Abstract
Molecular machines drive essential biological processes, with the component parts of these machines each contributing a partial function or structural element. Mitochondria are organelles of eukaryotic cells, and depend for their biogenesis on a set of molecular machines for protein transport. How these molecular machines evolved is a fundamental question. Mitochondria were derived from an α-proteobacterial endosymbiont, and we identified in α-proteobacteria the component parts of a mitochondrial protein transport machine. In bacteria, the components are found in the inner membrane, topologically equivalent to the mitochondrial proteins. Although the bacterial proteins function in simple assemblies, relatively little mutation would be required to convert them to function as a protein transport machine. This analysis of protein transport provides a blueprint for the evolution of cellular machinery in general.
Keywords: irreducible complexity, protein evolution, protein import, Caulobacter crescentus, TIM23 complex
Molecular machines drive essential biological processes, from protein synthesis and transport to genome maintenance, expression and inheritance (1, 2). Good examples include bacterial flagella (3, 4), the RNA polymerase holo-complex (5), and various protein transport machines that selectively transfer protein molecules across biological membranes (6). Proponents of Intelligent Design have argued that these sophisticated machines are “irreducibly complex,” with this standing as the proof that, at the molecular level, Darwin's principles of evolution cannot explain the complexity of living systems (7, 8). Our current investigation of the function and evolution of the protein transport machines in mitochondria provides an excellent, and perhaps unique, system to provide evidence that a sophisticated molecular machine can evolve from simpler components, in a process strictly adhering to Darwinian principles of evolution.
Mitochondria are essential organelles that provide energy to drive cellular processes and need to be constantly reproduced to ensure cells can divide and grow. Multiple lines of evidence show that the mitochondria in our cells evolved from intracellular bacteria (9–11), and that conversion of these intracellular bacteria into mitochondria required the evolution of protein transport machines (12). We proposed that simple “core” machines were established in the first eukaryotes by drawing on pre-existing bacterial proteins that had previously provided distinct functions. Subsequently, and in a step-wise process in keeping with Darwinian evolution, additional modules would have been added to the core machines to enhance their function. This proposition is supported by 3 findings: (i) that protein components found in bacteria are related in sequence to the components of mitochondrial protein transport machines, but (ii) that these bacterial proteins are not found as part of protein transport machines and (iii) that some apparently “primitive” organisms found today have protein transport machines that function with only one or few component parts.
Protein transport into mitochondria requires the action of the 4 membrane-embedded molecular machines: the TOM, TIM22, TIM23, and SAM complexes (10–13), each composed of up to 8 distinct protein subunits. However, bacteria do not import proteins across their outer and inner membranes, and the TOM and TIM23 complexes that provide this protein import function to mitochondria do not have counterparts in bacteria. The TIM23 complex is specifically responsible for protein transport across the mitochondrial inner membrane. This protein transport machine has been studied in yeast, in plants, and in humans, and is composed of a conserved set of protein subunits that associate together to form the molecular machine (13–15). Three of the subunits are both found in enough representative groups to suggest they are present in all eukaryotic organisms, and are essential for cell viability in yeast: (i) the Tim23 subunit, a simple transmembrane protein that forms the channel through which protein substrates pass into the mitochondrial matrix; (ii) Tim44, found on the inner face of the mitochondrial membrane where it interacts with both Tim23 and Hsp70, thereby docking the protein import motor to the Tim23 channel; and (iii) the Tim14/Pam18 subunit that interacts with several proteins in the TIM23 complex and directly stimulates the ATPase activity of Hsp70, thereby activating the motor to drive protein transport (10, 16–18). Here we show that α-proteobacteria have a protein of the Tim44 family that functions in membrane quality control and a Tim14/Pam18 protein that functions in a distinct process. Together with the LivH amino acid transporter, these component parts would have provided “pre-adaptation” to bacteria ahead of a need for protein import.
