Abstract
The histone H3 variant CENP-A is required for epigenetic specification of centromere identity through a loading mechanism independent of DNA sequence. Using multiphoton absorption and DNA cleavage at unique sites by I-SceI endonuclease, we demonstrate that CENP-A is rapidly recruited to double-strand breaks in DNA, along with three components (CENP-N, CENP-T, and CENP-U) associated with CENP-A at centromeres. The centromere-targeting domain of CENP-A is both necessary and sufficient for recruitment to double-strand breaks. CENP-A accumulation at DNA breaks is enhanced by active non-homologous end-joining but does not require DNA-PKcs or Ligase IV, and is independent of H2AX. Thus, induction of a double-strand break is sufficient to recruit CENP-A in human and mouse cells. Finally, since cell survival after radiation-induced DNA damage correlates with CENP-A expression level, we propose that CENP-A may have a function in DNA repair.
Keywords: chromatin, DNA repair
DNA repair in chromatin is thought to occur in a stepwise manner. Cells must recognize a damage event, and recruit repair and chromatin machinery to the site of damage. Damage signaling includes phosphorylation of ATM, which phosphorylates Chk2, H2AX, Nbs1, and many other proteins [reviewed in (1)]. Chromatin remodeling provides access to the damaged DNA [reviewed in (2, 3)]. According to current models, chromatin is restored some time after repair to the DNA is completed (4).
The reported kinetics of chromatin remodeling at sites of DNA damage span minutes to hours. Histone H2AX phosphorylation is absent from up to 6 kb on either side of a double-strand break, but spreads outward at least 40 kb on both sides (5, 6). After UV damage, new histone H3.1 appears approximately 30 min after damage (4), presumably due to reassembly after DNA repair. In contrast, bound H2B was detected surrounding double-strand breaks for up to 4 h, then replaced 10 h later (7). Finally, despite triggering DNA damage signaling, unprotected telomere free DNA ends do not induce detectable chromatin turnover at all (8). Thus, the extent and kinetics of histone turnover and replacement to sites of DNA damage are currently unclear.
Centromere protein A (CENP-A), a component of centromeric chromatin, is an essential histone H3 variant in all eukaryotic species examined to date. CENP-A is known to be required for (9, 10), and may be sufficient to promote (11, 12), centromere identity and assembly of the associated kinetochore protein complex, which mediates chromosome segregation during cell division. Since centromeric DNA sequences are not conserved in metazoans [reviewed in (13)], CENP-A presumably exerts its role in centromere specification via a sequence-independent mechanism. Previously, we observed that widespread DNA damage induced assembly of Xenopus CENP-A onto sperm DNA in cell-free egg extracts (14). Here we tested the hypothesis that localized DNA damage is sufficient to recruit human and mouse CENP-A in vivo.
Results
Endogenous CENP-A Recruited to DNA Damage Sites.
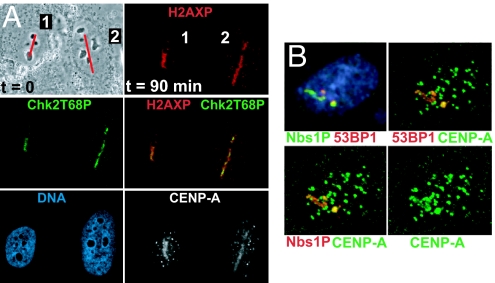
To generate double-strand breaks in DNA in living human cells, we used the second harmonic (532 nm) of a pulsed (12 ps) Nd:YAG laser (15). This strategy avoids local heating (15), and differs from approaches that use higher energy UV (337 nm) light, which produce damage not confined to the nuclear interior. We deliberately avoided presensitizing analogues such as BrdU, which can disrupt chromatin (16). After laser targeting along a 0.4 μm-wide line (Fig. 1A), phosphorylation of H2AX [reviewed in (17)] was routinely detected. As expected, no changes in nuclear structure were detected [DAPI, Fig. 1A and (15)]. Although H2AX phosphorylation can occur without DNA damage (18), an antibody against activated Chk2 (19, 20) detected a DNA damage-dependent epitope coincident with phosphorylated H2AX (Fig. 1A). Activated (Ser-1981 phosphorylated) ATM and Rad51, both thought to be bound primarily at double-strand breaks, were also detected (Fig. S1), as were phosphorylated Nbs1 and 53BP1 (Fig. 1B). CENP-A signals in laser targeted lines of interphase cells were observed in almost all cells (87% of 143b and 85% of HeLa, n = 30 and 20, respectively). In all cases, endogenous CENP-A was still detectable at centromeres (smaller foci; grayscale or green in Fig. 1), demonstrating that CENP-A is not removed from centromeres in response to DNA damage.
Fig. 1.
