Abstract
Mutations in the Nod2 gene are among the strongest genetic risk factors in the pathogenesis of ileal Crohn's disease, but the exact contributions of Nod2 to intestinal mucosal homeostasis are not understood. Here we show that Nod2 plays an essential role in controlling commensal bacterial flora in the intestine. Analysis of intestinal bacteria from the terminal ilea of Nod2-deficient mice showed that they harbor an increased load of commensal resident bacteria. Furthermore, Nod2-deficient mice had a diminished ability to prevent intestinal colonization of pathogenic bacteria. In vitro, intestinal crypts isolated from terminal ilea of Nod2-deficient mice were unable to kill bacteria effectively, suggesting an important role of Nod2 signaling in crypt function. Interestingly, the expression of Nod2 is dependent on the presence of commensal bacteria, because mice re-derived into germ-free conditions expressed significantly less Nod2 in their terminal ilea, and complementation of commensal bacteria into germ-free mice induced Nod2 expression. Therefore, Nod2 and intestinal commensal bacterial flora maintain a balance by regulating each other through a feedback mechanism. Dysfunction of Nod2 results in a break-down of this homeostasis.
Keywords: commensal bacteria, Crohn's disease, mouse, NLR
The intestinal mucosa is under continuous exposure to large numbers of commensal microorganisms and their products. The majority of intestinal microbiota reside in the large intestine, and more than 99% are composed of 4 major bacterial divisions: Bacteroides, Firmicutes, Proteobacteria, and Actinobacteria (1). The small intestine harbors significantly less bacteria, having loads of 103 and 107–8 in the proximal and terminal ileum, respectively, compared with 1011–12 in the colon (1). Under well-balanced steady-state conditions, interactions between the intestinal immune system and commensal bacteria elicit basal level immune responses. These responses regulate and protect the host from both pathogenic and non-pathogenic commensal bacteria. Changes in this homeostasis may influence susceptibility to or the progression of chronic inflammatory conditions in the intestine such as Crohn's disease (1, 2).
The pathogenesis of Crohn's disease seems to involve an inappropriate immune response and impaired epithelial barrier function (1–3). Genetic studies have revealed that an innate immune molecule, Nod2, is associated with susceptibility to Crohn's disease, highlighting the significance of innate immunity in the pathogenesis of inflammatory bowel diseases (4, 5). Nod2 belongs to the nucleotide-binding domain, leucine-rich repeat (NLR) family of cytoplasmic proteins (6, 7). Nod2 has been shown to respond to muramyl dipeptide, N-acetylmuramyl-l-alanyl-d-isoglutamine (MDP), a moiety of peptidoglycan consisting of N-acetylmuramyl-l-Ala-d-Glu (8, 9). This moiety is conserved in both Gram-positive and Gram-negative peptidoglycan, suggesting that Nod2 may detect a wide variety of bacteria (10). The Rip2 kinase is required for downstream signaling of Nod2 (11). Rip2 activates downstream signaling cascades, including NF-κB and MAP kinase cascades, resulting in the induction of immune response genes (11–13). Interestingly, Nod2-deficient mice have not been shown to develop spontaneous colitis (13, 14), suggesting that dysregulation of the Nod2 pathway alone is insufficient to induce Crohn's disease. Perhaps this finding is not surprising, because Crohn's disease is a multigenic disorder, and its pathogenesis is influenced by other factors including environment, altered immune regulation, and dysbiosis of commensal bacterial flora (1, 2, 15).
Although the exact mechanism by which Nod2 mutations contribute to Crohn's disease pathogenesis is still unclear, loss-of-function mutations in Nod2 have been suggested to alter host−microbial interactions through various mechanisms, including altered antimicrobial activity of Paneth cells in the terminal ileum (15). Nod2 is highly expressed in Paneth cells, which induce bacteria killing by secreting antibacterial compounds (16, 17). Nod2 mutations associated with Crohn's disease primarily predispose the development of ileal lesions (18) corresponding to the location of Paneth cells. Ileal Crohn's disease is characterized by a decrease in antimicrobial α-defensins produced by Paneth cells (19, 20). Studies comparing α-defensin levels in ileal Crohn's patients who did not have Nod2 mutation and in those who had any of the 3 major mutations (R702W, G908R, and 1007fs) associated with Crohn's disease showed no significant difference in α-defensin levels between the 2 groups (20). However, studies of individual mutations in Nod2 showed that the 1007fs mutation in Nod2 causes a decrease in the expression of α-defensin (19, 21). The level of α-defensin in ileostomy fluid is lower in patients who have Crohn's disease, and this difference is most marked in patients who have homozygous/compound heterozygous mutations in Nod2 (22). Expression of a subgroup of α-defensins is reduced in Nod2-defiient mice (13, 23). Therefore, impairment of Nod2 function may facilitate the entry of bacteria into epithelial cells because of defective regulation of defensin expression or of other antimicrobials and chemokines, resulting in impaired bactericidal capacity (13, 24).
