Abstract
Single-nucleotide polymorphism was used in the construction of an expressed sequence tag map of Aegilops tauschii, the diploid source of the wheat D genome. Comparisons of the map with the rice and sorghum genome sequences revealed 50 inversions and translocations; 2, 8, and 40 were assigned respectively to the rice, sorghum, and Ae. tauschii lineages, showing greatly accelerated genome evolution in the large Triticeae genomes. The reduction of the basic chromosome number from 12 to 7 in the Triticeae has taken place by a process during which an entire chromosome is inserted by its telomeres into a break in the centromeric region of another chromosome. The original centromere–telomere polarity of the chromosome arms is maintained in the new chromosome. An intrachromosomal telomere–telomere fusion resulting in a pericentric translocation of a chromosome segment or an entire arm accompanied or preceded the chromosome insertion in some instances. Insertional dysploidy has been recorded in three grass subfamilies and appears to be the dominant mechanism of basic chromosome number reduction in grasses. A total of 64% and 66% of Ae. tauschii genes were syntenic with sorghum and rice genes, respectively. Synteny was reduced in the vicinity of the termini of modern Ae. tauschii chromosomes but not in the vicinity of the ancient termini embedded in the Ae. tauschii chromosomes, suggesting that the dependence of synteny erosion on gene location along the centromere–telomere axis either evolved recently in the Triticeae phylogenetic lineage or its evolution was recently accelerated.
Keywords: dysploidy, linkage map, rice, sorghum, wheat
One of the intriguing attributes of plants is the great variation in genome size among related species. In the grass family, 1C genome size is 389 Mb in rice (Oryza sativa) (1) and 730 Mb in sorghum (Sorghum bicolor) (2) but it approaches 8,000 Mb in some diploid species in the tribe Triticeae (3, 4). The primary cause of these differences is the variation in the amount of repeated sequences present in a genome, principally transposable elements. Transposable elements filling the intergenic space are subjected to a remarkably rapid turnover rate in grasses (5), and a legitimate question is whether or not this rapid turnover rate impacts the stability of gene space. If it does, large plant genomes should show more rapid chromosome evolution, greater erosion of gene colinearity (synteny) along homoeologous chromosomes, and a higher rate of gene duplication and deletion compared with related small genomes. To learn whether these predictions have a material basis and to gain better understanding of the evolution of Triticeae genomes than is afforded by the existing restriction fragment length polymorphism (RFLP) and deletion bin maps (6–10), a high-resolution, expressed sequence tag (EST)-based genetic map of the Aegilops tauschii genome was constructed and compared with the rice (1) and sorghum (2) genome sequences. Because Ae. tauschii is the diploid ancestor of the D genome of common wheat (11, 12), a comparative, high-resolution genetic map will also be a valuable resource for wheat genetics and genomics.
Ae. tauschii and the rest of the Triticeae species are classified in the subfamily Pooideae of the grass family. In most phylogenetic reconstructions, Pooideae forms a clade with the subfamilies Ehrhartoideae (= Oryzoideae) and Bambusoideae (termed the BEP clade) (13). The BEP clade is a sister clade of the second major grass clade that contains subfamilies Panicoideae, which includes sorghum, Arundinoideae, Chlorideae, Centhothecoideae, Aristidoideae, and Danthoideae (termed the PACCAD clade). Triticeae genomes (x = 7) and rice genome (x = 12) appear more related to each other than each is to the sorghum genome (x = 10) (13–17). Phylogenetic reconstructions and comparisons of grass genome structure suggested that the basic chromosome number x was 12 in the common ancestor of Triticeae, rice, and sorghum (10, 13, 18).
The comparison of a large number of genes in a Triticeae genome with rice and sorghum allowed analysis of their linear order and reconstruction of events that resulted in the reduction of chromosome number from 12 to 7 in the Triticeae genomes. It also allowed comparison of the stability of gene space in large and small grass genomes and the erosion of synteny along the centrome–telomere axes of the ancient chromosome arms making up the chromosome arms of modern Triticeae genomes.
Results
Construction of the Ae. tauschii Genetic Map.
A total of 1,536 Illumina GoldenGate single-nucleotide polymorphism (SNP) assays were designed and multiplexed and used to genotype 572 F2 plants of the Ae. tauschii mapping population. Of these SNPs, 153 were not used because they did not detect SNP between the parents of the mapping population or the parents were not homozygous. Of the 1,383 assays used, 1,212 (87.6%) produced satisfactory data. The 1,212 SNPs were in 641 different genes. Additional polymorphisms were assayed with SNaPshot and RFLP (Table 1). Of the 878 loci mapped, 863 were ESTs (Table 1). A map produced from these data consisted of seven linkage groups (Table 1, SI Text, and Table S1) and had a maximum resolution of 0.087 cM.
Table 1.