Endogenous CENP-A localizes to sites of laser-induced damage, along with DNA repair markers phosphorylated H2AX, Chk2, Nbs1, and 53BP1. (A) (Top left) Phase contrast image of human osteosarcoma (143b) cells just before laser targeting along a line (red). (Other panels) Matched confocal immunofluorescent imaging 90 min (at 25 °C) after laser targeting. (Red) Phospho-histone H2AX; (green) Thr-68 phospho-Chk2; (blue) DNA detected with DAPI; (grayscale) endogenous CENP-A signal. Note the small foci in the CENP-A image are centromeres. Note that the cells are motile: the orientation of cell #2 changed in the 90 min after targeting. (B) HeLa cell 90 min. (at 25 °C) after laser targeting along a single line. (Blue) DNA, (red) 53BP1 or Nbs1 phospho-Ser-343, and (green) CENP-A. Other foci in the CENP-A image are centromeres.
Rapid Accumulation of GFP-CENP-A at Sites of DNA Damage.
To examine the kinetics of CENP-A targeting to sites of DNA damage, laser targeting was performed on two clonal human Hek293 cell lines inducibly expressing a GFP-CENP-A fusion protein (10) after FRT-mediated integration at a defined locus. Similar results were obtained with both lines. CENP-A mRNA increased approximately 7-fold within 24 h of induction (Fig. S2A), and the 44 kDa GFP-CENP-A protein accumulated to approximately the initial level of endogenous CENP-A, which was in turn reduced to about one third of its earlier level (Fig. S2B). The decrease in endogenous CENP-A may be due to competition between transfected and endogenous CENP-A for stabilization by CENP-A binding factors, as seen previously (3, 21–24).
Within 4 hours of induction, GFP-CENP-A (Fig. S2 C and D, green) colocalized with endogenous centromeres (detected using human anti-centromere autoantisera Fig. S2C, red), and was distributed throughout nuclei (Fig. S2D), consistent with prior reports (25, 26). After longer induction times (>24 h), all cells exhibited centromeric foci surrounded by a generalized nuclear signal. GFP signal was not removed by extraction with non-ionic detergents before fixation (Fig. S2, compare fixed cells in C with live cells in D). The generalized nuclear signal (Fig. S2D) represented full-length GFP-CENP-A, not a degraded form just containing GFP, since the majority of protein migrated at the expected molecular weight for GFP-CENP-A (≈45 kDa). Endogenous CENP-A (>80%) was retained in the pellet after washing with high salt and detergent (consistent with assembly into nucleosomes). In addition, while half the GFP-CENP-A was retained under these stringent conditions (Fig. S2E), the remainder was removed, consistent with a proportion of CENP-A outside of centromeric chromatin (11, 14, 21, 23, 26).
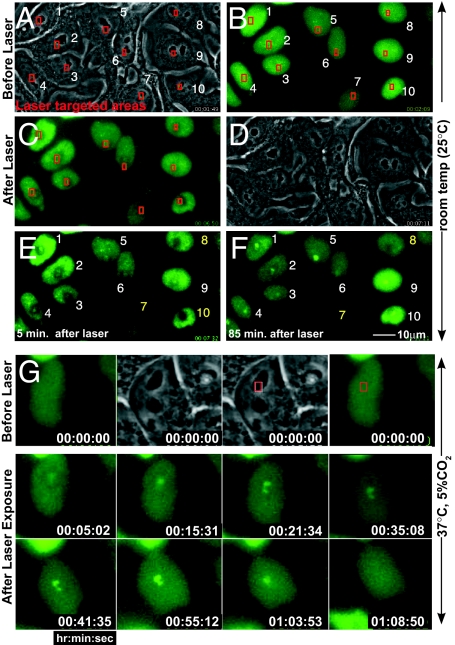
Timing of CENP-A recruitment to sites of DNA damage was determined after induction of GFP-CENP-A expression in the two clonal cell lines. Cells were visualized with phase-contrast and fluorescence microscopy, and areas were chosen for laser targeting (boxes in Fig. 2 A–C). Laser firing produced an initial approximately 1.5 μm2 photobleached area (Fig. 2 C and E), whereas GFP-CENP-A accumulated in a smaller (≈0.6 μm2) spot in the center, consistent with H2AX phosphorylation (see Fig. 4). This was expected since the multiphoton effect that creates double-strand breaks is limited to a smaller volume: comparable photobleaching was observed at approximately 6-fold lower laser doses, but this was insufficient to induce DNA damage, as reported earlier (15). Fluorescence recovery within the photobleached zone required at least 1 h (e.g., cell 8 in Fig. 2 E and F), much longer than the <1 s that would be expected for a soluble approximately 45-kDa protein (27), confirming that most nuclear GFP-CENP-A was not freely diffusible.
Fig. 2.