Results
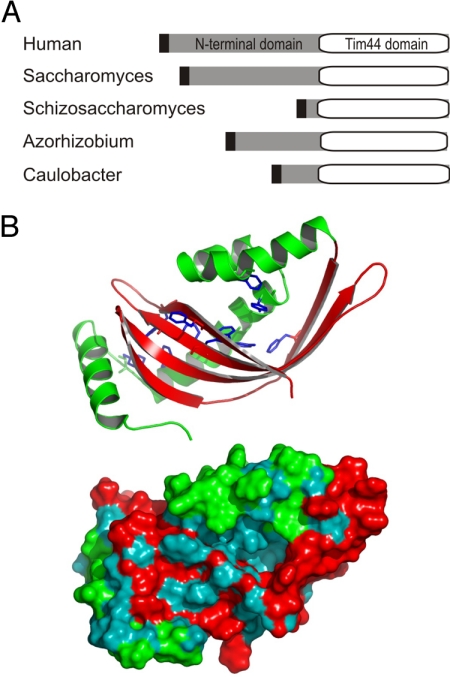
Analysis of genome sequence data shows Tim44 is found in all eukaryotes, with the proteins ranging in size from 25 kDa to 50 kDa (12). An N-terminal segment of variable length (3–30 kDa) is found and the characteristic “Tim44 domain” of approximately 20 kDa is always present (Fig. 1A). The characteristic domain of Tim44 binds lipids and might partially penetrate a monolayer patch of the mitochondrial inner membrane (19). The structure of the Tim44 protein from yeast and from humans has been solved: it shows that Tim44 has a deep hydrophobic pocket that might facilitate lipid binding.
Fig. 1.
Structure of TimA. (A) Representation of the domain organization in the Tim44 family of proteins. Black denotes the inner membrane targeting sequence for mitochondria (humans, Saccharomyces, Schizosaccharomyces) or bacteria (Azorhizobium, Caulobacter). (B) Model of TimA (CC3741) from C. crescentus based on the structures of human and yeast Tim44, represented as cartoon and a solvent-excluded surface. Hydrophobic residues are cyan and other residues are red or green. Aromatic residues (F104, F109, Y117, Y124, F144, F162, W211, F213, W224, and F228) are shown in stick representation. The structure was assessed with the Prosa2003, ProQ, and Verify3D quality scores (see Fig. S1).
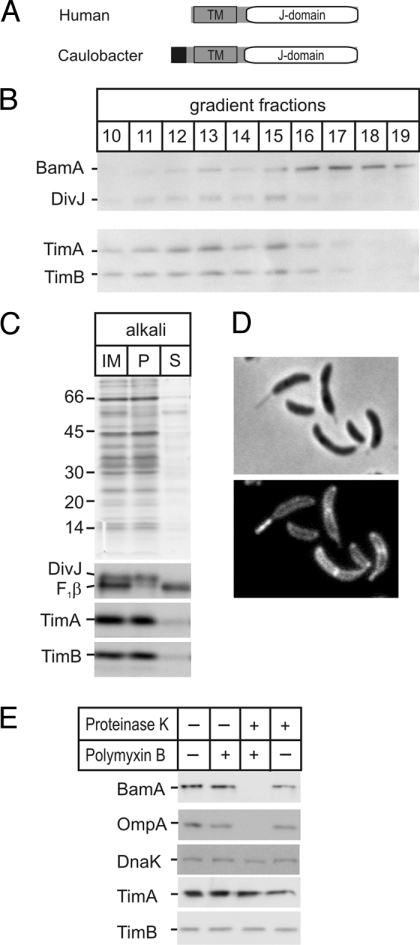
Species of α-proteobacteria, the bacterial group from which mitochondria evolved (7–9), have proteins with sequence similarity to the mitochondrial Tim44 (Fig. 1A) (10). The bacterial protein, which we call TimA, is sufficiently similar to the human/yeast Tim44 to allow structural modeling: Prosa2003, ProQ, and Verify3D quality scores show statistical significance [supporting information (SI) Fig. S1]. Modeling the bacterial protein on the structural coordinates of the yeast and human Tim44 suggests TimA also contains a hydrophobic pocket (hydrophobic residues are colored blue in Fig. 1B). In addition to the Tim44 homologue TimA, the genome of Caulobacter crescentus codes for a protein, which we have designated TimB (CC2164), with sequence similarity and equivalent domain structure to the mitochondrial Tim14/Pam18 protein family (Fig. 2A). Several sequence features: an N-terminal transmembrane segment and C-terminal J-domain, the signature HPD-X-GGS and 3 (rather than 4) helices in the J-domain characterizes the Tim14/Pam18 protein family, and all of these features are found in TimB from C. crescentus and all α-proteobacterial species for which genome data are available (Fig. S2).
