of interactions able to provide the wide range of coordinated cellular responses to estrogen.
Keywords: AIB1, autoregulatory feedback loop, primary transcript, processing
Upon 17-β-estradiol (E2) binding, estrogen receptors (ERs) mediate transcription by interacting directly to specific estrogen response elements (EREs) located in the promoter/enhancer region of its target genes or indirectly by tethering to nuclear proteins, such as AP1 and SP1 transcription factors (2–4). The cellular response to estrogen is highly regulated at multiple levels including transcription, RNA stability, and posttranslational modifications (5–8). Following treatment with E2, ERα transcription and mRNA stability is substantially reduced within 1 h of stimulation (7). Furthermore, E2–ERα interactions accelerate receptor degradation through the ubiquitin–proteasome pathway, an effect associated with its major coactivator AIB1 (8).
MicroRNAs (miRNAs) are a class of noncoding short RNAs, 21–24 nucleotides (nt) in length, that play a role in gene regulation. They downregulate expression of their target genes by base pairing to the 3′-UTR of target messenger RNAs (mRNAs) (9). During their biogenesis most miRNAs are transcribed as part of a longer transcript named pri-miRNA (10). These molecules are processed inside the nucleus by DROSHA, producing a pre-miRNA that is a 70-nt “imperfect” stem loop RNA actively transported into the cytoplasm. In the cytoplasm the pre-miRNA is cleaved by DICER, a dual processing event that releases a small double stranded RNA, about 22 nt in length. Here, nuclear processing activity is thought to be regulated at early stages of development and in a variety of tumor cells (11–13). There is also evidence of regulation at the next step, pre-miRNA precursor processing (14, 15). After formation of the small duplex RNA, only 1 strand is loaded onto a miRNA induced silencing complex (RISC). These RISCs, guided by their miRNA, interact with the 3′-UTR or sometimes with the coding region of target mRNAs, inhibiting protein translation or degrading the mRNA target (10).
Substantial data associate changes in miRNA activity with carcinogenesis and progression (16–19). The human mir-17–92 cluster is a polycistronic gene with a chromosomal location 13q31-q32 that encodes 6 miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92–1). Ancient duplications have given rise to 2 mir-17–92 paralogues in mammals: mir-106b-25 and mir-106a-363. Mir-17–92 is thought to be oncogenic in lung cancer and lymphomas (17, 20) or function as a tumor suppressor in breast cancer by downregulating AIB1 and/or cyclin D1 (21, 22). Furthermore, the genomic 13q31 area including mir-17–92 is correlated with loss of heterozygosity in breast cancer (23).
By a genome wide approach, we have elucidated the miRNAs regulated by ERα in breast cancer. Here, we show that among the few miRNAs upregulated by ERα, miR-18a encoded by the pri-mir-17–92, miR-19b encoded by both this primary transcript and its evolutionary paralogue pri-mir-106a-363, and miR-20b encoded by pri-mir-106a-363, downregulate ERα expression at the protein translational level, correlating the induction of these 2 genes during cell proliferation with a negative feedback loop. Remarkably, miR-20b also downregulates and targets the ERα coactivator AIB1. Since ERα can act as a ligand-activated oncogene, we suggest that the pri-mir-17–92 acts as a tumor suppressor in breast cancer, not only by downregulating cyclin D1 and AIB1 via the miR-17/20/106 family, but also by downregulation of ERα by miR-18, miR-19, and miR-17/20/106 members. For the first time we correlate ERα translational control by miRNAs as a further regulatory process involved in ERα transcriptional activity after ligand stimulation.
Results
ERα Induction Reveals pri-mir-17–92 Upregulation.