Rapid GFP-CENP-A accumulation at sites of DNA damage. (A–F) GFP-CENP-A cells before and after laser targeting at 25 °C. Phase contrast (A) before and (D) 5 min. after targeting the areas boxed in red. Epifluorescence images of GFP-CENP-A, immediately (B) before and (C) 4 or (E) 5 min. after initiating laser exposure. In most cells (numbered in white), GFP-CENP-A accumulated at the sites of targeting. [Cells #7, 8 and 10 (numbered in yellow) were bleached during laser targeting,] (F) Within 85 min, GFP-CENP-A formed foci within targeted regions. (G) Laser targeting as in (A–F), maintained at 37 °C after targeting: a CENP-A focus appears within ≈5 min after laser exposure, reaches its peak intensity ≈1 h after laser exposure, and then disappears approximately 15 min later. Timestamp represents hours:minutes:seconds.
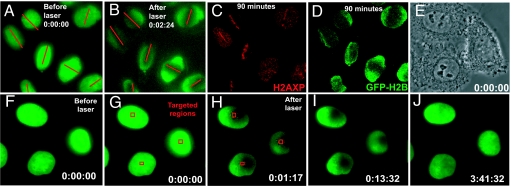
Fig. 4.
Histone H2B never accumulates in areas of laser-induced DNA damage. (A–D) Epifluorescence images of HeLa cells stably expressing YFP-H2B (A) before and (B–D) after laser exposure (red lines). (C) γ-H2AX and (D) YFP-H2B 90 min. after laser exposure. (E) Phase contrast image of HeLa cells stably expressing YFP-H2B before laser targeting. (F–J) YFP-H2B epifluorescence of the cells in (E) and (F and G) before or (H–J) after laser targeting. Red squares in (G and H) denote laser targeted areas. Timestamp is hours:minutes:seconds.
Damage consistently induced CENP-A foci in 71% of targeted cells (±10%, n = 176 interphase cells; for example, eight of 10 cells shown in Fig. 2), within an average of 5 min (±2 min, n = 82 interphase cells), including >30 experiments on separate days (n ≈ 100 cells per experiment). Once formed, each CENP-A focus remained stationary, and increased in intensity for about 1 h (Fig. 2F). Foci formed with identical frequency and kinetics at both room temperature (25 °C) and 37 °C. However, at 37 °C, GFP-CENP-A accumulations at targeted sites appeared, became brighter, and then were abruptly lost (e.g., between 63 min and 68 min in Fig. 2G). Foci were not lost at 25 °C, as some were still visible 18 h later. Cells with foci were never observed to enter mitosis.
The high frequency with which CENP-A accumulated at sites of laser exposure in asynchronous samples suggested that this can occur throughout the majority of interphase, as seen for endogenous CENP-A. Nuclear cross sectional area is known to correlate with cell cycle stage (28). Comparing the nuclear areas of cells that formed foci after laser exposure (n = 31, Fig. S3, red) with the areas of the randomly cycling cells surrounding them (n = 450; Fig. S3, green) confirmed that CENP-A focus formation was not restricted to a subset of interphase cell cycle stages. Conversely, no cell cycle stage (aside from mitosis) was refractory to CENP-A focus formation.
Finally, DNA damage induced focus formation was not a phenomenon limited to immortalized cells. GFP-tagged human CENP-A was transiently transfected into primary human fibroblasts, and laser-induced damage produced CENP-A foci with rapid kinetics (Fig. S4B).
Rapid Accumulation of GFP-CENP-A at Sites of I-SceI Cleavage.
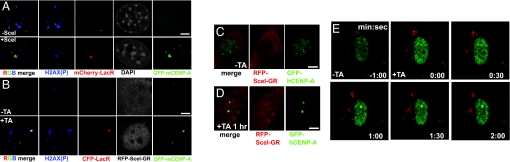
To further test whether a double-strand break is sufficient to recruit CENP-A to DNA, we used the site-specific endonuclease I-SceI (29) in a mouse NIH2/4 cell line (TM815 cells) which carries a single I-SceI target site flanked by LacI repeats (31). A GFP-tagged version of mouse CENP-A (GFP-mCENP-A) was constructed and expressed by transfection in these cells. As expected, GFP-mCENP-A was recruited to sites of laser-induced DNA damage with kinetics similar to GFP-tagged human CENP-A (GFP-hCENP-A) in human cells (Fig. S4A). After transient expression of I-SceI, a single double strand break, marked by the presence of phosphorylated histone H2AX, was generated at the lacI array [visualized using mCherry-lacR (red)] (Fig. 3A). In 47% of cells, GFP-mCENP-A (green) was recruited along with phosphorylated histone H2AX (blue) (Fig. 3A). (The less than 100% efficiency is expected in this triple transfection experiment.) Additionally, TM815 cells were co-transfected with GFP-mCENP-A and an RFP-tagged fusion of I-SceI with the glucocorticoid receptor (30). Before addition of the synthetic glucocoricoid triamcinolone acetonide (TA), RFP-I-SceI-GR was cytoplasmic, and GFP-mCENP-A was detected throughout nuclei (Fig. 3B). Within 1 h after TA addition, RFP-I-SceI-GR translocated into nuclei, and in 57% of the cells (n = 100) phosphorylated histone H2AX appeared at the I-SceI cleavage site (marked with CFP-LacR) along with a large GFP-mCENP-A focus (Fig. 3B).