Although the influence of Nod2 regulation of an antimicrobial peptide or other antimicrobial molecule in Crohn's disease pathology is an appealing idea, no experimental evidence directly links Nod2 deficiency with an imbalance in commensal flora, and the nature of the Nod2 regulatory effect on antimicrobials is not well understood. Here we show that Nod2 does indeed play a role in controlling commensal flora in the intestine. In comparison with wild-type controls, Nod2-deficient mice were found to have increased amounts of commensals as well as reduced capability to clear newly colonizing bacteria.
Results
The Nod2 Ligand Induces Bacteria-Killing Activity in Intestinal Crypt Secretions.
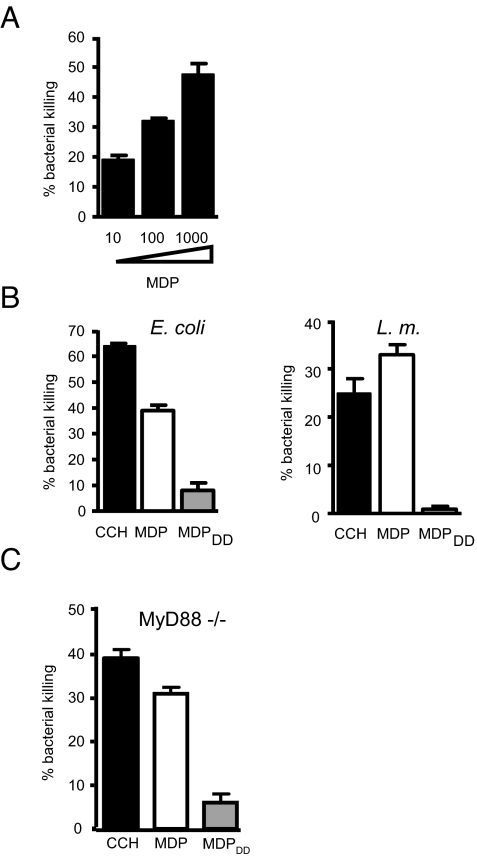
Bacterial components have been shown to induce secretions with bactericidal activity from terminal ileum crypts (25). Because Nod2 is highly expressed in both mouse and human ileal crypts, we first investigated whether the Nod2 ligand, MDP, can induce bacteria killing (13, 16, 17). Crypts isolated from the terminal ileum of C57BL/6 mice were stimulated with MDP, and the supernatant was collected 30 min later. Bacteria-killing activity in the supernatant was assessed by incubation with Escherichia coli, which subsequently were plated on LB plates to count viable bacterial numbers. MDP-induced secretions from mouse intestinal crypts were capable of bactericidal activity against E. coli in a dose-dependent manner, in agreement with previous observations of MDP-induced bacteria-killing activity using a Salmonella typhimurium virulence-impaired phoP mutant strain (Fig. 1A) (25). It is known that chirality in the peptide chain of MDP is critical for Nod2 activation: MDP can activate Nod2, whereas chiral isomers such as N-acetylmuramyl-d-alanyl-d-isoglutamine (MDPDD) and N-acetylmuramyl-l-alanyl-l-isoglutamine (MDPLL) cannot (10). To assess the specificity of MDP for the induction of bacteria killing in this assay, we used inactive MDP chiral isomers. As a positive control, we used a muscarinic receptor agonist, carbamylcholine (CCH), that selectively modulates intracellular Ca2+ concentrations and induces secretion of antimicrobials from Paneth cells (26). Although both CCH and MDP could induce bacteria killing, the inactive chiral isomer MDPDD could not (Fig. 1B). Assessing bactericidal activity toward Listeria monocytogenes, we found that MDP but not MDPDD could induce killing of this Gram-positive bacterium (Fig. 1B). These studies suggest that only the active Nod2 ligand, MDP, can induce bacteria killing of both Gram-positive and Gram-negative bacteria. Although MDP is not a Toll-like receptor (TLR) ligand, there is crosstalk between TLR and Nod2 signaling (2, 10, 15). However, MDP-induced secretion is not mediated by TLRs, because the active Nod2 ligand, MDP, and CCH, but not the inactive chiral isomer of MDP, could induce bacteria-killing activity in MyD88-deficient crypts (Fig. 1C).
Fig. 1.
Nod2 ligand induces bacteria-killing activity in intestinal crypt secretions. (A) Crypts were isolated from the terminal ileum of C57BL/6 mice. We stimulated 2,000 crypts with MDP for 30 min in iPIPES buffer at concentrations of 10, 100, and 1000 μg/mL. Secretions were mixed with E. coli at 1 × 103 cells for 1 h at 37 °C, and bacteria killing was measured by colony counting of serial dilutions. Reduced bacterial numbers in comparison with nonstimulated controls were plotted as percent killing. (B) Crypts were stimulated with 1 μM CCH, 1 mg/mL MDP, and 1 mg/mL MDPDD for 30 min. Bactericidal activity of crypts secretions was determined against E. coli and Listeria monocytogenes by incubating secretions and bacteria for 1 h. (C) Crypts from MyD88-deficient mice were stimulated, and bactericidal activity against E. coli was measured as in (B). Error bars represent standard deviation. Data are representative of 2 or 3 independent experiments.