Summary of the Ae. tauschii genetic maps
| Chromosome | Length, cM | No. loci mapped | Illumina | SNaPshot | RFLP | Sequencing | SSR |
|---|---|---|---|---|---|---|---|
| 1D | 180.5 | 148 | 63 | 2 | 83 | 0 | 0 |
| 2D | 186.9 | 96 | 71 | 2 | 19 | 4 | 0 |
| 3D | 197.0 | 188 | 137 | 2 | 47 | 2 | 0 |
| 4D | 127.4 | 107 | 88 | 4 | 15 | 0 | 0 |
| 5D | 171.2 | 87 | 61 | 15 | 8 | 0 | 3 |
| 6D | 149.3 | 119 | 101 | 2 | 16 | 0 | 0 |
| 7D | 154.5 | 133 | 120 | 0 | 13 | 0 | 0 |
| Total | 1166.8 | 878 | 641 | 27 | 201 | 6 | 3 |
The position of each of the seven centromeres was inferred from the allocation of loci into chromosome arms (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi) and locations of loci on the genetic map reported here. Each centromere was located into an interval flanked by either single or several loci (Table S1) that were immediately distal to the interval containing the centromere.
Orthologous and Paralogous Relationships Among the Ae. tauschii, Rice, and Sorghum Chromosomes.
Among the 863 EST loci, 66% and 64% shared order along the chromosome with genes along sorghum and rice pseudomolecules, respectively (Fig. S1 and Table S1). Paired t test (P = 0.11) failed to show that synteny of the Ae. tauschii chromosomes with rice was greater than with sorghum. In addition to these clearly defined relationships, each Ae. tauschii chromosome or its section showed weaker colinearity with an additional rice and sorghum chromosome or chromosome section (for details see SI Text, Fig. S1, Table S1, and Table S2). An entire rice or sorghum chromosome or its section was also simultaneously colinear with two different Ae. tauschii chromosomes (Fig. S2). As done by Salse et al. (10), the more-related chromosomes will be called orthologous and the less-related chromosomes will be called paralogous. Orthologues are corresponding rice, sorghum, and Triticeae chromosomes that diverged from a single chromosome of the common ancestor of rice, sorghum, and the Triticeae. Paralogues are duplicated chromosomes that originated by paleotetraploidy that preceded radiation of the grass family (19, 20).
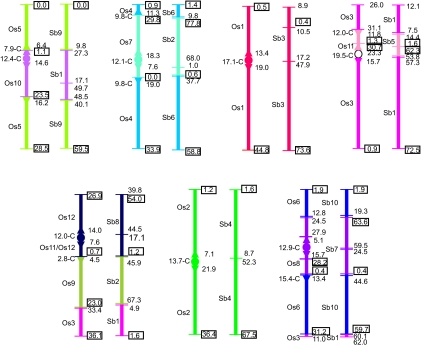
Analyses of orthologous relationships along each Ae. tauschii chromosome shed light on the evolution of the seven chromosomes making up the Triticeae genomes. The summary in Fig. 1, which is based on data in Table S1, shows that chromosomes 3D and 6D were each orthologous along their entire lengths to a single rice chromosome, chromosomes 1D, 2D, and 7D to two complete rice chromosomes, and chromosome 4D to one complete and one incomplete rice chromosome. Chromosome 7D additionally contained a small segment at the tip of the long arm translocated from an interstitial position in chromosome 4D. Chromosome 5D was orthologous to two complete rice chromosomes, Os9 and Os12, and to a portion of Os3 translocated from 4DS to the end of the 5DL arm. Taking into account the 4DS–5DL translocation and neglecting the small 4DL–7DL translocation, chromosomes 4D and 5D also corresponded to two rice orthologues. These syntenic relationships suggested that Ae. tauschii chromosomes 1D, 2D, 4D, 5D, and 7D originated by fusions of two ancestral chromosomes.
Fig. 1.
Orthologous relationships of Ae. tauschii chromosomes (from left to right) 1D–4D (top row) and 5D–7D (bottom row) with rice chromosomes Os1–Os12 and sorghum chromosomes Sb1–Sb10. Ae. tauschii chromosomes are subdivided into arbitrarily colored sections reflecting relationships to rice and sorghum orthologues. Break points and telomeres are indicated by horizontal bars. The coordinates (in Mb) on rice and sorghum pseudomolecules of the first and last gene mapped within an Ae. tauschii section are to the right of each chromosome and are boxed if they are within 2 Mb of the rice and sorghum termini. The solid circles are active Ae. tauschii centromeres unequivocally corresponding to specific rice centromeres, and the open circle is an active Ae. tauschii centromere that could correspond to the centromere of either of two rice chromosomes flanking it. The locations of sorghum centromeric synteny gaps are indicated by single horizontal bars. Triangles symbolize inferred locations of centromeres lost from Ae. tauschii chromosomes. The Mb positions of centromeres (designated C) on rice pseudomolecules are specified to the left of each chromosome.
Each Ae. tauschii chromosome was also paralogous to one or more chromosomes in rice and sorghum. Orthologous and paralogous relationships detected within and among the three genomes are summarized in Table 2.
Table 2.