Fig. 3.
Rapid GFP-CENP-A accumulation at double-strand breaks induced by I-SceI cleavage in human and mouse cells. (A) Mouse cells carrying an I-SceI target site co-integrated with lacO repeats (30) after transient transfection to express (green) GFP-mCENP-A and (red) mCherry-lacR and (top) without or (bottom) with HA-tagged I-SceI. (Blue) γ-H2AX; (grayscale) DNA detected with DAPI. (B) The same mouse cell line as in (A), transiently co-transfected to express (green) GFP-mCENP-A, (red) CFP-lacR and (grayscale) RFP-I-SceI-GR and (top) without TA or (bottom) after TA addition for 1 h. (Blue) γ-H2AX. (C–E) Human cells carrying a single SceI target site on chromosome 10 and multiple SceI target sites on chromosome 6 were transiently co-transfected to express GFP-CENP-A and RFP-I-SceI-GR. (C and D) Cells imaged for (red) RFP-SceI-GR or (green) GFP-hCENP-A and (C) without or (D) 1 h. after addition of TA, which induces nuclear accumulation of RFP-I-SceI-GR to cleave the DNA. (E) Timelapse images of a single cell transfected as in (C and D) immediately before and after addition of TA. Timestamp is minutes:seconds.
GFP-hCENP-A was also recruited to double stranded DNA breaks in a diploid human cell line carrying I-SceI sites at two loci (31). Transient co-transfection (Fig. 3C) was used to express GFP-hCENP-A and RFP-I-SceI-GR (30). Before addition of TA, RFP-I-SceI-GR was detected in the cytoplasm, and GFP-hCENP-A was detected throughout the nucleus, with visible foci at centromeres (Fig. 3C). After addition of TA, RFP-I-SceI-GR became nuclear within 1 h and GFP-hCENP-A formed one or two new foci in 75% of the cells (n = 100 per experiment, repeated three times). Cells with two foci displayed one larger focus and one smaller focus (Fig. 3D), as expected for this cell line with multiple target sites for cleavage on chromosome 6 and a single site on chromosome 10 (31). Remarkably, GFP-hCENP-A formed a focus as rapidly as 1 min after addition of TA (Fig. 3E). Taken together, these results demonstrate that CENP-A is rapidly recruited to defined DNA double-strand breaks.
Histones H3.1 and H2B Do Not Accumulate at Double-Strand Breaks.
Next, we tested whether other histones accumulated at sites of DNA damage. Despite CENP-A accumulation and H2AX phosphorylation at sites of laser targeting (Fig. 1), neither YFP-H2B (n = 72 cells per experiment, repeated 3 times) (Fig. 4) nor YFP-H3.1 (n = 75 cells per experiment; see Fig. S5) accumulated in targeted areas in live or fixed cells. Instead, both YFP-H2B (Fig. 4 and Fig. S6) and YFP-H3.1 (Fig. S5) expressing cells gradually recovered fluorescence in the laser targeted areas, with similar kinetics (3–4 h, n = 10 cells each), consistent with a previous study of chromatin reassembly of fluorescently tagged core histones (32). Neither YFP-H2B nor YFP-H3.1 focal accumulations were observed, even after fixation and staining with anti-GFP antibodies to detect photobleached YFP-tagged histones. Moreover, even using transient transfection to produce a majority of histone H3.1 as a GFP-tagged protein, no GFP-H3.1 foci were ever observed after laser targeting (n = 63 cells; Fig. S7A).
The Centromere-Targeting Domain of CENP-A (the CATD) Can Drive Histone H3 to Sites of DNA Damage.
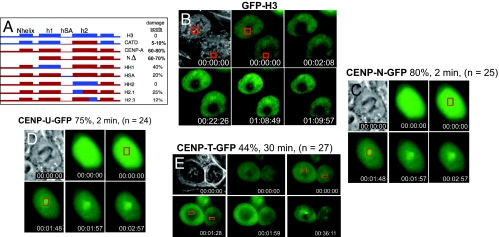
It was reported previously that substitution into histone H3.1 of the CENP-A centromere Targeting Domain, or CATD, the central 31 aa portion of CENP-A (the last 6 residues of α helix1, all of loop 1, and all of α helix 2) is sufficient to promote assembly of chimeric histone H3.1 to centromeres (33). Since CENP-A accumulated at sites of DNA damage in a majority of cells, but histone H3.1 never did, we tested whether this centromere targeting domain would cause accumulation of chimeric H3.1 at sites of DNA damage. Genes encoding a panel of CENP-A:H3.1 chimeric proteins (26, 33) were tagged with GFP and expressed in HeLa and HCT116 cells using transient transfection. The CATD was able to drive recruitment of histone H3.1 to sites of DNA damage, albeit at lower frequency than wild-type CENP-A (Fig. 5 A and B). The complementary mutations in CENP-A, replacing parts of the CATD with the analogous sequences of histone H3.1, reduced or abolished targeting to sites of laser exposure (Fig. 5A, HSA, HH2, H2.1, and H2.3), collectively identifying the α2 helix (mutant HH2) as the most critical region. Thus, recruitment to sites of DNA damage and assembly at centromeres utilizes a common targeting motif within CENP-A.