Nod2 Is Required for the Bactericidal Activity of Crypt Secretions of the Terminal Ileum.
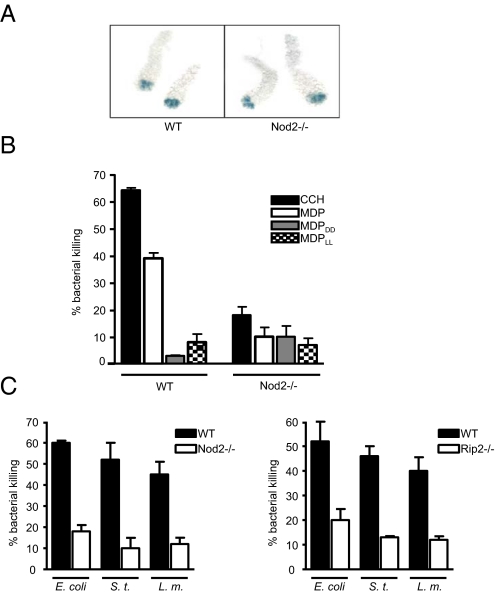
Next, we examined the effect of the Nod2 deletion on the bactericidal activity of crypt secretions. We observed no visible morphological defects in crypts from Nod2-deficient mice, indicating that Nod2 deficiency does not interfere with normal development of Paneth cells (Fig. 2A). We performed bactericidal assays using Nod2-deficient crypts stimulated with MDP. We found that in addition to the inactive isomers, MDPDD and MDPLL, MDP could not induce effective bacteria-killing activity in Nod2-deficient crypts (Fig. 2B). This phenotype was observed even for the nonspecific inducer of secretion, CCH, suggesting that Nod2 may have a general effect on secretion and/or Paneth cell composition not specific to MDP-induced signaling (Fig. 2B). Furthermore, we performed killing assays on 3 different bacteria, E. coli, S. typhimurium, and Listeria monocytogenes, using secretions from wild-type and Nod2-deficient crypts stimulated with CCH. Our data showed that the ability of Nod2-deficient crypts to kill both Gram-negative and Gram-positive bacteria was reduced significantly (Fig. 2C). Similarly, crypts deficient in Rip2, a downstream kinase of Nod2, also were reduced in their ability to kill bacteria (Fig. 2C). Therefore, Nod2 is necessary for crypts to function effectively in inducing bacteria killing. The broad spectrum of antimicrobial activity dependent on Nod2 suggests that Nod2 may have a significant effect on commensal flora in the gut.
Fig. 2.
Nod2 is required to induce bactericidal activity in crypt secretions of the terminal ileum. (A) Crypts were isolated from the terminal ileum of wild-type and Nod2-deficient mice. Granules within Paneth cells were stained with 0.25% amido black. (B) We stimulated 2,000 crypts from wild-type and Nod2-deficient mice with CCH (1 μM), MDP, MDPDD, and MDPLL (1 mg/mL), and secretions were subjected to a bactericidal activity assay against E. coli. (C) Crypts from wild-type and Nod2-deficient mice (Left) or from wild-type and Rip2-deficient mice (Right) were stimulated with CCH as in (B), and secretions were tested for bactericidal activity against E. coli, S. typhimurium, and Listeria monocytogenes. Error bars represent standard deviation. Data are representative of 5 independent experiments.
Nod2 Is Required for the Regulation of Commensal Bacteria Flora in the Terminal Ileum.
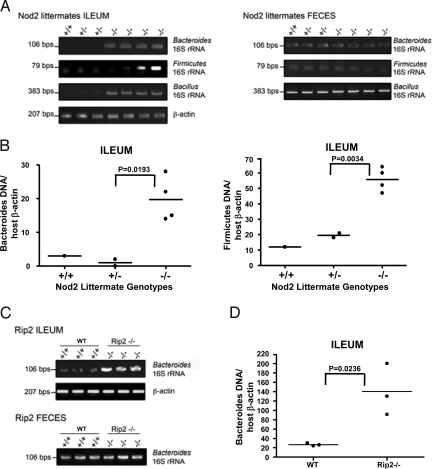
To test if Nod2 has any effect on commensal microbiota in vivo, we examined bacterial flora in Nod2-deficient mice. Commensal bacterial flora are affected by a multitude of factors: genetic background, diet, parents, siblings, sex partners, and interaction between commensal and pathogenic bacteria (27). To investigate whether Nod2 affects commensal bacteria, we decided to eliminate as many of the variable factors as possible. First, we backcrossed Nod2-deficient mice to the C57BL/6 background for more than 12 generations to generate a congenic background. Second, we re-derived the mice into specific pathogen-free (SPF) conditions by transferring embryos obtained by hysterectomy into sterile foster female recipients. The mice were maintained in isolated barrier units thereafter. In addition to regular SPF species, rodent opportunistic pathogenic bacteria such as Helicobacter or Clostridium, which can be found in many animal facilities in the United States, were excluded from this animal colony. Terminal ilea were isolated from Nod2 wild-type, -heterozygous, and -deficient mice. Ileal contents were removed, and DNA was isolated from the segments. The isolated DNA includes both host DNA, and DNA from commensal bacteria attached to the intestinal wall. We chose to test the genus Bacteroides, which is the most abundant genus of commensal bacteria in mice and humans (28), as well as Firmicutes, a grouping containing genera such as Bacillus and Clostridium. We analyzed Bacteroides loads in feces and terminal ileum using genus-specific 16S ribosomal RNA