Summary of main orthologous and paralogous relationships among Ae. tauschii, rice, and sorghum chromosomes or their sections
| Ae. tauschii | Rice | Sorghum |
|---|---|---|
| 1D (3D + 4D) | Os5 (Os1) + Os10 (Os3) | Sb9 (Sb3) + Sb1 (Sb1) |
| 2D (6D + 4D) | Os4 (Os2) + Os7 (Os3) | Sb6 (Sb4) + Sb2 (Sb1) |
| 3D (1D) | Os1 (Os5) | Sb3 (Sb9) |
| 4D (1D + 5D) | Os3 (Os7 + Os10) + Os11 (Os12) | Sb1 (Sb2 + Sb1) + Sb5 (?) |
| 5D (4D + 7D+?) | Os12 (Os11)+ Os9 (Os8) + Os3 (?) | Sb8 (Sb5) + Sb2 (Sb7) + Sb1 (?) |
| 6D (7D + 2D) | Os2 (Os6 + Os4) | Sb4 (Sb10 + Sb6) |
| 7D (6D + 5D + 4D) | Os6 (Os2) + Os8 (Os9) + Os3 (?) | Sb10 (Sb4) +Sb7 (Sb2) + Sb1 (?) |
Across each row, chromosomes in bold are orthologous to complete chromosomes or their sections but paralogous to chromosomes or their sections in parentheses. Chromosomes in parentheses are orthologous across species but paralogous to those in bold. For instance, chromosome 1D consists of two sections orthologous to Os5 and Os10 and Sb9 and Sb1; Os5 is orthologous to Sb9 and Os10 to Sb1. The two 1D sections are paralogous to sections of 3D and 4D, Os1 and Os3 and Sb3 and Sb1. The section of 3D is orthologous to sections of Os1 and Sb3 and the section of 4D is orthologous to sections of Os3 and Sb1. Chromosome Sb1 is paralogous to itself, which is caused by the fusion of orthologues of Os10 with Os3 in the sorghum genome, making up Sb1. In the chromosome 4D row, chromosomes Os7 and Os10 and sections of Sb2 and Sb1 are paralogous to Os3 and Sb1. The results imply that a section of Os7 is orthologous to 1D, for which there is no evidence. The anomaly is caused by the paralogous relationship of a section of 1D to a section of Os3 which, in turn, is paralogous to two chromosomes, Os7 and Os10, within the rice genome (20).
We arrived at the same conclusion as others (10, 13, 18) that the common ancestor of Triticeae, rice, and sorghum had 12 chromosomes (see SI Text for our reasoning). This number was reduced to x = 10 in sorghum and x = 7 in Triticeae.
Mechanism of Dysploid Reduction from x = 12 to x = 7 in Triticeae and x = 10 in Sorghum.
Dysploidy is a change in the basic chromosome number (x) of a genome without concomitant loss or gain of genes (21) The elucidation of dysploid reduction of the basic chromosome number in Triticeae was made possible by locating the sites of the seven active and five lost Triticeae centromeres, the present and past chromosome termini, and their relationships to those in rice and sorghum (Fig. 1). In every Ae. tauschii chromosome, the interval containing the active centromere always included the location of the centromere in the orthologous rice chromosome (Table S1). Loci bracketing a centromere in Ae. tauschii were several Mb apart in the orthologous rice pseudomolecule. For example, the centromere of Os1, which is orthologous to 3D, was at 17.1 Mb (http://rice.plantbiology.msu.edu/pseudomolecules/centromere.shtml). Markers flanking the centromeric interval in 3D were separated by a gap in synteny of 5.6 Mb in the Os1 pseudomolecule (Fig. 1). The same markers were separated by a gap of 30.4 Mb in the orthologous Sb3 pseudomolecule (Fig. 1). Similar gaps in synteny were observed across all active and lost centromeres in the Ae. tauschii chromosomes (Fig. 1). Rationale supporting the locations of active and lost centromeres and present and past termini for each Ae. tauschii chromosome can be found in SI Text. The average gap in synteny was 10.3 Mb across rice orthologous regions corresponding to the active centromeres (Fig. 1). This average gap size did not statistically differ (P = 0.86, t test) from an average gap size of 9.3 Mb across the inferred locations of lost centromeres (Fig. 1). The average synteny gap across all centromeres was 9.9 Mb in the rice pseudomolecules. Similar results were obtained for synteny gaps across the active and lost Ae. tauschii centromeres in comparison with sorghum pseudomolecules. The average gap in synteny was 34.5 Mb across active Ae. tauschii centromeres, which was similar (P = 0.37, t test) to an average gap size of 32.8 Mb across the inferred, lost centromeres. The average size of the gap in synteny across all centromeres was 33.6 Mb. The 3.3-fold expansion of the gaps in sorghum compared with rice is consistent with the observation that size difference between the rice and sorghum genomes is almost entirely accounted for by great expansions of centromeric heterochromatic regions in sorghum (2). The fact that the average synteny gap sizes across the active centromeres were similar to those across the inferred locations of lost centromeres supports the argument that the locations of the lost centromeres in Ae. tauschii chromosomes were inferred correctly.
A total of 27 major translocation breakpoints were identified in the Ae. tauschii chromosomes; 22 took place in the telomeric or centromeric regions and 5 were interstitial. The translocation breakpoints resulted in 12 fusion points (Fig. 1). Nine were fusions of telomeric with telomeric, telomeric with centromeric, and centromeric with centromeric breakpoints and three were fusions involving an interstitial breakpoint.