Fig. 5.
CENP-A recruitment to sites of DNA damage requires its centromere-targeting domain (CATD) and recruits centromeric nucleosome-associated factors CENP-N and CENP-U. (A) Schematic of CENP-A/H3 chimeric proteins and frequencies at which each is recruited to DNA damage foci. (B–E) Human HCT116 cells were transiently transfected. DNA damage was induced by laser targeting, and frequencies of focal accumulation at site of DNA damage were measured. (B) GFP-H3CATD. (C) GFP-CENP-N. (D) GFP-CENP-U. (E) GFP-CENP-T.
Other Centromeric Proteins Are also Recruited to Sites of DNA Damage.
Since CENP-A recruitment to sites of DNA damage and centromeres requires the same domains, we examined whether other centromeric proteins were also recruited to sites of DNA damage. GFP-tagged expression constructs for CENP-N, CENP-U, CENP-T, and CENP-M (22, 34, 35) were each transiently transfected into HCT116 cells, and these cells were subjected to laser exposure. In most (80%) targeted cells, CENP-N-GFP accumulated very rapidly (within 2 min) at sites of DNA damage (Fig. 5C). CENP-U-GFP was recruited to these sites of DNA damage with comparable kinetics (2 min) and as frequently (75%), although the foci were consistently smaller than those formed by CENP-N-GFP (Fig. 5D). CENP-T-GFP accumulated less often (44%) at sites of DNA damage, and only became visible later (30 min on average; Fig. 5E). In contrast, CENP-M-GFP did not accumulate at sites of DNA damage, and its recovery within the bleached areas was very rapid (Fig. S7B)., consistent with freely diffusing protein (27). Taken together, these results demonstrate that, in addition to CENP-A, components of the CENP-ANAC, including CENP-N and CENP-U, are rapidly recruited to sites of DNA damage, while others (e.g., CENP-T) assemble after CENP-A, consistent with the order of assembly at centromeres (34–36).
CENP-A Recruitment to Double-Strand Breaks Is Enhanced by NHEJ but Independent of H2AX.
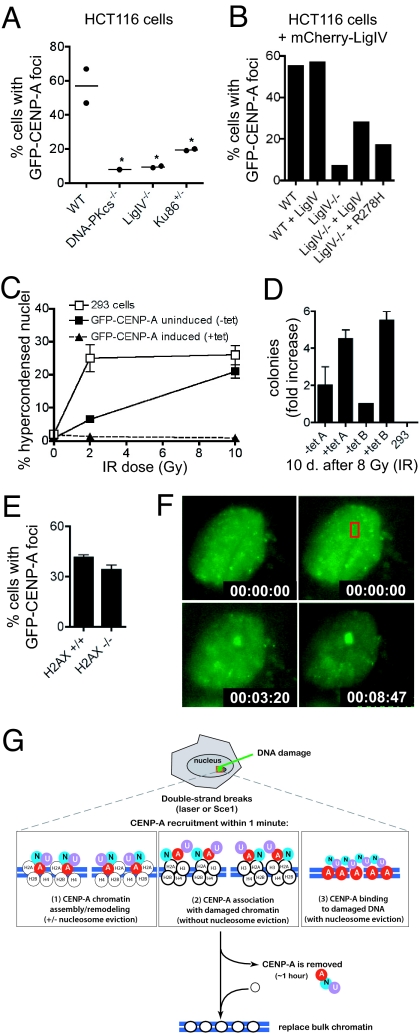
In mammalian cells, non-homologous end joining (NHEJ) is thought to be the predominant pathway for double-strand break repair (37). Early steps in NHEJ involve binding of the Ku86/Ku70 heterodimer to free DNA ends, followed by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and eventual ligation by DNA ligase IV. To measure CENP-A accumulation after laser-induced damage, GFP-CENP-A was transiently transfected into HCT116 cell lines lacking various components of the NHEJ pathway (38, 39). Laser exposure induced GFP-CENP-A foci at high frequency in parental (WT) HCT116 cells (57 ± 10%; Fig. 6A), whereas foci occurred approximately 5-fold less frequently in HCT116 derivatives deficient in Ligase IV−/− or DNA-PKcs−/− (10%; Fig. 6A). In HCT116 cells heterozygous for Ku86 (Ku86+/−), CENP-A accumulation was detected at an intermediate frequency (19%; Fig. 6A). Together, these data suggested that the highest frequency of CENP-A accumulation at sites of damage correlates with activity of the NHEJ DNA repair pathway.