gene primers that detect most strains of Bacteroides. In the terminal ileum, Bacteroides were barely detectable in Nod2 wild-type and -heterozygous mice (Fig. 3A), in agreement with a previous report indicating that only a small number of bacteria exist in this segment of the normal, healthy intestine (29). However, significant amounts of Bacteroides as well as Firmicutes and Bacillus were detected in Nod2-deficient terminal ilea (Fig. 3A). The loads of Bacteroides and Firmicutes were quantified by real-time PCR using the 16S RNA gene-specific primers and normalized by real-time PCR data for the host β-actin gene, confirming the increased bacterial loads in the terminal ilea of Nod2-deficient mice (Fig. 3B). There was no significant difference in Bacteroides loads in feces in any of the Nod2 genotypes (Fig. 3A, Right), probably because the expression of Nod2 is localized mainly to the crypts of the terminal ileum (16, 17), and any Nod2-dependent control of microbiota is likely to be lost in the lower colon. The same observations were made with the ileum from Rip2-deficient mice by semiquantitative PCR (Fig. 3C) and quantitative real-time PCR (Fig. 3D), confirming the relevance of the Nod2-Rip2 pathway. Overall, our data show that Nod2 is essential for the control of both Gram-positive and Gram-negative commensal microbiota in the terminal ileum.
Fig. 3.
Nod2 is required for the regulation of commensal bacteria flora in the terminal ileum. (A) Terminal ilea were isolated from littermates of Nod2+/+, Nod2+/−, and Nod2−/− mice, and DNA samples were prepared. Fecal samples from the same animals were collected, and DNA was isolated. Bacteroides, Firmicutes, and Bacillus in the terminal ileum and feces were detected by PCR using primers for species-specific 16S rRNA genes. Primers for β-actin were used to show loading control of host genomic DNA. Data are representative of 3 independent littermate sets. (B) Bacteroides and Firmicutes in the terminal ileum in Nod2+/+, Nod2+/−, and Nod2−/− mice were quantified by real-time PCR using primers for species-specific 16S rRNA genes. Data were normalized by real-time PCR data for the β-actin gene in host genomic DNA. The p-values were determined by Student's t test. (C) Commensal bacterial flora in the terminal ileum of wild-type and Rip2-deficient mice were detected as (A). (D) Bacteroides in the terminal ileum in Rip2-deficient mice were quantified as (B).
Nod2 Suppresses De Novo Colonization of Opportunistic Pathogens in the Terminal Ileum.
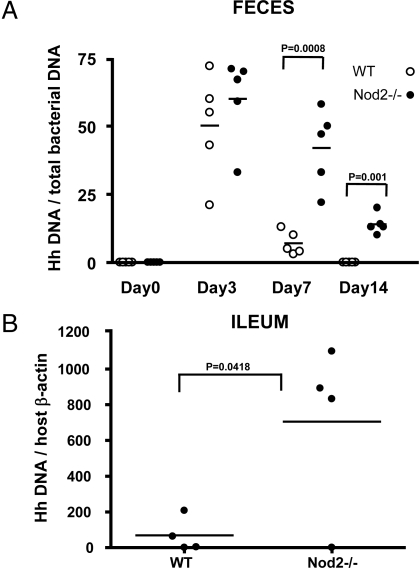
Both pathogenic and nonpathogenic commensal bacteria enter the intestine via an oral route. Therefore, we tested whether Nod2 has the ability to prevent de novo colonization of pathogenic bacteria in addition to the ability to control commensal microbiota at steady state. We colonized Nod2-deficient mice with the opportunistic pathogen, Helicobacter hepaticus. H. hepaticus, a Gram-negative microaerophilic bacterium, is a common mouse intestinal commensal in most animal facilities in the United States and colonizes the intestinal crypts of the cecum and colon, establishing a life-long infection (30). Wild-type and Nod2-deficient mice were inoculated with H. hepaticus via oral gavage, and feces samples were collected at days 0, 3, 7, and 14 after inoculation to isolate DNA samples. The presence of H. hepaticus was examined by quantitative real-time PCR using specific primers for the H. hepaticus 16S RNA gene and normalized by the total amount of bacteria DNA in the samples. Although wild-type mice were able to clear H. hepaticus rapidly at days 7 and 14, clearance of the bacteria was significantly delayed in Nod2-deficient mice (Fig. 4A). Furthermore, increased numbers of H. hepaticus were detected in the terminal ilea of Nod2-deficient mice 14 days after inoculation (Fig. 4B). This finding indicates that, in addition to its ability to restrict numbers of commensal microbiota in the terminal ileum at steady states, Nod2 can protect the host from colonization by some pathogenic bacteria in the intestine.
Fig. 4.