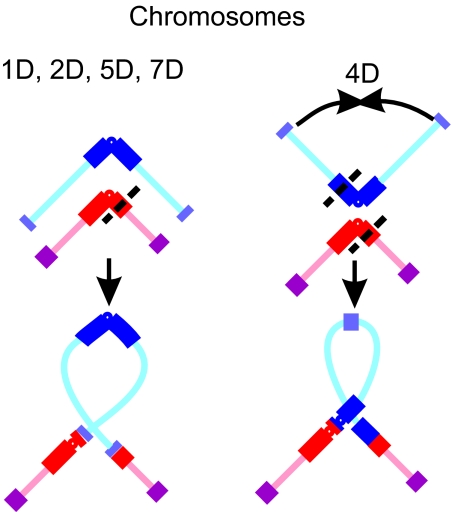
Chromosomes 1D, 2D, and 7D each evolved via insertion of an entire chromosome by its telomeres into a break in the centromeric region of another chromosome (Fig. 2). In 7D, the inserted chromosome underwent several inversions (Table S3) resulting in both termini of the Triticeae orthologue of Os8 being on the same side of the centromere (Fig. 1). The same mechanism very likely also generated chromosome 5D, in which a chromosome corresponding to Os12 was likely inserted into the centromere of the subacrocentric chromosome corresponding to Os9. The Os9 short arm and its centromeric region contain few genes (1). Assuming that the Triticeae orthologue was similar, the Os9 short arm genes may have escaped detection at the tip of 5DS or been deleted. Variation on this theme is present in chromosome 4D, in which the telomeres of the Triticeae orthologue of Os11 fused and its broken centromeric region was inserted into the Triticeae orthologues of Os3 (Fig. 2). Whether this took place in one or two steps cannot be discerned from the present data.
Fig. 2.
Insertional model of dysploid reduction of basic chromosome number. Centromeres are indicated by opened circles, and heterochromatin is indicated by boxes. A break is indicated by a dashed line.
Another variation on the latter theme was observed in sorghum chromosomes Sb1 and Sb2 (for details see SI Text). Chromosome Sb1 originated by the insertion of the entire orthologue of Os10 to the centromeric region of Os3. Before the insertion, however, a pericentric translocation of an Os10 long-arm segment to the short arm facilitated by telomere–telomere fusion took place (Fig. S3). The same event occurred in Sb2, in which an insertion of the Os9 orthologue was accompanied or preceded by pericentric translocation of a distal segment of the Os9 long arm to the short arm and telomere–telomere fusion (Fig. S3).
A feature common to all of these dysploid events is a single break in the centromeric region of a chromosome into which another chromosome is inserted. The insertion that generated Sb2 shows that the centromeric break can take place anywhere in the centromeric region, including a region containing genes. Assuming that the centromere location in the sorghum orthologue of Os7 was the same as in modern Os7, there are many genes between the point of insertion of Os9 orthologue and the Os7 centromere (see SI Text and Fig. S3). The insertion that generated Ae. tauschii chromosome 7D may have also taken place in the centromeric region not in the centromere. The insertion point is separated by two genes from the presumed location of the ancient Os6 centromere. However, the region was subjected to pericentric inversions and other rearrangements and other interpretations are possible (see SI Text).
A by-product of the insertion of a chromosome by its telomeres into the centromeric region of another chromosome is maintenance of the centromere–telomere polarity of chromosome arms in the new chromosome. The ancestor of rice and Triticeae had 24 chromosome arms. After the reduction to 14 arms in Triticeae, 21 of the ancient arms in Ae. tauschii have the same centromere–telomere orientation as in rice.
Relationship Between the Locations of Genes on the Centromere–Telomere Axis and Synteny.
Ancient chromosome arms have either terminal or interstitial positions in the chromosome arms of modern Triticeae genomes. Because gene synteny in wheat is eroded more in the distal portions of modern chromosome arms then in proximal portions (22–25), it is therefore of interest to compare erosion of synteny along the centromere–telomere axes of ancient arms forming the modern Ae. tauschii chromosomes.
To assess synteny along the terminal arms, 13 terminal chromosome segments (the distal segment of 2DS was not investigated because of its short length) were divided into proximal and distal halves on the basis of the numbers of loci on the genetic map. The numbers of syntenic genes between Ae. tauschii and orthologous arms in rice and sorghum were counted in each half. The average synteny with both rice and sorghum chromosomes was significantly higher in the proximal half than in the distal half of the 13 Ae. tauschii chromosome segments (Table 3).
Table 3.