Fig. 2.
Location and topology of TimA and TimB. (A) Domain structure of Tim14 (41, 42) and TimB. (Black, signal sequence; TM, transmembrane domain.) The “J-domain” (31) that interacts with Hsp70 is shown (detailed sequence analysis provided in Fig. S2). (B) Membranes were fractionated on sucrose gradient and analyzed by SDS/PAGE and immunoblotting. (C) Inner membrane vesicles (IM) were extracted with alkali and the pellet (P) and supernatant (S) fractions analyzed by Coomassie staining (Upper) and immunoblots for TimA, TimB, the integral membrane protein DivJ, and peripheral membrane protein F1β. (D) Fluorescence microscopy of the OmpA-mCherry strain shows the periplasmic location of the protein. (E) Caulobacter cells were incubated without (−) or with (+) polymyxin B and proteinase K as indicated and then analyzed by SDS/PAGE and immunoblotting.
To determine whether the topologies of TimA and TimB are identical to the mitochondrial proteins Tim44 and Tim14, antibodies were raised to TimA and TimB and used to analyze the proteins in fractionated bacterial cell extracts. We developed a method to separate outer and inner membranes of C. crescentus on sucrose gradients, and found TimA and TimB co-purify with the inner membrane protein DivJ (20) and distinctly from the outer membrane protein BamA (21) (Fig. 2B). Extraction of inner membrane vesicles with alkali shows both TimA and TimB are integral membrane proteins (Fig. 2C). To determine the membrane orientation of TimA and TimB, we developed a “mitoplasting” assay. A strain of C. crescentus was prepared in which an mCherry epitope was attached to the peptidoglycan-binding domain of the OmpA-like protein (CC3229), with fluorescent microscopy confirming the periplasmic location of mCherry (Fig. 2D). After selective permeabilization of the outer membrane by Polymyxin B, proteinase K was added to degrade the outer membrane protein BamA and the OmpA-mCherry fusion in the periplasm, and shows that neither TimA nor TimB are exposed to the periplasm (Fig. 2E).
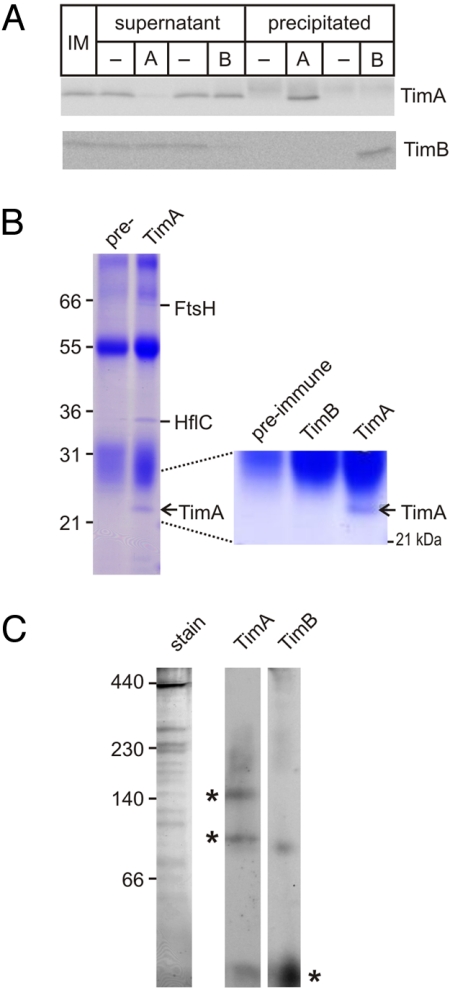
Although both TimA and TimB are located in the inner membrane and facing the cytoplasm, they do not associate with each other. Antibodies raised to TimA and TimB were used in immunoprecipitation analysis: antibodies that recognize TimA do not co-precipitate TimB, and antibodies that recognize TimB do not co-precipitate TimA (Fig. 3A). The immunoprecipitations were scaled up to identify directly any protein partners of either TimA or TimB. Coomassie blue staining of the immunoprecipitations confirmed that TimB does not associate with TimA (Fig. 3B) and identified 2 proteins that are co-immunoprecipitated with TimA (Fig. 3B). In Escherichia coli, the ATP-dependent protease FtsH has been shown to form a complex with HflC (22), wherein HflC differentially regulates FtsH degradation of substrates (23–25). Although mitochondria have AAA-proteins (related to FtsH) and prohibitins (related to HflC), neither of these proteins associate with Tim44 in the TIM23 complex of mitochondria. Fig. 3C shows that blue native PAGE resolves 2 stable complexes of approximately 100 kDa and 150 kDa containing TimA. We speculate based on subunit sizes that these represent TimA:HflC and TimA:HflC:FtsH (Fig. 3C).