Nod2 suppresses colonization of pathogenic commensal bacteria in the terminal ileum. (A) H. hepaticus (5 × 108/mouse) was inoculated into wild-type and Nod2-deficient mice via gastric gavage. Fresh feces samples were collected from mice at 0, 3, 7, and 14 days after inoculation, and DNA was purified. H. hepaticus colonization was quantified by real-time PCR using H. hepaticus-specific primers and 100 ng of purified DNA per reaction. Data were normalized by real-time PCR data for Eubacteria using the bacteria 16S RNA gene primer sets, which detect all bacterial strains. The p-values were determined by Student's t test. (B) Terminal ilea were isolated from wild-type and Nod2-deficient mice, and DNA samples were prepared. H. hepaticus colonization was quantified by real-time PCR using H. hepaticus-specific primers and 100 ng of purified DNA per reaction. Data were normalized by real-time PCR data for the β-actin gene in host genomic DNA. The p-values were determined by Student's t test.
Nod2 and Rip2 Gene Expression Are Dependent on Commensal Flora.
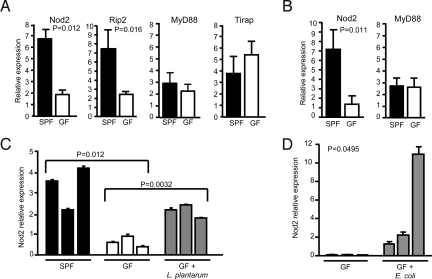
In normal hosts, commensal bacteria activate a sequential program of homeostatic responses of the immune system (1). Because Nod2 expression is inducible upon stimulation with bacterial products such as LPS (31), it is feasible that Nod2 expression may be induced by the presence of commensal bacterial flora in the intestine. To test this possibility, we generated germ-free (GF) mice by re-deriving BALB/c mice into GF conditions and compared the expression of Nod2 in these mice with that in mice under conventional SPF conditions at the age of 2 months. Nod2 expression was significantly reduced in mice under GF conditions (Fig. 5A). This difference in Nod2 expression was maintained even after mice aged, because we observed similarly reduced Nod2 expression in 5-month-old mice under GF conditions (Fig. 5B). Likewise, the expression of a downstream kinase, Rip2, was significantly reduced in GF mice, indicating that Rip2 expression also is dependent on commensal bacteria (Fig. 5A). In contrast, the expression of adaptor molecules for TLRs, MyD88, and Tirap/Mal was independent of commensal bacteria (Fig. 5 A and B). These data suggest that commensal bacteria play an important role in inducing the Nod2-Rip2 signaling pathway. To confirm further that commensal bacteria can induce Nod2 expression, GF mice were monoassociated with the Gram-positive nonpathogenic commensal bacterium, Lactobacillus plantarum, and were kept in a gnotobiotic isolator for 1 month. The expression of Nod2 was significantly increased by complementation with Lactobacillus plantarum (Fig. 5C). To test if Gram-negative bacteria also can restore the expression of Nod2, GF mice were monoassociated with the nonpathogenic, probiotic E. coli strain Nissle 1917. The expression of Nod2 also was induced by complementation with E. coli (Fig. 5D). Collectively these results indicate that Nod2 expression is regulated by commensal bacterial flora.
Fig. 5.
Nod2 and Rip2 gene expression are dependent on commensal flora. (A) BALB/c mice were re-derived into GF conditions, and mRNA was extracted from the terminal ileum of 2-month-old BALB/c mice under SPF (n = 3) and GF (n = 3) conditions. The expression of Nod2, Rip2, MyD88, and Tirap/Mal was examined by quantitative real-time PCR. (B) The expression of Nod2 and MyD88 was examined as in (A) in the terminal ileum of 5-month-old BALB/c mice under SPF (n = 3) and GF (n = 3) conditions. (C) Two-month-old BALB/c mice raised under GF conditions were colonized with Lactobacillus plantarum and incubated for 1 month. Nod2 expression in the terminal ileum was examined by quantitative real-time PCR, together with 3-month-old BALB/c mice housed under SPF and GF conditions as controls. (D) Nod2 expression in the terminal ileum was examined as in (C) in 3-month-old GF mice and GF mice colonized with probiotic E. coli strain Nissle 1917 for 1 month. Each bar represents data from a single mouse. The p-values were determined by Student's t test.
Discussion
In this study, we have shown that Nod2 is important for the regulation of commensal microbiota in the intestine. This regulation appears to be dependent on the downstream kinase Rip2, because Rip2-deficient mice also failed to regulate commensal bacteria in the terminal ileum (Fig. 3B). Altered commensal flora in Nod2-deficient mice is not a result of inflammation, because Nod2-deficient mice lack spontaneous intestinal inflammation (13, 14). Collectively, these data show a critical role for the Nod2-Rip2 pathway in regulating homeostasis between bacterial flora and innate immunity. Interestingly, the expression of Nod2 and Rip2 is regulated by the presence of commensal microbiota (Fig. 5). Therefore, a controlled balance exists between Nod2 and the commensal bacteria, creating a negative feedback loop in which commensal bacteria positively regulate Nod2 and associated signaling molecules, which in turn negatively regulate commensal flora. Human and animal model studies have supported the hypothesis that dysbiosis of the commensal microbiota may underlie the pathogenesis of Crohn's disease (1). Moreover, Nod2 mutations in Crohn's disease are linked mainly to ileal involvement of the disease (18). Therefore, it is tempting to speculate that mutations in the Nod2 gene in Crohn's disease are associated with changes in the composition and load of the commensal microbiota in the terminal ileum that may facilitate disease progression and pathology.