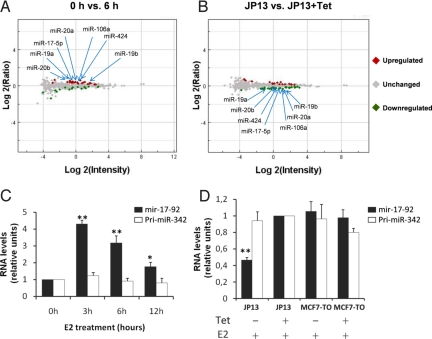
To determine whether ERα regulates the expression of miRNAs, E2 was added to MCF-7 cells and miRNA chip hybridization was performed to elucidate early (0–3 h) and delayed (>6 h) regulation of miRNAs by ERα. As a control we used an MCF-7-TO (MCF-7-Tet-Off)-derived cell line, JP13, that conditionally overexpresses a protein composed of the zinc finger transcriptional repressor PLZF fused to ERα (PLZF-ERα), acting as a dominant-negative that inhibits expression of estrogen-regulated genes and estrogen-stimulated growth of MCF-7 cells (24). Following E2 stimulation and before microarray hybridization, we assessed the reliability of the system using quantitative real time PCR (RT-qPCR) to reveal expression levels of the ERα-regulated gene GREB1 [supporting information (SI) Fig. S1A and ref. 25]. GREB1 expression was reduced in PLZF-ERα cell lines treated without doxytetracycline (Tet) compared to cell lines that do not express the fusion protein (Fig. S1B). Although following array analysis we did not reveal any miRNAs with expression changes greater than 2-fold comparing 0 h to 3, 6, and 12 h (P < 0.05) (Table S1), we found that those miRNAs that increased following E2 in MCF-7 cells decreased in the JP13-Tet-Off system used as a control (Fig. 1A and B and Table S1). The ERα-upregulated miRNAs were generated by the processing of 3 paralogous primary miRNAs: pri-mir-17–92, pri-mir-106a-363, and pri-mir-106b-25 (Fig. 1 A and B, Table S1 and Fig. S2).
Fig. 1.
Pri-mir-17–92 is increased by E2 and decreased by overexpression of PLZF-ERα. (A) MCF-7 cell lines underwent E2 stimulation (10 nM) after 72 h of hormone deprivation. After total RNA extraction and labeling we used a microarray platform containing probes for 470 human miRNAs. After hybridization and scanning, raw data were imported into the Rosetta Resolver system for analysis. A P < 0.01 was used as cut-off for identification of miRNAs upregulated or downregulated between 0 h versus 6 h. (B) JP13 cells were cultured in the presence or absence of Tet for 72 h, followed by the addition of 10 nM E2 for 24 h before microarray analysis. Once again a P < 0.01 was used as cut-off for identification of miRNAs downregulated in JP13 + Tet versus JP13 − Tet. (C) MCF-7 cells were maintained in DMEM (minus phenol red) supplemented with 10% charcoal-dextran FBS for 3 days and then were either left untreated or treated with 10 nM E2 for the indicated time periods. After total RNA extraction, expression of pri-mir-17–92 and pri-miR-342 was analyzed by RT-qPCR using SYBR green and normalized to GAPDH. (D) JP13 and MCF-7-TO cells were cultured in the presence or absence of Tet for 72 h, followed by the addition of 10 nM E2 for 24 h. Once again, after total RNA extraction, expression of pri-mir-17–92 and pri-miR-342 were analyzed by RT-qPCR using SYBR green and normalized to GAPDH. The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM. For RT-qPCR data, the asterisk indicates P < 0.05 in comparison to time 0 h, the double asterisk represents P < 0.005 in comparison to time 0 h. P values were obtained using a 2-tailed Student's t-test.
To confirm the change of expression detected by the microarray, we performed RT-qPCR choosing those miRNAs modulated between 1.2- to 2-fold in cells treated with E2 and those repressed similarly by PLZF-ERα in JP13 cell lines. Firstly, we examined the expression of the unprocessed pri-mir-17–92 and family members. Pri-mir-17–92 appeared upregulated within 3 h of E2 treatment reaching a 4- to 5-fold change in comparison to 0 h (Fig. 1C) defining it as a new early ERα-regulated gene. Levels of expression were significantly repressed by PLZF-ERα (Fig. 1D). Pri-mir-342, a negative control, showed no changes (Fig. 1 C and D), and we obtained the same results normalizing the value of expression for GAPDH, for the snRNA U6 and for the snoRNA U47. We excluded from this analysis the paralogous pri-mir-106b-25 because the fold change of the miRNAs encoded by it were considered too low by our pre-established criteria (Table S1). Furthermore, levels of expression of the pri-mir-106a-363 in MCF-7 cells appeared too low to detect (we were not able to amplify it using 7 different sets of primers from 7 genomic regions). However, it is known that in the P493–6 B cell line that although there is c-MYC-regulated expression of typical miRNAs encoded by both pri-mir-17–92 and pri-mir-106a-363, it is possible to detect the pri-mir-17–92 but not pri-mir-106a-363, indicating that the latter could either be less expressed or alternatively processed more rapidly (26). Furthermore, miR-424, miR-450, and miR-542–3p located within 6 kb of the same genomic region, appeared significantly upregulated by E2 and significantly repressed by PLZF-ERα in a perfectly reciprocal manner (Table S1). We also observed a subset of miRNAs that were subtly downregulated by the E2-ERα complex from 0 to 12 h, and the majority of these belonged to the miR-181 family (Table S1).