Fig. 3.
TimA and TimB do not associate in a membrane complex. (A) Inner membrane vesicles (IM) solubilized with DDM were subject to immunoprecipitation assays with pre-immune serum (−) or antiserum recognizing TimA (A) or TimB (B). The immuno-depleted supernatants and the precipitates were analyzed by SDS/PAGE and immunoblotting with the sera as shown. (B) Immunoprecipitations from inner membrane vesicles (1 mg total protein) using either pre-immune serum (pre-) or immune serum raised to TimA were analyzed by SDS/PAGE and Coomassie staining, with the identity of the precipitated proteins determined by MS. (C) Inner membrane vesicles were solubilized with the detergent DDM and analyzed by blue native PAGE and immunoblotting for TimA and TimB. The Coomassie blue–stained gel (stain) is also shown.
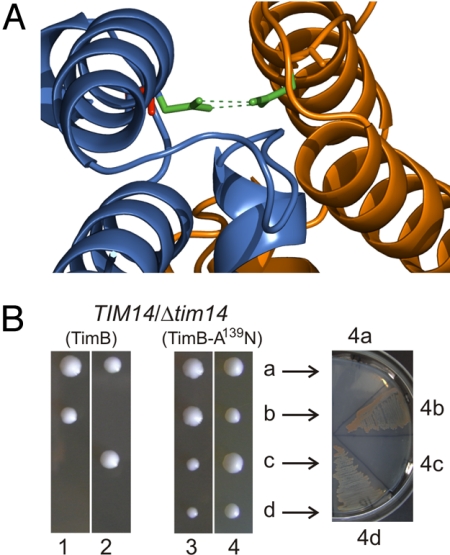
Immunoprecipitations with antibodies against TimB did not identify any partner proteins, and blue native PAGE shows TimB is predominantly found in a monomeric form at approximately 20 kDa (Fig. 3C). To test the hypothesis that a bacterial TimB could be converted to a mitochondrial Tim14, we analyzed the crystal structure (2GUZ) for interactions between the Tim14 and Tim16 subunits of the TIM23 complex. An asparagine residue in Tim14 forms a pair of hydrogen bonds with an asparagine in Tim16 (green, Fig. 4A). This critical asparagine is not conserved in bacterial TimB proteins. Pair-wise sequence comparisons suggest the TimB from the α-proteobacterium Parvularcula bermudensis (26) is most closely related to the yeast Tim14 (36% identity through the J-domain), and we engineered a copy with the point mutation A139N for expression in yeast (Fig. 4B). Heterozygous TIM14/Δtim14 cells were transformed to express either TimB or TimB(A139N), the cells were then induced to undergo meiosis, and the resulting haploid spores dissected and allowed to grow. Δtim14 yeast cells expressing TimB(A139N) are viable: the His+ cells on semisynthetic medium lacking histidine correspond to the Δtim14 haploids (Fig. 4B). This growth complementation demonstrates that minimal mutation is required to convert a bacterial TimB to function in the TIM23 complex.