MyD88 also is known to control intestinal infection by pathogenic bacteria such as Listeria monocytogenes or vancomycin-resistant Enterococcus through an increased expression of antimicrobials (32, 33). Interestingly, Paneth cell-intrinsic expression of MyD88 has little effect on luminal microbial numbers. Rather, it limits the translocation and dissemination of microbes across the mucosal barrier by triggering expression of MyD88-dependent, Nod2-independent antimicrobials such as RegIIIγ, RegIIIβ, CRP-ductin, and RELMβ (23). We have shown that MyD88 expression is not inducible by commensal flora, whereas the expression of both Nod2 and Rip2 is dependent on commensal bacteria. Because both MyD88 and Nod2 have the ability to protect the intestine from infection by certain pathogenic bacteria, these data suggest that these 2 innate immune pathways may have similar but distinct functions. We could speculate that the primary function of the Nod2-Rip2 pathway is to regulate commensal flora at a steady state and to suppress de novo colonization of pathogenic bacteria such as H. hepaticus, whereas MyD88 may have a more prominent role in protection against invading bacteria.
The exact mechanism by which Nod2 contributes to the control of commensals in the intestine is not understood. It is possible that Nod2 regulates commensal flora with multiple mechanisms. First, Nod2 may regulate commensal flora through the bacteria-killing activity of ileal crypt secretions as shown in this study (Fig. 2). Crypts from Nod2-deficient mice have an intrinsic defect in their ability to kill bacteria; therefore, Nod2 may have the ability to control the function of ileal crypts directly. Second, Nod2 may regulate commensal flora via activity of the adaptive immune system. It recently has been shown that epithelial expression of Nod1, another NLR protein, induces lymphoid tissue genesis in the intestine via mouse β-defensin 3− and CCL20-mediated signaling through the chemokine receptor CCR6 (34). Because Nod1 and Nod2 are similar proteins and share early signaling pathways, it is reasonable to expect that Nod2 may regulate commensal flora with a similar mechanism. Third, it is possible that Nod2 expression in the myeloid lineage cells may contribute indirectly to the regulation of microbiota (35). Nod2 is expressed in myeloid cells such as monocytes, macrophages, and dendritic cells (2, 17, 35). Their activation may contribute to a stronger immune surveillance system via the development of mature intestinal lymphoid tissues by stimulating lymphocytes with cytokines and/or co-stimulatory molecules. Fourth, it is feasible that, in addition to Paneth cells, other intestinal epithelial cells may contribute to the regulation of commensal bacteria (24, 36). Nod2 also is expressed in non-Paneth epithelial cells, although at a lower level (2, 17, 24), and activation of intestinal epithelial cells may inhibit bacterial growth directly via β-defensin/chemokine secretion or indirectly by promoting lymphoid tissue genesis via chemokines. It is likely that Nod2 expression in different cells or tissues may be necessary to respond to a variety of commensals and pathogens. It is inviting to speculate that, depending on the microbe's site and mode of infections, Nod2 and other members of the innate immune system employ specific and varied defense mechanisms to control bacterial flora, from the genesis of lymphoid tissue in the intestine to the secretion of Paneth cells in the terminal ileum.
In conclusion, the results of this study reveal an important role for Nod2 in regulating commensal bacteria. In this pathway, a controlled balance exists between Nod2 and commensal bacteria, creating a negative feedback loop. Therefore, normal functionality of Nod2 in the intestine is important in maintaining the homeostasis between microbiota and the host immune system.
Materials and Methods
Mouse Strains.
C57BL/6 mice were purchased from Taconic Farms. Nod2- and Rip2-deficient mice (kindly provided by Richard Flavell, Yale University), and MyD88-deficient mice (kindly provided by Shizuo Akira, Osaka University) were backcrossed to C57BL/6 for 12 generations and re-derived into SPF conditions and maintained in isolated barrier units thereafter at Taconic Farms. Long-term colonies of GF BALB/c mice were established using re-derivation by a Caesarean section and maintained in flexible plastic isolators. Fecal samples from GF mice and swabs from the inner surface of the isolators were cultured under both aerobic and anaerobic conditions, and smears were stained with Gram staining and fluorescence dyes on a weekly basis and before the experiment to verify the continued sterility of the colony. Both GF and SPF BALB/c mice were maintained on a sterile experimental diet (37). GF BALB/c mice were monoassociated at 2 months of age with cultured Lactobacillus plantarum (ATTCC BAA-793, kindly provided by Ursula Wiedermann, Medical University of Vienna) or probiotic E. coli strain Nissle 1917 (Ardeypharm) by gastric gavage. Bacterial monoassociation and the absence of contamination by other bacterial species was confirmed by periodic aerobic and anaerobic culturing of stool samples. GF, SPF, and monoassociated BALB/c mice were maintained in the Gnotobiotic Laboratory at the Institute of Microbiology, Czech Academy of Sciences, Novy Hradek, Czech Republic.