Next, we performed RT-qPCR for miR-18a, miR-19a, miR-19b, miR-20a, miR-92 (derived from pri-mir-17–92), miR-19b, miR-20b, miR-92 (from pri-mir-106a-363), miR-424, and miR-181b; the 2 techniques showed an overall correlation (Fig. S3 A–I and Table S1). Comparing the low levels of expression of the miRNAs to the higher levels of the pri-miRNA after stimulation, there was negative regulation of miRNA biogenesis, following transcriptional induction by ERα (Fig. 1C and Fig. S3).
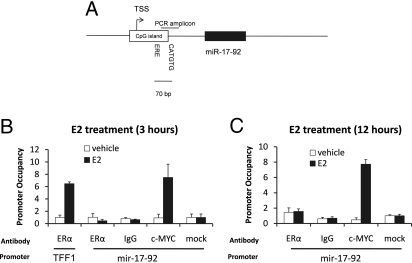
C-MYC Directly Regulates the pri-mir-17–92 upon Estrogenic Stimulation.
It has already been demonstrated that pri-mir-17–92 is transcriptionally regulated by c-MYC in the P493–6 B cell line during the G1–S cellular transition phase (26). Since c-MYC mRNA is upregulated by ERα within 1 h of E2 treatment in breast cancer cells (27), c-MYC could contribute to the increased transcription of the pri-mir-17–92 upon E2 stimulation. Interestingly, we observed a half site conserved ERE 70 bp upstream of the c-MYC consensus site (E-box) of the mir-17–92 promoter (Fig. 2A and Fig. S4). Using cycloheximide (CHX), we demonstrated that new protein synthesis is not required exclusively for pri-mir-17–92 expression (Fig. S5) and because it has been demonstrated that estrogen responsive genes can contain both ERα and c-MYC binding elements located within close proximity [13–214 bp within the promoter and regulated by both transcription factors in an E2-dependent manner (28)], we performed chromatin immunoprecipitation (ChIP) assays for both ERα and c-MYC: coprecipitated DNA was analyzed by amplifying the genomic region containing both consensus sites (Fig. 2A and Fig. S4) by real time PCR (Fig. 2 B and C). Although TFF1, a known estrogen-regulated gene, is confirmed here as regulated by ERα (Fig. 2B), we observed only c-MYC interacting with the mir-17–92 promoter region analyzed (Fig. 2 B and C). We demonstrated that c-MYC is recruited to the mir-17–92 promoter in breast cancer cells upon E2 stimulation.
Fig. 2.
c-MYC directly regulates the pri-mir-17–92 upon estrogenic stimulation. (A) Schematic representation of the mir-17–92 cluster genomic region. Both the c-MYC binding site and a putative ERE half site are indicated. (B) MCF-7 cells were maintained in estrogen-free medium for 3 days (starvation) and then either left untreated (vehicle) or treated with 10 nM E2 for 3 h after which ChIP was performed, followed by real time PCR. The c-MYC interaction site genomic region is presented. (C) After starvation, MCF-7 cells were treated with E2 for 12 h before ChIP.
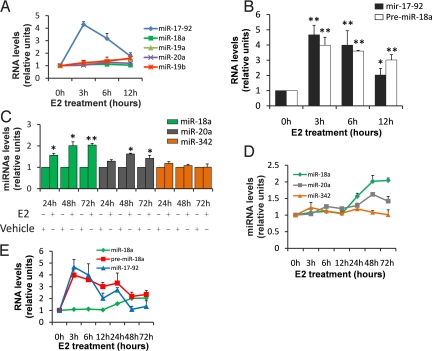
Pri-mir-17–92 Is Negatively Regulated Following DROSHA Cleavage Prolonging miRNA Maturation over Time.