Fig. 4.
TimB can be converted to function in mitochondrial protein import. (A) Model of PbTimB-A139N (blue) binding to Tim16 (orange) based on the published Tim14-Tim16 interaction (2GUZ). In Tim14, the asparagine residue (green) forms a pair of hydrogen bonds with an asparagine in Tim16. In CcTimB and PbTimB, the native residue at this position is an alanine (red). (B) TimB from C. crescentus and P. bermudensis were engineered for expression in yeast by adding an N-terminal mitochondrial targeting sequence and transmembrane domain. A point mutation was introduced at residue 139 of PbTimB (TimB-A139N) to replace the alanine with an asparagine residue. After transformation with plasmids carrying the engineered TimB constructs, Tim14/Δtim14 yeast cells were sporulated. Tetrads were dissected: 2 spores of tetrads 1 and 2 formed viable colonies; 4 spores of tetrads 3 and 4 formed viable colonies. Arrows indicate subculturing the cells from tetrad 4 to measure growth phenotype, verifying that the 2 smaller colonies are Δtim14 cells kept viable by PbTimB(A139N).
Discussion
Bacteria have several amino acid and peptide transporters that could have served as a primitive protein transport channel, and previous sequence analyses have made the case that the mitochondrial protein transport channel Tim23 was derived from LivH-type amino acid transporters (13, 27). Here we show that α-proteobacteria also have proteins related in sequence to the other 2 ubiquitous components of the TIM23 complex, Tim44 and Tim14. These newly described proteins, TimA and TimB, function in distinct protein complexes in bacteria, yet evolved to serve as modules of a protein transport machine in mitochondria (Fig. 4). We suggest that the evolution of a protein transport pathway into mitochondria required only that the LivH amino acid transporter could accept polymers of amino acids (i.e., proteins); even if this were an inefficient process initially, it would be a starting point on which Darwinian selection could act. Point mutations in a short segment required for interaction of the TimA protein with LivH would provide a docking point for the bacterial Hsp70, which is the direct homologue of the protein transport motor (15, 28, 29). Point mutations that favored interaction of TimB with LivH would provide proximity of TimB stimulation to the motor's otherwise low-level activity. This model agrees with Jacob's proposition of evolution as a “tinkerer,” building new machines from salvaged parts (30). With these 3 bacterial proteins cooperating as subunits of a primitive transport machine, a step-wise evolution of the more sophisticated mitochondrial TIM complex would be enabled.
Molecular machines have been described as being of irreducible complexity (7, 8). But could a single component of the machine function in the absence of the others to provide even inefficient protein transport? Although searches of genomes have not found a species of eukaryote in which the LivH/Tim23-type channel is present in the absence of Tim44 and Tim14 subunits, equivalent studies on the TOM complex in the outer mitochondrial membrane have provided just such a proof of principle.
Like the TIM23 complex in the inner membrane, the TOM complex in the outer mitochondrial membrane is composed of multiple components (Fig. 4). Three essential and ubiquitous subunits are found: the Tom40 channel and Tom22 and Tom7 subunits (12). In yeast and other fungi, in which much of the biochemical analysis of the TOM complex has been undertaken, additional Tom5 and Tom6 subunits are also found as part of the core TOM complex, and additional import receptors are sometimes present to maximally enhance protein transport (15). Exhaustive analysis of the genome sequence of one group of organisms, the microsporidia, shows that they have lost the Tom22, Tom5, Tom6, and Tom7 components from their core TOM complex, and have only the Tom40 channel subunit (31). Microsporidia are parasites that have evolved from the lineage that gave rise to fungi, but have subsequently reduced their genomes to code for a smaller set of cellular components (31, 32). The simplified TOM complex in microsporidians provides an excellent example of how the first, simple protein import machines might have functioned.