Fig. 6.
CENP-A in function in DNA repair independent of H2AX, and possible models for CENP-A assembly at sites of DNA repair. (A) HCT116 cells generated by targeted disruption of NHEJ genes were transiently transfected to express GFP-CENP-A; frequencies of focus formation were measured after laser exposure. [WT (n = 47 cells); DNA-PKcs−/− (n = 39 cells); LigIV−/− (n = 57 cells); Ku86+/− (n = 38 cells); P = 0.0202 (one-way ANOVA)]. (B) Frequencies of GFP-CENP-A recruitment to sites of laser-induced DNA damage in Ligase IV−/− cells transfected to express mCherry-LigaseIV or mCherry-LigaseIV-R278H. (C) Percentages of cell death following 0 Gy, 2 Gy, and 10 Gy irradiation (done in triplicate) in cells with a stably integrated, GFP-CENP-A gene, either with or without tetracyline-mediated gene induction. Cell death was scored by small, bright nuclei [using Hoechst] and quantified 24 h later by automated microscopy (n = 2,500–5,000 cells per sample). Error bars show the standard devia tion. (D) Cell survival in a colony formation assay after 8 Gy irradiation of parental 293 cells or inducible GFP-CENP-A cells. (E) Frequency of (transiently transfected) GFP-mCENP-A recruitment to sites of laser-induced DNA damage in wild-type and H2AX null MEFs. (F) GFP-mCENP-A recruitment to focal DNA damage (red box) in an H2AX null cell. (G) Model for CENP-A recruitment to sites of DNA damage. Laser-mediated damage or Sce1 cleavage creates DNA double-strand breaks. CENP-A, CENP-N and CENP-U are recruited within 1–2 min. Three possible modes of CENP-A binding are shown (see Discussion).
To further test this hypothesis, GFP-tagged Ligase IV was transiently transfected into WT or Ligase IV−/− HCT116 cells (Fig. S8 A and B). In the absence of DNA damage, GFP-Ligase IV localized throughout the nucleus, with weak GFP signal in the cytoplasm, consistent with a previous report of Ligase IV localization by immunofluorescence (40). After laser targeting, the majority of GFP-Ligase IV was bleached, indicating that it is highly dynamic. GFP-Ligase IV rapidly formed a bright focus at the site of laser exposure in almost all WT (Fig. S8A; 96%, n = 55) and Ligase IV−/− cells (Fig. S8B, 88%, n = 57). Similar foci were detected with a GFP-tagged mutant, R278H, reported to have reduced (5–10% of WT) catalytic activity (40, 41), in both WT (Fig. S8C, 94%, n = 36) and Ligase IV−/− cells (Fig. S8D, 94%, n = 50).
Next, constructs encoding wild-type or mutant Ligase IV fused to mCherry (instead of GFP) were transiently co-transfected along with GFP-CENP-A into HCT116 WT and Ligase IV−/− cells, and double-positive cells were laser targeted (Fig. 6B). As expected, expressing mCherry-Ligase IV did not increase the frequency of GFP-CENP-A foci in WT cells after laser exposure (57%; n = 20), but did increase the frequency in Ligase IV−/− cells (28%; n = 29), while mCherry-Ligase IV-R278H produced an intermediate effect (17%; n = 12) (Fig. 6B).
The observation that CENP-A is recruited to sites of damage so rapidly, and apparently even in the absence of Ligase IV activity, raised the possibility that CENP-A recruitment might participate in DNA repair. Since CENP-A depleted cells or cells unable to load CENP-A are non-viable (24, 42), to test whether CENP-A could promote cell survival after DNA damage, inducible GFP-hCENP-A cells were exposed in triplicate to three doses of ionizing radiation (0 Gy, 2 Gy, and 10 Gy), with or without induction of GFP-hCENP-A. Parental 293 cells (Fig. 6C, open boxes) displayed 26% hypercondensed nuclei, indicative of cell death, within 24 h of 2 Gy, whereas cells induced to express GFP-hCENP-A (triangles) were protected (only 2% hypercondensed nuclei). Uninduced cells exhibited intermediate sensitivity at 2 Gy (5% hypercondensed nuclei), probably due to basal GFP-hCENP-A expression (see below). Both the parental and uninduced GFP-hCENP-A cells exhibited extensive cell death after 10 Gy (Fig. 6C, boxes).