Reagents and Bacterial Strains.
CCH, MDP (N-acetylmuramyl-l-alanyl-d-isoglutamine), and MDPLL (N-acetylmuramyl-l-alanyl-l-isoglutamine) were purchased from Sigma and were resupended in endotoxin-free water. MDPDD (N-acetylmuramyl-d-alanyl-d-isoglutamine) was obtained from InvivoGen. The strains used in bacteria-killing assays were E. coli DH5alpha, S. typhimurium 1433, and Listeria monocytogenes 1403s. All strains were grown in LB broth at 37 °C.
Crypt Isolation from Mouse Small Intestine.
Mouse intestinal crypts were isolated as described (25) with some modifications. Briefly, 10 cm of mouse small intestine measured from the terminal ileum was cut longitudinally and washed in cold PBS. Intestines were cut in 2-cm fragments and shaken in a 15-mL conical tube (BD Biosciences) in 30 mM EDTA in Dulbecco's PBS, pH 7.4, for 5 min. Washes were repeated 4 times for a total of 5 fractions, and crypts were spun down and rapidly replaced into Dulbecco's PBS. Fractions were observed under a light microscope to identify crypt-enriched fractions. Mouse intestinal crypts were fixed on glass coverslips with 10% formalin and stained for 30 min at room temperature with 0.25% amido black dye. Crypts were washed briefly with PBS, and coverslips were put onto glass slides for microscopy.
Bacteria-Killing Assays.
Bactericidal assays were performed as described (25). Crypts from crypt-enriched fractions were counted in a Bright-Line Hemacytometer (VWR), and numbers were normalized to 1000–2000 crypts/well in 50 μL of iPIPES (10 mM Pipes, 150 mM NaCl). Crypts were stimulated with 1 μM CCH, 1 μg/mL LPS, and 1 mg/mL MDP, MDPDD, or MDPLL for 30 min at 37 °C. Crypts were spun down, and supernatants were collected. Supernatants were incubated with 1 × 103 cfu of bacteria for 1 h at 37 °C, and cells were plated in serial dilutions up to 10−2. Plates were incubated overnight at 37 °C, and bacterial colonies were counted. Percent killing was calculated by the following formula:
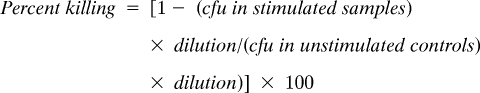 |
H. hepaticus Colonization.
H. hepaticus (ATCC 51448, kindly provided by David Schauer at MIT) was cultured on trypticase soy agar 5% sheep blood agar plates (BD Biosciences) at 37 °C in an anaerobic gas chamber (BD Biosciences) flled with a gas mixture of CO2 5%, H2 5%, N2 90%. After 2 days, a suspension of H. hepaticus in Brucella broth was made to a concentration of 5 × 108 cells/mouse, and mice were gastrically gavaged at this concentration. To confirm colonization, fresh fecal samples were collected from the mice at the indicated days after gavage.
Isolation of Bacterial DNA from Stool and Small Intestine.
DNA from mouse feces was isolated using the Qiagen DNA stool isolation kit according to the manufacturer's instructions. DNA from mouse small intestine was isolated by treating 1-cm sections of terminal ileum in lysis buffer [100 mM NaCl, 20 mM Tris (pH7.6) 10 mM EDTA, 0.5% SDS, 0.4 mg/mL proteinase K] overnight at 55 °C. DNA was precipitated using a high-salt/ethanol precipitation method and was washed extensively with 70% ethanol.
Detection of Mouse Commensal Bacteria.
H. hepaticus was detected from DNA isolated from stool samples using primers specific for H. hepaticus 16s rRNA (38). Bacteroides-, Firmicutes-, and Bacillus-specific 16s rRNA primers (39–41) were used to analyze commensal flora attached to the walls of the mouse terminal ileum. Primer pairs used in quantitative PCR analysis are as follows: H. hepaticus (5′-GCATTTGAAACTGTTACTCTG-3′) (5′-CTGTTTTCAAGCTCCCCGAAG-3′); Bacteroides (5′-GAGAGGAAGGTCCCCCAC-3′) (5′-CGCTACTTGGCTGGTTCAG-3′); Firmicutes (5′-GCTGCTAATACCGCATGATATGTC-3′) (5′-CAGACGCGAGTCCATCTCAGA-3′); Bacillus (5′-GCGGCGTGCCTAATACATGC-3′) (5′-CTTCATCACTCACGCGGCGT-3′).
Real-Time Quantitative PCR Analysis.