Numbers and percentages (in parentheses) of genes showing synteny in the proximal and distal halves of the most distal segments of Ae. tauschii chromosomes orthologous to rice and sorghum chromosomes
| Ae. tauschii chromosome arm | No. loci | Synteny with rice |
Synteny with sorghum |
||
|---|---|---|---|---|---|
| Proximal half | Distal half | Proximal half | Distal half | ||
| 1DS | 85 | 21 (49.4) | 5 (11.7) | 19 (44.7) | 6 (14.1) |
| 1DL | 41 | 15 (73.2) | 12 (58.5) | 14 (68.3) | 12 (58.5) |
| 2DL | 47 | 21 (89.4) | 12 (51.0) | 21 (89.4) | 12 (51.0) |
| 3DS | 62 | 22 (71.0) | 19 (61.3) | 21 (67.7) | 18 (58.1) |
| 3DL | 124 | 53 (95.5) | 31 (50.0) | 59 (95.2) | 30 (48.4) |
| 4DS | 21 | 10 (95.2) | 7 (66.7) | 7 (66.7) | 7 (66.7) |
| 4DL | 66 | 25 (75.6) | 21 (63.6) | 24 (72.7) | 22 (66.7) |
| 5DS | 34 | 10 (58.8) | 7 (41.2) | 10 (58.8) | 5 (29.4) |
| 5DL | 18 | 6 (66.7) | 8 (88.9) | 5 (55.5) | 9 (100.0) |
| 6DS | 51 | 18 (70.6) | 13 (51.0) | 18 (70.6) | 13 (51.0) |
| 6DL | 66 | 27 (81.8) | 22 (66.7) | 28 (84.8) | 22 (66.6) |
| 7DS | 37 | 15 (81.1) | 11 (59.5) | 13 (70.3) | 12 (64.9) |
| 7DL | 51 | 17 (66.7) | 12 (47.1) | 17 (66.7) | 12 (55.6) |
| Mean | 20.0 (74.2) | 13.8 (55.2) | 19.7 (70.1) | 13.8 (55.6) | |
| P | 0.004 | 0.02 | |||
Synteny was also assessed along the ancient chromosome arms in interstitial positions in the Ae. tauschii chromosome arms 1DL, 2DS, 4DS, 4DL, 5DS, 5DL, 7DS, and 7DL. The 4D section orthologous to Os11 was subdivided into two ancient arms, which were analyzed separately. The numbers of Ae. tauschii loci syntenic with those in rice and sorghum were similar in the distal and proximal halves (Table 4).
Table 4.
Numbers and percentages (in parentheses) of genes showing synteny with rice and sorghum in the proximal and distal halves of chromosome arms embedded in other chromosomes in Ae. tauschii
| Ae. tauschii chromosome arm | No. loci | Synteny with rice |
Synteny with sorghum |
||
|---|---|---|---|---|---|
| Proximal half | Distal half | Proximal half | Distal half | ||
| 1DL | 21 | 5 (47.6) | 8 (76.2) | 4 (38.1) | 9 (65.7) |
| 2DS | 27 | 9 (66.7) | 14 (100.0) | 12 (88.9) | 11 (81.5) |
| 4DS (ancient S) | 8 | 4 (100.0) | 4 (100.0) | 2 (50.0) | 2 (50.0) |
| 4DS (ancient L) | 7 | 4 (100.0) | 3 (85.7) | 3 (85.7) | 3 (85.7) |
| 5DL | 27 | 12 (88.9) | 8 (59.2) | 12 (88.9) | 8 (59.2) |
| 7DS | 34 | 16 (94.1) | 16 (94.1) | 14 (82.4) | 13 (76.5) |
| 7DL | 10 | 5 (100.0) | 5 (100.0) | 4 (80.0) | 3 (60.0) |
| Mean | 7.8 (87.4) | 8.3 (88.4) | 7.3 (73.4) | 7.0 (71.2) | |
| P | 0.71 | 0.78 | |||
Structural Changes in Relation to Genome Size.
A total of 50 structural changes were identified: 31 inversions, of which 3 were pericentric and 28 paracentric, and 19 translocations (Table S1 and Table S3). Most of the translocations resulted in the translocation of entire chromosomes accompanying the dysploid reduction. Of the paracentric inversions, 3 involved 2 loci, 13 involved 3–5 loci, 8 involved 6–10 loci, and 7 involved 11 or more loci (Table S3). Paracentric inversions involving only two or three loci could be errors in map construction. Seven such inversions were included in the data (Table S3). Evidence for inversions BE426301P–BE444599 in 2D, BF478716S–BE585724S in 5D, and BQ161010– BE637570P in 6D was based on a single cross-over each. These inversions are therefore tentative. The remaining four were based on two or more cross-overs. Of the 50 translocations and inversions, 40 originated in the Ae. tauschii lineage, 8 in the sorghum lineage, and 2 in the rice lineage. In the Ae. tauschii lineage, paracentric inversions (24) outnumbered pericentric inversions (3) and translocations (13). Three paracentric inversions and five translocations were detected in the sorghum lineage, and one translocation and one paracentric inversion were detected in the rice lineage. The average length of paracentric inversions in the Ae. tauschii lineage expressed in terms of their lengths in rice pseudomolecules was 1.3 Mb. Because the Ae. tauschii genome is approximately an order of magnitude larger than is the rice genome, the average paracentric inversion in the Ae. tauschii genome was estimated to be 13 Mb long.
Discussion
Map Construction.
Of the 1,383 multiplexed GoldenGate assays used for genotyping, 87.6% generated satisfactory results, and only 0.002% of a total of 365,000 genotype calls did not withstand manual validation and were removed. The success rate was comparable to that obtained in soybean (89%) (26) but was higher than in other plant mapping populations (67% to 77.1%) (27, 28). The lengths of the individual linkage groups constructed from these genotyping data were within the range expected for Triticeae linkage maps and showed no map expansion.
Orthologous and Paralogous Relationships Between the Ae. tauschii, Rice, and Sorghum Chromosomes.
Comparative studies of wheat–rice chromosome relationships based on the wheat EST deletion bin maps (29) revealed orthologous relationships between wheat and rice chromosomes (8, 10). Each of the relationships was confirmed by the Ae. tauschii genetic map. The same orthologous relationships with the rice chromosomes were also revealed by barley EST comparative maps (9) (http://harvest.ucr.edu). A failure to detect an Os11 orthologous region in the 4H chromosome in ref. 9 was the main difference.