Remarkably, the pri-mir-17–92 expression is striking compared to the miRNAs that are produced by its processing (miR-17, miR-18a, miR-19a, miR-19b, and miR-20a), indicative of modulation of miRNAs biogenesis at the posttranscriptional level (Figs. 1C, 3A, and Fig. S3). A primary transcript undergoes a dual processing event, the first in the nucleus by DROSHA (pre-miRNA production), the second in the cytoplasm by DICER. To define the step(s) of miRNA biogenesis in which regulation occurs, we measured levels of the pri-miR-17–92-derived pre-miR-18a after E2 treatment. DROSHA pri-mir-17–92 cleavage to pre-miR-18a was not a regulatory or “rate-limiting” step here because both were induced at similar levels (Fig. 3B). However, the primers used to amplify the pre-miR-18a also amplify pri-mir-17–92. Therefore, to establish that we could distinguish between pri- and pre-miRNA, we stimulated the cell lines with E2 and then separated the small RNA fraction from the large RNA fraction. We used the large RNA fraction to measure pri-mir-17–92 and the small RNA fraction to measure pre-miR-18a (Fig. S6). As a further control we measured the pri-mir-17–92 from the small RNA fraction without obtaining any amplification product. These data demonstrated that pri-mir-17–92 is induced by the E2–ERα complex, then it is processed by DROSHA releasing the pre-miR-18a, but the passage between pre-miR-18a and miR-18a is attenuated until at least 12 h following initial E2 stimulation. Furthermore, using RT-qPCR, we found that both miR-18a and miR-20a mature forms increase their levels of expression from 24 to 72 h after E2 stimulation (Fig. 3 C and D). Analyzing the levels of the pri-mir-17–92 and the pre-miR-18a from 0 to 72 h, we observed that pri-mir-17–92 is transcriptionally upregulated after 3 h, then DROSHA promptly processes the pri- to the pre-miR-18a, whereas the formation of the mature form from the pre-miR-18a is delayed (Fig. 3E). In addition while the miR-18a levels start to increase at 24 h, both pri-mir-17–92 and pre-miR-18a levels decline, indicative of the processing delay we observed (Fig. 3E).
Fig. 3.
Pri-mir-17–92 is negatively regulated following DROSHA cleavage prolonging miRNA maturation over time. (A) Comparison of the levels of expression between pri-mir-17–92 (normalized to GAPDH) and miRNAs encoded from this cluster (normalized to U47). The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM. (B) After starvation expression levels of both pri-mir-17–92 and pre-miR-18a has been analyzed by RT-qPCR using SYBR green and normalized to U6 snRNA followed E2 treatment as indicated. The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM. The asterisk indicates P < 0.05 in comparison to time 0 h, the double asterisk represents P < 0.005 in comparison to time 0 h. P values were obtained using a 2-tailed Student's t-test. (C) After starvation expression levels of miR-18a, miR-20a, and miR-342 were analyzed by RT-qPCR and normalized to U47 snRNA. The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM. The asterisk indicates P < 0.05 in comparison to vehicle treatment, the double asterisk represents P < 0.005 in comparison to vehicle treatment. P values were obtained using a 2-tailed Student's t-test. (D) Representation of miR-18a, miR-20a, and miR-342 levels (normalization to U47 snoRNA) from 0 to 72 h of E2 treatment by RT-qPCR. The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM. (E) Representation of the miR-18a (normalization to U47), pre-miR-18a and pri-mir-17–92 levels (normalization to U6) from 0 to 72 h of E2 treatment. The mean of 3 experiments each performed in triplicate are presented, error bars represent SEM.
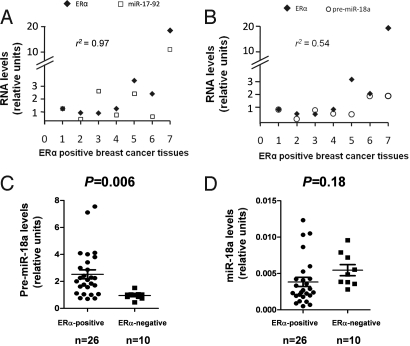
Pri-mir-17–92 Expression Is Correlated with ERα Levels in ERα-Positive Primary Breast Cancers.