To test whether cells could continue dividing after radiation in a CENP-A-dependent manner, clonogenic survival assays were performed. After 8 Gy, parental 293 cells were uniformly killed, but induction of GFP-hCENP-A promoted cell survival and sustained colony growth (Fig. 6D). Intermediate numbers of surviving colonies were observed with the two independent clonal lines in the absence of tetracycline-mediated induction of GFP-hCENP-A. Survival was accompanied by increased levels of CENP-A (detected as increased GFP fluorescence), (Fig. S9) presumably from transient stabilization of GFP-hCENP-A.
Finally, to test whether CENP-A recruitment requires histone H2AX (5, 6), GFP-mCENP-A was transiently transfected into mouse embryonic fibroblasts (MEFs) from H2AX null mice (43) and cells were laser targeted after 48 h (Fig. 6 E and F). Absence of H2AX did not affect GFP-mCENP-A focus formation, with similar frequencies of rapid recruitment in laser targeted WT (43 ± 3%; n = 65) and H2AX null (32 ± 4%; n = 72) MEFs.
Discussion
Our observations using laser-induced DNA damage and I-SceI cleavage now establish that CENP-A accumulates at double strand DNA breaks in normal and immortalized human and mouse cells. Recruitment of CENP-A, CENP-N and CENP-U occurs within 1–2 min, a timescale as fast as that seen for phosphorylation of H2AX or other components of DNA repair [e.g., frequencies at which each is recruited to DNA damage foci. (B–E) Human HCT116 cells were transiently transfected. DNA damage was induced by laser targeting, and frequencies of focal accumulation at site of DNA damage were measured. (B) GFP-H3CATD. (C) GFP-CENP-N. (D) GFP-CENP-U. (E) GFP-CENP-T.ATM, Nbs1; (44)]. CENP-A recruitment is transient, disappearing again within approximately 1 h, and it is independent of H2AX. A comparison with CENP-A spot size and intensity at centromeres, estimated to contain approximately 5,000–15,000 CENP-A nucleosomes (24, 45), suggests that CENP-A spreads outward surrounding a double-strand DNA break.
Mammalian CENP-A assembles into nucleosomes in vitro (24, 46, 47) and has been found in nucleosomes of asynchronous cells (22, 42, 48, 49). It has also been proposed that CENP-N interacts with CENP-A in nucleosomal form (36). We would caution, however, that it has not been established whether DNA damage-dependent CENP-A loading represents assembly into nucleosomes. At least three models for CENP-A binding are plausible (Fig. 6G). First, CENP-A could be assembled into nucleosomes (Fig. 6G, left). If so, the failure of H2B-GFP to re-accumulate at these damage foci would require that CENP-A must be assembled along with H2B, H2A, and H4 from the original (photobleached) nucleosomes or an adjacent (photobleached) pool. A specific possibility is the selective removal of H3 (with or without H4) from individual nucleosomes and their replacement with CENP-A or CENP-A/H4. Second, CENP-A (and CENP-N and CENP-U) could bind on top of existing chromatin (Fig. 6G, middle). Third, a final alternative (Fig. 6G, right) is that DNA repair occurs in a nucleosome-free zone, as proposed previously (6). This would require removing histone H3-containing nucleosomes and recruiting CENP-A bound in a non-nucleosomal form that lacks H2A and H2B, as has been proposed for budding yeast centromeres (50–52).
While it is clear that CENP-A recruitment to double-strand breaks occurs in both human and mouse cells, including primary cells, the function of CENP-A at double-strand breaks has not been defined. Our demonstration that CENP-A is recruited to double-strand breaks independent of H2AX suggests that it is not part of the damage checkpoint. In contrast, the cell-cycle independence of CENP-A recruitment, rapid kinetics, and correlation with Ligase IV catalytic activity, suggest that CENP-A may be recruited to double-strand breaks along with components of the NHEJ pathway, and raises the possibility that CENP-A participates in DNA repair.
Finally, when added to our earlier demonstration that CENP-A assembly at centromeres of Xenopus sperm requires DNA repair activities stockpiled in egg cytosol (14), CENP-A recruitment with some of its centromeric partners to double strand DNA breaks suggests a mechanism for the formation of new centromeres (neocentromeres), which are only rarely detected as stable events (51). Since the timing of CENP-A disappearance from double-strand breaks correlates with the timing of DNA repair, our evidence provides the initial support for a model in which CENP-A is recruited along with repair machinery, and removed as repair is completed. CENP-A retention at these sites along with CENP-N and CENP-U would provide the nucleus for a new centromere, but only after an extremely rare convergence of permissive conditions. These conditions could include (1) failure to complete DNA repair and remove CENP-A, (2) coincidental timing of DNA damage and centromeric chromatin replication, the latter of which has been argued to occur only during G1 (53), or (3) defective DNA damage checkpoint signaling, allowing cells to enter mitosis prematurely.
Materials and Methods
GFP-CENP-A stable inducible cell lines were generated based on a published plasmid (10) and commercial Flp-In Hek293 cells (Invitrogen). Transient transfections were performed in various cell lines (22, 31, 38, 39) using Lipofectamine2000 (Invitrogen). Laser methods were essentially as recently described (15). Additional materials and methods are discussed in SI Text.