Yu et al. (20) identified 18 major duplications within the rice genome and confirmed a hypothesis that the duplications originated by paleotetraploidy (19). Because most of these duplications predate the radiation of grasses (19, 20) they must also be present in the Triticeae genomes. They were detected in both comparisons of the wheat deletion bin maps (10, 30) and the Ae. tauschii genetic map and were responsible for the paralogous relationships between Ae. tauschii, rice, and sorghum chromosomes.
Genome Size and the Rate of Genome Evolution.
Of 50 different inversions and translocations, two and eight were allocated to the rice and sorghum lineages, respectively, and 40 were allocated to the Ae. tauschii lineage. Because divergence of the BEP and PACCAD clades likely preceded the divergence of Ehrhartoideae and Pooideae, the structural changes that took place after the divergence of the two clades but before the divergence of Ehrhartoideae and Pooideae would therefore be incorrectly allocated to the Panicoideae lineage, resulting in overestimation of the number of changes in sorghum. Although the number of structural changes was only slightly higher in sorghum than in rice, there was an order of magnitude more of them in the Triticeae lineage compared with rice and sorghum, indicating that the large sizes of Triticeae genomes are accompanied by accelerated genome evolution. The relationship must be investigated in more grass species and if substantiated it may have important consequences for the understanding of the role of genome size and repeated sequences in genome evolution. A process that accelerates structural genome evolution also likely accelerates gene evolution by duplication, translocations, and deletion of individual genes. An increase in the number of interchromosomal duplications was observed in RFLP maps of diploid species with large genomes, such as einkorn wheat and barley (≈30% duplicated loci), compared with those with small genomes, such as rice (5.6% duplicated loci) and common bean (8.9% duplicated loci) (31). Gene duplications and translocations are important because they can lead to the evolution of new genes (32–34).
The average paracentric inversion in the Ae. tauschii genome was estimated to be 13 Mb long, which is equivalent to ≈3.6 cM. Because of the small sizes of most of the paracentric inversions and the low resolution of the previous Triticeae comparative maps, most paracentric inversions may have previously escaped detection in comparative mapping in the tribe. Structural variation may therefore be more common among Triticeae genomes than currently assumed.
Insertional Dysploidy.
The Triticeae basic chromosome number evolved via the loss of five functional centromeres. Four of them correspond to those of rice chromosomes Os4, Os5, Os6, and Os9 and the fifth could correspond to either that of Os3 or Os11.
The classical “dislocation hypothesis” of dysploid reduction (35) postulates two independent translocations that translocate the euchromatic portion of each arm to other chromosomes. The heterochromatic centromeric region is subsequently lost. Because the process is hypothesized to occur via two independent translocations, the two arms could potentially end up anywhere in the genome and in any orientation.
This is not what was found here. In four of the five chromosomes, both arms were translocated to the same chromosome and were in the original orientation. The likelihood of that happening four times by chance is exceedingly small. The presence of both arms in the same derived chromosome and in the original orientation suggests that each dysploid reduction originated by a single translocation event. In all five derived Triticeae chromosomes, one chromosome was inserted in a single step into the centromeric region of another chromosome. Three dysploid reductions from x = 12 to x = 9 in finger millet (Eleusine coracana) were reported to have taken place by an insertion of a chromosome into the centromeric region of another chromosome (36). Two dysploid reductions in sorghum from x = 12 to x = 10 have also taken place by this process (see SI Text). Finger millet, sorghum, and Triticeae are members of three different grass subfamilies and the two major grass clades. The taxonomical range of the genomes and the fact that all dysploid events in them took place via this process suggest that insertional dysploidy has been a dominant evolutionary mechanism in the grass family.
An insertion of a complete chromosome into a centromeric region is likely to result in a dicentric chromosome. Wheat dicentric chromosomes are stably transmitted because one centromere becomes inactive (37). If this were of a general occurrence in grasses, insertional dysploidy would generate functionally monocentric chromosomes.
The five Triticeae insertions, three finger millet insertions, and two sorghum insertions involved different chromosome combinations and must therefore be independent of each other. Of the 10 dysploid events only one (Sb1) involved paralogues. There is therefore no evidence that homoeology caused by paleotetraploidy of grasses contributed to this process.
Consequences of Dysploid Reduction for the Evolution of Gene Space Along Chromosome Arms.
In wheat and related diploid species, gene deletions and gene duplications preferentially take place in distal, high-recombination regions of chromosomes (22, 24, 38). Synteny between chromosomes is therefore eroded faster in distal, high-recombination chromosome regions than in proximal, low-recombination regions (22, 23, 25).
The terminally located ancient chromosome arms in the Ae. tauschii chromosomes showed greater erosion of synteny in their distal regions than in their proximal regions as expected. In contrast, interstitially located ancient chromosome arms did not show this relationship. This finding suggests that the synteny-erosion pattern observed in the Ae. tauschii and wheat genomes may have evolved recently and may be an attribute of Triticeae genomes. Whether or not the pattern is related to accelerated genome evolution in large grass genomes, and/or the enhancement of the recombination gradient, or some other cause remains an important question for the understanding of grass genome evolution.