There is no question that molecular machines are remarkable devices, with independent modules capable of protein substrate recognition, unfolding, threading, and translocation through membranes (1). Nonetheless, the complexity of these machines is not irreducible. At a defined point in evolutionary time, when the early eukaryotes carried in their cytoplasm intracellular, symbiotic bacteria, there were no mitochondria and therefore no mitochondrial protein transporters. Subsequent to that point in time, TOM and TIM23 complexes arose in eukaryotic organisms, with surveys of modern organisms showing them to have evolved to varying degrees of sophistication. Darwin wrote, “if it could be demonstrated that any complex organ existed which could not possibly have been formed by numerous successive slight modifications my theory would absolutely break down” (33). Even viewed at the molecular level, evidence suggests that the theory holds.
Materials and Methods
Molecular Cell Biology Methods for C. crescentus and Yeast.
Detailed methods for growth, genetic manipulation, purification of membranes and raising of marker antisera are provided in the SI Methods.
Immunoprecipitation.
Inner membrane vesicles (10 μg protein) were solubilized in dodecylmaltoside (DDM), at a ratio of 10:1 DDM:protein, in ACA750 buffer, and 3 μL of sera (TimA, TimB, or pre-immune sera) was added and incubated for 1 h at 4 °C, with constant rotation. Protein A agarose beads (Santa Cruz Biotechnology) were pre-absorbed with 10 μg solubilized membrane vesicles before addition to the sample of inner membrane vesicles and sera. Incubation was continued for 1 h. The solution was centrifuged and unbound protein collected, and beads washed twice with ACA750/0.1% DDM. Bound protein was released from the protein A agarose beads by boiling in Laemmli buffer. Unbound and bound fractions were analyzed on a 12% SDS/PAGE followed by immunoblotting with TimA and TimB sera.
Large-scale immunoprecipitation followed the previously detailed procedure with the following modifications: 60 μL of sera was cross-linked to protein A agarose with dimethyl pimelimidate hydrochloride before addition of 1 mg solubilized inner membrane vesicles; after 2 h, incubation unbound proteins were removed by 6 washes of 0.1% DDM in ACA750 buffer and a final wash of PBS solution before analysis by SDS/PAGE. Precipitated protein bands were excised from the gel and peptides identified by MS as previously described (34).
Blue Native PAGE.
Inner membrane vesicles (10 μg protein) were solubilized in DDM, at ratios of detergent to protein between 2:1 and 22.5:1. Ratios of 10:1 to 22.5:1 showed no change in complexes of interest, and a ratio of 22.5:1 DDM to inner membrane vesicles is shown in Fig. 3A. BN-PAGE used a 9% to 16% separating gel and 4% stacker with buffers and gel compositions as previously described (35). Gels were stained with Coomassie (10% (vol/vol) acetic acid, 45% (vol/vol) methanol, 0.25% (wt/vol) Coomassie R250, or transferred to PVDF with a TransBlot Semidry Transfer cell (Bio-Rad) for 90 min at 10 V.
Sequence Analysis and Structural Modeling.
The sensitive homology and fold detection program HHpred was used to identify both yeast and human Tim44 proteins (E-values 5.2E-38 and 1E-39, respectively) as structural templates for TimA. A comparative model of TimA from C. crescentus (gi:13425513, residues 79–234, excluding the transmembrane region) was built using Modeller version 9v2 (36) and the multiple sequence alignment produced by HHpred, with the yeast (pdb:2FXT) and human (pdb:2CW9) Tim44 structures as templates. Model quality was assessed using the Prosa2003 (37, 38) ProQ (39), and Verify3D (40) quality scores. Hidden Markov model analysis was as previously described (10).
Supplementary Material
Acknowledgments.
We thank Erin Lim, Joel Selkrig, and Pavel Dolezal for contributions to the early studies toward this project and for critical discussion of the work; and thank Sri Harsha Ramarathinam and Tony Purcell for expert mass spectrometry analyses. This work was supported by grants from the Australian Research Council and National Health & Medical Research Council (T.L., V.A.L., and R.A.S.). S.C. is a recipient of a Melbourne International Research Scholarship and a Melbourne International Fee Remission Scholarship from the University of Melbourne. A.J.P. is supported by an National Health & Medical Research Council Training Fellowship; S.P. is supported by a PEW Latin American Fellowship; and T.L. is a Federation Fellow of the Australian Research Council. The laboratory of C.J.-W. is supported by the National Institutes of Health via grants GM065835 and GM076698.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908264106/DCSupplemental.