To evaluate ERα modulation of the pri-mir-17–92 at the physiologic level, we examined a correlation between ERα mRNA and pri-mir-17–92, and ERα mRNA and pre-miR-18a, in breast cancer tissues by RT-qPCR. Levels of pri-mir-17–92 were correlated with ERα mRNA in tissues (r2 = 0.97, P = 0.0002, Fig. 4A), further indicating that ERα regulates the expression of this primary miRNA. However pre-miR-18a was less correlated with ERα (r2 = 0.54, P = 0.21, Fig. 4B). Next, we addressed whether pre-miR-18a, miR-18a, and miR-20a were differentially expressed in primary breast cancer tissues, comparing the average expression levels between ERα-positive and -negative tumors. Pre-miR-18a levels were significantly higher in ERα-positive tumors (2.52 ± 0.30) compared with negative tumors (0.90 ± 0.08, P = 0.006, Fig. 4C), supporting our data. Moreover, expression levels of miR-18a showed no significant differences between the 2 groups of samples (Fig. 4D), indicating that impaired pre-miR-18a processing to miR-18a occurs in tumors.
Fig. 4.
ERα modulates pri-mir-17–92 in breast cancer tissues. (A) Expression levels of ERα and pri-mir-17–92 (Pearson correlation 0.97) or (B) pre-miR-18a (Pearson correlation 0.54) was measured by RT-qPCR in ERα-positive breast cancers. (C) RT-qPCR showed that expression levels of pre-miR-18a are significantly higher in ERα-positive than in ERα-negative tumors (unpaired, 2-tailed Student's t-test P = 0.006). Error bars represent SEM. (D) RT-qPCR showed that expression levels of miR-18a are not different between ERα-positive and ERα-negative tumors (unpaired, 2-tailed Student's t-test P = 0.18). Error bars represent SEM.
MiR-18a, miR-20b, and miR-19b Negatively Modulate the ERα Transcriptional Activity After Estrogen Stimulation.
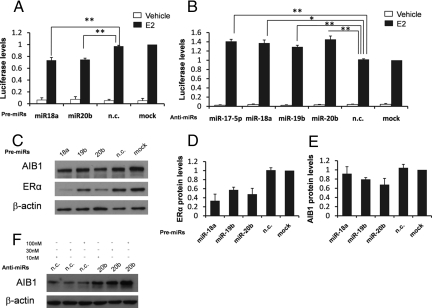
Using the available miRNA target prediction software [TargetScan (29), Pictar (30), and Pita (31)], we observed whether ERα is a potential target of some or all of these miRNAs. Surprisingly, we found that miR-18, miR17/20/106, and miR-19 family members were predicted to target ERα. To experimentally validate this prediction, we chose miR-18a encoded by pri-mir-17–92, miR-19b encoded by both pri-mir-17–92 and the pri-mir-106a-363, and miR-20b encoded by the pri-mir-106a-363 (Fig. S2). First, we addressed whether these miRNAs influence ERα transcriptional activity. MELN cells (MCF-7 cells, stably transfected with a luciferase reporter gene under the control of an ERE using the β-globin promoter) were transfected with pre-miR-18a, pre-miR-20b, and pre-miR-negative control (pre-miR-n.c.). E2-stimulated reporter activity was significantly reduced when MELN cells were transfected with pre-miR-18a and pre-miR-20b, whereas the level of induction was not affected by pre-miR-n.c. (Fig. 5A). Remarkably, anti-miR-18a, anti-miR-20b, and anti-miR-19b molecules able to silence their miRNA function significantly increased reporter activity (Fig. 5B). The effect of miRNA silencing on luciferase reporter activity was similar to treatment with anti-miR-17–5p, previously reported to reduce the transcriptional activity of ERα by downregulating the coactivator AIB1 (21) (Fig. 5B).
Fig. 5.