Materials and Methods
Genetic Map Construction.
Ae. tauschii accessions AS75 (collected in Shaanxi, China, by C. Yen, Sichuan Agricultural University, Yaan, Sichuan, China) and AL8/78 (collected in Armenia by V. Jaaska, University of Estonia, Tartu, Estonia) were crossed, and DNA was isolated (39) from 572 F2 progeny. A total of 1,536 SNPs were genotyped in the 572 plants with multiplexed GoldenGate SNP assays (Illumina) and 26 SNPs were genotyped in a subset of 174 plants with SNaPshot (Applied Biosystems). RFLP at 201 loci were genotyped as described previously (40). For details of SNP genotyping with GoldenGate and SNaPshot see SI Text.
Graphical genotypes of the 572 plants were arranged so that the number of double cross-overs and singleton loci was minimized. Each cross-over was validated by re-examining genotyping data. Questionable data were excluded. Genetic maps were then constructed with JoinMap4.0 with an initial logarithm of odds score of 10. After a final order of loci was reached, a representative marker from each group of cosegregating markers (termed a recombination block) was selected and a map was constructed by using the Kosambi mapping function (41).
Orthologous and Paralogous Chromosome Relationships.
A homology search was performed between FASTA sequences of wheat ESTs or EST contigs (http://wheat.pw.usda.gov/cgi-bin/westsql/contig.cgi) and O. sativa (IRGSP pseudomolecules Build04) and S. bicolor (http://genome.jgi-psf.org/Sorbi1/Sorbi1.download.ftp.html) genome sequences using an e-10 cutoff. Progression of starting nucleotides along a pseudomolecule paralleling the progression of the genetic map was used as evidence of colinearity. Colinearity was statistically tested by computing the correlation (r) of positions of loci on the genetic map and starting nucleotides on the rice and sorghum pseudomolecules (Table S1). Only chromosomes or chromosome regions with seven or more loci were used in correlation analyses. Regions with large inversions were subdivided into two groups on the basis of gross gene order, and correlations were computed separately for each group of loci. Correlations were computed separately also for translocated regions.
To determine which rice and sorghum pseudomolecule was orthologous and which was paralogous to a specific Ae. tauschii chromosome, the lengths of the concatenated sequences detected by BLASTN were summed and ranked according to their total length. The pseudomolecule (or its portion) with the highest rank was declared orthologous to an Ae. tauschii chromosome and that with second highest rank was declared paralogous (for details see SI Text and Table S2).
Inversions and Translocations.
The order of recombination blocks along an Ae. tauschii linkage group was compared with the order along the orthologous rice and sorghum pseudomolecules. If the order in the rice and sorghum pseudomolecules differed from the order along the Ae. tauschii linkage group, it was assumed that the structural change took place in the Ae. tauschii lineage. If Ae. tauschii and sorghum showed the same order and rice showed a different order, it was assumed that a structural change took place in the rice lineage, and if rice and Ae. tauschii showed the same order and sorghum showed a different order, it was assumed that the change occurred in the sorghum lineage.
Relationship Between Synteny and Location of Loci on the Centromere–Telomere Axis of Ancient Chromosome Arms.
Arms of Ae. tauschii chromosomes 1D, 2D, 4D, 5D, and 7D were subdivided into segments corresponding to the arms of the two (three in the case of 5D) ancient chromosomes making up these five Ae. tauschii chromosomes. Loci in the distal segments of these five chromosomes and both arms of 3D and 6D were divided into distal and proximal halves. The numbers of colinear loci in rice and sorghum pseudomolecules were counted, including those in inverted regions. The significance of the difference between the two halves was assessed by paired t test, using the numbers of syntenic genes in each half as paired variables.
To assess the relationship between synteny and gene location in the ancient chromosome arms embedded in Ae. tauschii chromosome arms 1DL, 2DL, 4DS, 4DL, 5DL, 5DS, and 7DL, each of the interstitial ancient arms was subdivided: half juxtaposed to the ancient terminus and half juxtaposed to the ancient centromere. The numbers of colinear loci in the two halves on rice and sorghum orthologues were counted and significance was assessed as above.
Supplementary Material
Acknowledgments.
We thank C. M. Nicolet and V. Rashbrook (DNA Technologies Core at the University of California Davis Genome Center) for performing the Golden Gate assays, and Professor Peter Langridge (Australian Centre for Plant Functional Genomics, University of Adelaide, Adelaide, Australia) and Patrick S. Schnable (Center for Plant Genomics, Department of Agronomy, Iowa State University, Ames, Iowa) for advising this project. This work was supported by National Science Foundation Grant DBI-0321757.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908195106/DCSupplemental.