Supplementary Material
Acknowledgments.
We thank the M.W.B. laboratory (University of California, San Diego) and Beth Baber (University of California, San Diego and Salk Institute) for laser sofeware and equipment support.; Annette Ochoa, David Chung and the Willert Laboratory for cell culture reagents and technical assistance (University of California, San Diego); Ainhoa Mielgo and Jeff Lindquist for reagents (University of California, San Diego); Beth Weaver (University of Wisconsin-Madison) for cells; Dan Foltz (University of Virginia), Ben Black (University of Pennsylvania) and Xiaodong Huang (University of California, San Diego) for cells and plasmids; David Chen (UT Southwestern), Scott Stuart (University of California, San Diego) and Kevin Sullivan (National University of Ireland, Galway) for plasmids and antibodies; Kevin Harvey (EMD Biosciences, San Diego), Matt Weitzman (Salk Institute) and Kyoto Yokomori (University of California, Irvine) for antibodies; Eric Hendrickson and York Marahrens (University of Minnesota), Matt Thayer (Oregon Health & Science University) and Andre Nussenzweig (National Institutes of Health) for cell lines. We thank Samarendra Mohanty and Suzanne Genc, Beckman Laser Institute, University of California, Irvine, for help with laser dosimetry and the dual objective method. This work has been supported by a grant from the National Institutes of Health Grant GM 074150 (to D.W.C.). Partial support for M.W.B. came from a grant from the Air Force Office of Scientific Research Grant FA9550-04-1-0101, and from the Beckman Laser Institute Foundation. D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908233106/DCSupplemental.
References
- 1.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Bao Y, Shen X. SnapShot: Chromatin remodeling complexes. Cell. 2007;129:632. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 5.Berkovich E, Monnat RJ, Jr, Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- 6.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodeling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 8.Wu P, de Lange T. No overt nucleosome eviction at deprotected telomeres. Mol Cell Biol. 2008;28:5724–5735. doi: 10.1128/MCB.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugimoto K, Fukuda R, Himeno M. Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct Funct. 2000;25:253–261. doi: 10.1247/csf.25.253. [DOI] [PubMed] [Google Scholar]
- 11.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 13.Murphy TD, Karpen GH. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- 14.Zeitlin SG, Patel S, Kavli B, Slupphaug G. Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair (Amst) 2005;4:760–772. doi: 10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Kong X, et al. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37(9):e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miki K, Shimizu M, Fujii M, Hossain MN, Ayusawa D. 5-Bromouracil disrupts nucleosome positioning by inducing A-form-like DNA conformation in yeast cells. Biochem Biophys Res Commun. 2008;368:662–669. doi: 10.1016/j.bbrc.2008.01.149. [DOI] [PubMed] [Google Scholar]
- 17.Fillingham J, Keogh M-C, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 18.Chadwick BP, Lane TF. BRCA1 associates with the inactive X chromosome in late S-phase, coupled with transient H2AX phosphorylation. Chromosoma. 2005;114:432–439. doi: 10.1007/s00412-005-0029-1. [DOI] [PubMed] [Google Scholar]
- 19.Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 20.Tsvetkov L, Xu X, Li J, Stern DF. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J Biol Chem. 2003;278:8468–8475. doi: 10.1074/jbc.M211202200. [DOI] [PubMed] [Google Scholar]
- 21.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 25.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dundr M, Misteli T. Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol. 2003;13:13.15. doi: 10.1002/0471143030.cb1305s18. [DOI] [PubMed] [Google Scholar]
- 28.Fidorra J, Mielke T, Booz J, Feinendegen LE. Cellular and nuclear volume of human cells during the cell cycle. Radiat Environ Biophys. 1981;19:205–214. doi: 10.1007/BF01324188. [DOI] [PubMed] [Google Scholar]
- 29.Watabe H, Iino T, Kaneko T, Shibata T, Ando T. A new class of site-specific endodeoxyribonucleases. Endo.Sce I isolated from a eukaryote, Saccharomyces cerevisiae. J Biol Chem. 1983;258:4663–4665. [PubMed] [Google Scholar]
- 30.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet. 2005;14:2813–2827. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- 32.Kimura H, Cook PR. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 34.Hellwig D, et al. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J Biophotonics. 2008;1:245–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- 35.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 38.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 41.Riballo E, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 42.Okada M, Hori T, Fukagawa T. The DT40 system as a tool for analyzing kinetochore assembly. Subcell Biochem. 2006;40:91–106. doi: 10.1007/978-1-4020-4896-8_7. [DOI] [PubMed] [Google Scholar]
- 43.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 44.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 46.Conde e Silva N, et al. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 47.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obuse C, et al. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 49.Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 51.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 52.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.