MiR-18a, miR-19b, and miR-20b suppress ERα-mediated signaling. (A) Luciferase activity in MELN cells untransfected or transiently transfected for 48 h with pre-miR-18a, pre-miR-20b, and pre-miR-n.c. in the absence or presence of 10 nM of E2 for 24 h. (B) Luciferase activity in MELN cells transiently transfected for 48 h with anti-miR-17–5p, anti-miR-18a, anti-miR-19b, anti-miR-20b, anti-miR-n.c. or untransfected in the absence or presence of 10 nM of E2 for 24 h. (C) Western blot showing ERα, AIB1, and β-actin in MCF-7 cells untransfected or transiently transfected with pre-miR-18a, pre-miR-19b, pre-miR-20b, and pre-miR-n.c. (D) Densitometric analysis of ERα Western blot shown in C normalized to β-actin. (E) Densitometric analysis of AIB1 Western blot (shown in C normalized to β-actin. The mean of 3 independent experiments are presented, error bars represent SEM. (F) Western blots showing AIB1 and β-actin in MCF-7 cells transfected with anti-miR-n.c. and anti-miR-20b at 10, 30, and 100 nM concentrations. One representative experiment from 3 independent experiments is shown.
Mir-17–5p, miR-106b, and miR-20a are able to negatively regulate AIB1 protein translation by a direct interaction with the 3′-UTR of AIB1 mRNA (21, 22, 32). Because we observed that miR-17/20/106 and the miR-18 family members potentially target ERα, we evaluated whether the reduction in ERα transcriptional activity induced by miR-20b overexpression was the result of the contemporary negative regulation of AIB1 and ERα and in addition, whether the reduction in ERα transcriptional activity induced by overexpression of miR-18a was the result of a reduction of ERα protein levels. To address if these miRNAs negatively regulate either ERα and/or AIB1, we overexpressed pre-miR-18a, pre-miR-19b, pre-miR-20b, and pre-miR-n.c. and measured protein levels. ERα was markedly reduced by the overexpression of all 3 premiRs analyzed in comparison to either untransfected or pre-miR-n.c. transfected cells although the reduction with pre-miR-19b was less pronounced (Fig. 5 C and D). Furthermore, miR-20b downregulated AIB1 because transfection of pre-miR-20b into MCF7 cells reduced AIB1 protein levels (Fig. 5 C and E). On the other hand, the transfection of anti-miR-20b increased AIB1 (a dose–response was also observed here; Fig. 5F). Because a reduction in either ERα or AIB1 mRNA levels after transfection of precursors was not observed, it appears likely that this regulation occurs at the protein translation step (Fig. S7 A and B).
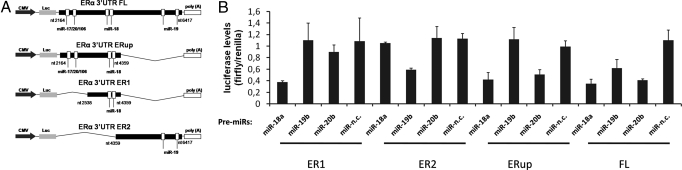
To confirm whether these miRNAs directly target ERα, we inserted into the luciferase reporter vector 4 fragments of the 3′-UTR of ERα: the full length (FL, containing all of the putative miRNAs interaction sites), the first half part of the 3′-UTR (ERup for miR-17/20/106 and miR-18), a fragment containing just the putative miR-18 family interaction sites (ER1), and finally a fragment corresponding to the second half section, containing the miR-19 family interaction sites (ER2) (Fig. 6A). Transfection of miR-18a, miR-19b, and miR-20b, derived from pri-mir-17–92 and pri-mir-106a-363, were used to investigate direct interactions with the 3′-UTR of ERα constructs and we demonstrated that these miRNAs profoundly downregulate luciferase activity for the constructs containing miRNA interaction sites, but not for the ones in which these sites are absent (Fig. 6B). This indicates direct targeting of ERα by a number of miRNAs derived from these paralogous primary miRNAs. We did not observe any downregulation of luciferase reporter activity upon miR-17–5p overexpression, according to a recent report (33).
Fig. 6.
ERα is directly regulated by miRNA-3′-UTR interaction. (A) Representation of the 4 different lengths of ERα 3′-UTR cloned in the pMIR-REPORT lucifearse vector and miRNAs interaction sites. (B) Luciferase activity from cells cotransfected with pre-miR-18a, pre-miR-19b, pre-miR-20b, pre-miR-n.c., and different lengths of DNA fragments corresponding to the ERα 3′-UTR. Firefly luciferase was normalized for transfection levels to Renilla luciferase as indicated in experimental procedures.