References
- 1.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 2.Paterson AH, et al. The sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 3.Bennett MD. DNA amount, latitude, and crop plant distribution. Environ Exp Bot. 1976;16:93–108. [Google Scholar]
- 4.Vogel KP, Arumuganathan R, Jensen KB. Nuclear DNA content of perennial grasses of the Triticeae. Crop Sci. 1999;39:661–667. [Google Scholar]
- 5.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurata N, et al. Conservation of genome structure between rice and wheat. Bio/Technology. 1994;12:276–278. [Google Scholar]
- 7.Devos KM, Gale MD. Comparative genetics in the grasses. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- 8.Sorrells ME, et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein N, et al. A 1,000-loci transcript map of the barley genome: New anchoring points for integrative grass genomics. Theor Appl Genet. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. [DOI] [PubMed] [Google Scholar]
- 10.Salse J, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kihara H. Discovery of the DD analyzer, one of the ancestors of Triticum vulgare (Japanese) Agric Hortic (Tokyo) 1944;19:13–14. [Google Scholar]
- 12.McFadden ES, Sears ER. The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered. 1946;37:81–89. 107–116. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- 13.Barker NP, et al. Phylogeny and subfamilial classification of the grasses (Poaceae) Ann Missouri Bot Gard. 2001;88:373–457. [Google Scholar]
- 14.Salamin N, Hodkinson TR, Savolainen V. Building supertrees: An empirical assessment using the grass family (Poaceae) Syst Biol. 2002;51:136–150. doi: 10.1080/106351502753475916. [DOI] [PubMed] [Google Scholar]
- 15.Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K. Whole chloroplast genome comparison of rice, maize, and wheat: Implications for chloroplast gene diversification and phylogeny of cereals. Mol Biol Evol. 2002;19:2084–2091. doi: 10.1093/oxfordjournals.molbev.a004033. [DOI] [PubMed] [Google Scholar]
- 16.Caetano-Anolles G. Grass evolution inferred from chromosomal rearrangements and geometrical and statistical features in RNA structure. J Mol Evol. 2005;60:635–652. doi: 10.1007/s00239-004-0244-z. [DOI] [PubMed] [Google Scholar]
- 17.Leebens-Mack J, et al. Identifying the basal angiosperm node in chloroplast genome phylogenies: Sampling one's way out of the Felsenstein zone. Mol Biol Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- 18.Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. Plos Genet. 2007;3:1254–1263. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA. Structure and evolution of cereal genomes. Curr Opin Genet Dev. 2003;13:644–650. doi: 10.1016/j.gde.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, et al. The genomes of Oryza sativa: A history of duplications. Plos Biol. 2005;3:266–281. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieger R, Michaelis A, Green MM. Glossary of Genetics and Cytogenetics. Berlin: Springer; 1968. [Google Scholar]
- 22.Akhunov ED, et al. The organization and rate of evolution of the wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 2003;13:753–763. doi: 10.1101/gr.808603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhunov ED, et al. Synteny perturbations between wheat homoeologous chromosomes by locus duplications and deletions correlate with recombination rates along chromosome arms. Proc Natl Acad Sci USA. 2003;100:10836–10841. doi: 10.1073/pnas.1934431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvorak J, Akhunov ED. Tempos of deletions and duplications of gene loci in relation to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics. 2005;171:323–332. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.See DR, et al. Gene evolution at the ends of wheat chromosomes. Proc Natl Acad Sci USA. 2006;103:4162–4167. doi: 10.1073/pnas.0508942102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyten DL, et al. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor Appl Genet. 2008;116:945–952. doi: 10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 27.Pavy N, et al. Enhancing genetic mapping of complex genomes through the design of highly multiplexed SNP arrays: Application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics. 2008 doi: 10.1186/1471-2164-9-21. 10.1186/1471-2164-1189-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckert AJ, et al. High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L.) Tree Genet Genomes. 2009;5:225–234. [Google Scholar]
- 29.Qi LL, et al. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics. 2004;168:701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valarik M, Linkiewicz A, Dubcovsky J. A microcolinearity study at the earliness per se gene Eps-Am1 region reveals an ancient duplication that preceded the wheat–rice divergence. Theor Appl Genet. 2006;112:945–957. doi: 10.1007/s00122-005-0198-6. [DOI] [PubMed] [Google Scholar]
- 31.Dubcovsky J, et al. Genetic map of diploid wheat, Triticum monococcum L, and its comparison with maps of Hordeum vulgare L. Genetics. 1996;143:983–999. doi: 10.1093/genetics/143.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan L, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhunov ED, Akhunova AR, Dvorak J. Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol Biol Evol. 2007;24:539–550. doi: 10.1093/molbev/msl183. [DOI] [PubMed] [Google Scholar]
- 34.Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stebbins GL. Chromosomal Evolution in Higher Plants. London: Edward Arnold; 1971. [Google Scholar]
- 36.Srinivasachary , Dida MM, Gale MD, Devos KM. Comparative analyses reveal high levels of conserved colinearity between the finger millet and rice genomes. Theor Appl Genet. 2007;115:489–499. doi: 10.1007/s00122-007-0582-5. [DOI] [PubMed] [Google Scholar]
- 37.Sears ER, Camara A. A transmissible dicentric chromosome. Genetics. 1952;37:125–135. doi: 10.1093/genetics/37.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dvorak J, Yang Z-L, You FM, Luo MC. Deletion polymorphism in wheat chromosome regions with contrasting recombination rates. Genetics. 2004;168:1665–1675. doi: 10.1534/genetics.103.024927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvorak J, McGuire PE, Cassidy B. Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988;30:680–689. [Google Scholar]
- 40.Luo MC, Yang ZL, Dvorak J. Position effects of ribosomal RNA multigene loci on meiotic recombination in wheat. Genetics. 1998;149:1105–1113. doi: 10.1093/genetics/149.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1943;12:172–175. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.