References
- 1.Chiu W, Baker ML, Almo SC. Structural biology of cellular machines. Trends Cell Biol. 2006;16:144–150. doi: 10.1016/j.tcb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Pallen MJ, Gophna U. Bacterial flagella and type III secretion: case studies in the evolution of complexity. Genome Dyn. 2007;3:30–47. doi: 10.1159/000107602. [DOI] [PubMed] [Google Scholar]
- 4.Pallen MJ, Matzke NJ. From The Origin of Species to the origin of bacterial flagella. Nat Rev Microbiol. 2006;4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 5.Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 7.Behe M. The challenge of irreducible complexity. Nat Hist. 2002;111:74. [Google Scholar]
- 8.Miller K. The flaw in the mousetrap. Nat Hist. 2002;111:75. [Google Scholar]
- 9.Muller M, Martin W. The genome of Rickettsia prowazekii and some thoughts on the origin of mitochondria and hydrogenosomes. Bioessays. 1999;21:377–381. doi: 10.1002/(SICI)1521-1878(199905)21:5<377::AID-BIES4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Emelyanov VV. Mitochondrial connection to the origin of the eukaryotic cell. Eur J Biochem. 2003;270:1599–1618. doi: 10.1046/j.1432-1033.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 11.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 12.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 13.Murcha MW, et al. Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 2007;143:199–212. doi: 10.1104/pp.106.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfanner N, Chacinska A. The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim Biophys Acta. 2002;1592:15–24. doi: 10.1016/s0167-4889(02)00260-4. [DOI] [PubMed] [Google Scholar]
- 15.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 16.van der Laan M, Rissler M, Rehling P. Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res. 2006;6:849–861. doi: 10.1111/j.1567-1364.2006.00134.x. [DOI] [PubMed] [Google Scholar]
- 17.Bolender N, et al. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mokranjac D, Neupert W. Energetics of protein translocation into mitochondria. Biochim Biophys Acta. 2008;1777:758–762. doi: 10.1016/j.bbabio.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Weiss C, et al. Domain structure and lipid interaction of recombinant yeast Tim44. Proc Natl Acad Sci USA. 1999;96:8890–8894. doi: 10.1073/pnas.96.16.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatsos X, et al. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol Rev. 2008;32:995–1009. doi: 10.1111/j.1574-6976.2008.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kihara A, Akiyama Y, Ito K. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 1996;15:6122–6131. [PMC free article] [PubMed] [Google Scholar]
- 23.Kihara A, Akiyama Y, Ito K. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J Mol Biol. 1998;279:175–188. doi: 10.1006/jmbi.1998.1781. [DOI] [PubMed] [Google Scholar]
- 24.van Bloois E, et al. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 2008;582:1419–1424. doi: 10.1016/j.febslet.2008.02.082. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 26.Cho JC, Giovannoni SJ. Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the alpha-Proteobacteria. Int J Syst Evol Microbiol. 2003;53:1031–1036. doi: 10.1099/ijs.0.02566-0. [DOI] [PubMed] [Google Scholar]
- 27.Rassow J, et al. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- 28.Schiller D, et al. Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol Cell Biol. 2008;28:4424–4433. doi: 10.1128/MCB.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfanner N, Truscott KN. Powering mitochondrial protein import. Nat Struct Biol. 2002;9:234–236. doi: 10.1038/nsb0402-234. [DOI] [PubMed] [Google Scholar]
- 30.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 31.Waller RF, et al. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell. 2009;8:19–26. doi: 10.1128/EC.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burri L, et al. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc Natl Acad Sci USA. 2006;103:15916–15920. doi: 10.1073/pnas.0604109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell AW, et al. Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J Immunol. 2001;166:1016–1027. doi: 10.4049/jimmunol.166.2.1016. [DOI] [PubMed] [Google Scholar]
- 35.Stenberg F, et al. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 38.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 41.Davey KM, et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43:385–393. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mokranjac D, Sichting M, Neupert W, Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.