Discussion
In this study we were able to classify miRNAs upregulated by estrogen as the members encoded by the paralogous transcripts pri-mir-17–92 and pri-mir-106a-363. For individual miRNAs small changes were observed, but as multiple mature molecules derived from these primary transcripts target ERα and/or AIB1 this increases both the overall level of the miRNAs regulating these 2 proteins after E2 induction and the effects of silencing; it is known that multiple molecules affecting a single target increase their inhibitory effect (34).
Changes in pri-mir-17–92 were significantly greater than the miRNAs derived from it, implicating inhibition during miRNA biogenesis: DROSHA cleavage of pri- to pre-miRNAs occurred rapidly, indicating that this step is not rate limiting (Fig. S8). Such regulation however has been described regarding let-7 family members during stem cell differentiation: LIN-28 is able to interact with pri-let-7 and/or pre-let-7 impairing its processing (12, 15). It has also been reported that c-MYC downregulates let-7 maturation increasing the transcription of LIN-28b in P493–6 B cell lines (35). Furthermore, it has been shown that the RNA binding protein KSRP, interacting with DICER, promotes the biogenesis of a subset of miRNAs comprising miR-20a and miR-106a in both HeLa and NIH 3T3 cells (36). Because we did not observe any estrogen-mediated upregulation of LIN-28 and/or any expression of LIN-28b, or any estrogen-mediated downregulation of DICER and/or KSRP in our models, this indicates that these factors are not responsible. Our data indicate that pri-mir-17–92 (not only let-7), is regulated after induction. Additionally, many expression studies note discordance in the levels of mature miRNAs derived from polycistronic precursors. Although regulation of pri-mir-17–92-derived microRNAs could not be explained by the candidate factors we tested, the apparent prevalence of regulated miRNA maturation strongly suggests involvement of additional RNA binding proteins in this process.
The importance of miRNA activity in breast cancer biology is also highlighted by the finding that a number of miRNAs show a differential expression between ERα positive and ERα negative breast cancers (37, 38). We demonstrated that pri-mir-17–92 expression is highly correlated with the level of ERα in breast cancers, and that pre-miR-18a derived from DROSHA-pri-mir-17–92 cleavage is also significantly more expressed in ERα-positive compared to ERα-negative tumors. This indicates that a specific increase of this pri-miRNA also occurs in physiologic conditions. It is interesting that miR-18a produced by pri-mir-17–92 is not expressed preferentially in ERα-positive tumors. This further suggests that ERα-positive tumors escape the inhibitory targeting of ERα caused by miRNAs by in turn downregulating DICER processing of those miRNAs during tumor progression. Here we demonstrate that the factors implicated in attenuation of miRNA processing are also active in cancer tissues themselves.
The modulation of the pri-mir-17–92 by ERα appears mediated by the c-MYC oncogene by its direct interaction with the mir-17–92 promoter. It has been reported that c-MYC directly downregulates the expression of a set of miRNAs in B cells (39). Because we have not observed any reduction of those after estrogenic stimulation, we conclude that the upregulation of pri-mir-17–92 through ERα-c-MYC is specific to breast cells.
By forming a complex with several coactivators or corepressors, ERα transcriptionally modulates several genes implicated in cell proliferation and apoptosis such as BCL2, c-MYC, and cyclin D1. AIB1, SRC1, and TIF2 belong to the same family of coactivators that interact and collaborate with ERα in the transcriptional regulation of target genes (40). MiR-17–5p and miR-20a encoded by pri-mir-17–92, and the homologue miR-106b, downregulate the translation of AIB1 (21, 22, 32). Because following E2-mediated upregulation: (i) miR-18a, miR-19b, and miR-20b downregulate ERα and (ii) miR-20a, miR-17–5p, miR-106a, and miR-20b downregulate AIB1, we conclude that both primary transcripts are implicated in the regulation of ERα transcriptional activity upon estrogenic stimulation. Several studies have indicated that after estrogenic induction, both ERα and AIB1 are rapidly downregulated. This attenuation occurs at transcriptional, posttranscriptional, and posttranslational levels (5–8). We propose here the translational regulation by miRNAs as a further step of ERα transcriptional activity attenuation after estradiol-mediated ERα activation. Interestingly, this regulation occurs especially at a later time and in a negative feedback loop because DICER pri-mir-17–92 processing appeared inhibited after early ERα upregulation (Fig. S8).