RNA from terminal ileum sections of GF or SPF BALB/c mice was isolated using TRIzol reagent (Invitrogen) and was ethanol precipitated. Contaminating DNA was removed using the Turbo DNA-free kit (Ambion), and cDNA synthesis was performed using the SuperScript III cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. Real-time quantitative PCR analysis was performed using 200 ng cDNA and the Quanta BioSciences Sybr Green qPCR mix (VWR) on an Applied Biosciences ABI 7700 real-time machine. Primer pairs used in quantitative PCR analysis are as follows: Nod2 (5′-CGACATCTCCCACAGAGTTGTAATCC-3′) (5′-GGCACCTGAAGTTGACATTTTGC-3′); Rip2 (5′-CGACATCTCCCACAGAGTTGTAATCC-3′) (5′-GGATGTGTAGGTGCTTCACTG-3′); TIRAP (5′-CCTCCTCCACTCCGTCCAA-3′) (5′-CTTTCCTGGGAGATCGGCAT-3′); β-actin (5′-GCTGTGCTGTCCCTGTATGCCTCT-3′) (5′-CTTCTCAGCTGTGGTGGTGAAGC-3′); MyD88 (5′-TTTGGGAGGTAGAGGCAGGAG AAT-3′) (5′-GGTAGGGGATGGGGGAGGTAGG- 3′).
Acknowledgments.
The authors thank T. B. Meissner and M. Delhase for helpful discussions and R. S. Blumberg for critical reading of the manuscript. This work was supported by Grants R01DK074738, R03TW6833, and P30DK034854 from the National Institutes of Health (to K.S.K.), the Crohn's and Colitis Foundation of America (K.S.K.), and by Grant 303/06/0974 from the Czech Science Foundation (to H.T.-H.). K.S.K. is a recipient of the Investigator Award from the Cancer Research Institute and the Claudia Adams Barr Award, and T.P.-O. is a recipient of the Benacerraf Memorial Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Wilmanski JM, Petnicki-Ocwieja T, Kobayashi KS. NLR proteins: Integral members of innate immunity and mediators of inflammatory diseases. J Leukocyte Biol. 2008;83:13–30. doi: 10.1189/jlb.0607402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting JP, et al. The NLR gene family: A standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 9.Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 10.Traub S, von Aulock S, Hartung T, Hermann C. MDP and other muropeptides—direct and synergistic effects on the immune system. Journal of Endotoxin Research. 2006;12:69–85. doi: 10.1179/096805106X89044. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 12.Li J, et al. Regulation of IL-8 and IL-1beta expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 14.Pauleau AL, Murray PJ. Role of Nod2 in the response of macrophages to toll-like receptor agonists. Mol Cell Biol. 2003;23:7531–7539. doi: 10.1128/MCB.23.21.7531-7539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nature Reviews Immunology. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 16.Ogura Y, et al. Expression of NOD2 in Paneth cells: A possible link to Crohn's ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lala S, et al. Crohn's disease and the NOD2 gene: A role for Paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 18.Gasche C, Grundtner P. Genotypes and phenotypes in Crohn's disease: Do they help in clinical management? Gut. 2005;54:162–167. doi: 10.1136/gut.2003.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simms LA, et al. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 21.Bevins CL, Stange EF, Wehkampl J. Decreased Paneth cell defensin expression in ileal Crohn's disease is independent of inflammation, but linked to the NOD2 1007fs genotype. Gut. 2009;58:882–883. [PubMed] [Google Scholar]
- 22.Elphick D, Liddell S, Mahida YR. Impaired luminal processing of human defensin-5 in Crohn's disease: Persistence in a complex with chymotrypsinogen and trypsin. Am J Pathol. 2008;172:702–713. doi: 10.2353/ajpath.2008.070755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaishnava S, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–208563. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begue B, et al. Microbial induction of CARD15 expression in intestinal epithelial cells via toll-like receptor 5 triggers an antibacterial response loop. J Cell Physiol. 2006;209:241–252. doi: 10.1002/jcp.20739. [DOI] [PubMed] [Google Scholar]
- 25.Ayabe T, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 26.Satoh Y, Habara Y, Ono K, Kanno T. Carbamylcholine- and catecholamine-induced intracellular calcium dynamics of epithelial cells in mouse ileal crypts. Gastroenterology. 1995;108:1345–1356. doi: 10.1016/0016-5085(95)90681-9. [DOI] [PubMed] [Google Scholar]
- 27.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence in situ hybridization study in mice. World Journal of Gastroenterology. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foltz CJ, et al. Spontaneous inflammatory bowel disease in multiple mutant mouse lines: Association with colonization by Helicobacter hepaticus. Helicobacter. 1998;3:69–78. doi: 10.1046/j.1523-5378.1998.08006.x. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez O, et al. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J Biol Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 32.Brandl K, et al. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 35.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nature Reviews Immunology. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenstiel P, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- 37.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflammatory Bowel Diseases. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 38.Dieleman LA, et al. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun. 2000;68:5107–5113. doi: 10.1128/iai.68.9.5107-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Layton A, et al. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol. 2006;72:4214–4224. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deloris AA, et al. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–1104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- 41.Chen ML, Tsen HY. Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. Journal of Applied Microbiology. 2002;92:912–919. doi: 10.1046/j.1365-2672.2002.01606.x. [DOI] [PubMed] [Google Scholar]