Methods
MiRNA Microarray.
Isolated RNA was labeled using the Agilent labeling kit following the manufacturer's instruction (Agilent Technologies). The Agilent human (V1) miRNA microarray platform, containing probes for 470 human (and 64 viral miRNAs from the Sanger database v9.1), was used to perform miRNA expression profiling.
RT-qPCR Assay.
For RT-qPCR assays, cDNA was synthesized from 1 μg of purified Dnase-treated RNA by the SuperScript III First-Strand cDNA synthesis system (Invitrogen); RT-qPCR was performed on a 7900HT Thermocycler using the Power SYBR green PCR master mix (both from Applied Biosystems). For detection of mature miRNAs, the TaqMan MicroRNA assay kit (Applied Biosystems) was used. Sequences of primers used are provided in Table S2.
ChIP.
Cross-linked chromatin was prepared from MCF-7 cells as described previously with minor modifications (43). Aliquots of 20 μg were incubated overnight with 2 μg of c-Myc (sc-764) and ERα (sc-543) antibodies (Santa Cruz Biotechnology) or without (mock controls) in a total volume of 1 mL and immunoprecipitated. Triplicate samples of 5 μL of immunoprecipitated genomic DNA were amplify by real time PCR. Values are expressed as fold of enrichment with respect to input DNA. Primer sequences used in this assay are listed in Table S2.
Supplementary Material
Acknowledgments.
We thank Simak Ali, Jesus Gil, Justin Sturge, and Ernesto Yagüe for their critical reading of the manuscript. We are indebted to the advice, expertise, and time of Carmelo Ferrai, Dan Stoicescu, and Joshua T. Mendell. We thank Laki Buluwela for the JP13 cell lines. We are grateful to the Breast Cancer Campaign for a small pilot grant supporting this work and to the family of Terry Cadbury and Lord David Alliance, CBE.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906947106/DCSupplemental.
References
- 1.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 2.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kushner PJ, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 4.Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- 5.Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–2330. [PubMed] [Google Scholar]
- 6.Read LD, Greene GL, Katzenellenbogen BS. Regulation of estrogen receptor messenger ribonucleic acid and protein levels in human breast cancer cell lines by sex steroid hormones, their antagonists, and growth factors. Mol Endocrinol. 1989;3:295–304. doi: 10.1210/mend-3-2-295. [DOI] [PubMed] [Google Scholar]
- 7.Saceda M, et al. Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol Endocrinol. 1988;2:1157–1162. doi: 10.1210/mend-2-12-1157. [DOI] [PubMed] [Google Scholar]
- 8.Shao W, Keeton EK, McDonnell DP, Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc Natl Acad Sci USA. 2004;101:11599–11604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 16.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 17.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 21.Hossain A, Kuo MT, Saunders GF. Mir-17–5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Z, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eiriksdottir G, et al. Mapping loss of heterozygosity at chromosome 13q: Loss at 13q12–q13 is associated with breast tumour progression and poor prognosis. Eur J Cancer. 1998;34:2076–2081. doi: 10.1016/s0959-8049(98)00241-x. [DOI] [PubMed] [Google Scholar]
- 24.Buluwela L, et al. Inhibiting estrogen responses in breast cancer cells using a fusion protein encoding estrogen receptor-alpha and the transcriptional repressor PLZF. Gene Ther. 2005;12:452–460. doi: 10.1038/sj.gt.3302421. [DOI] [PubMed] [Google Scholar]
- 25.Rae JM, et al. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 26.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 27.Santos GF, Scott GK, Lee WM, Liu E, Benz C. Estrogen-induced post-transcriptional modulation of c-myc proto-oncogene expression in human breast cancer cells. J Biol Chem. 1988;263:9565–9568. [PubMed] [Google Scholar]
- 28.Cheng AS, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 32.Petrocca F, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol. 2009;29:3783–3790. doi: 10.1128/MCB.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Chang TC, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 38.Mattie MD, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 41.Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol Cell Biol. 2009;29:1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.