Abstract
Toll-like receptors (TLRs) are major receptors that enable inflammatory cells to recognize invading microbial pathogens. MicroRNAs are small non-coding RNAs that play important regulatory roles in a variety of biological processes. In this study, we found that a microRNA, miR-147, was induced upon stimulation of multiple TLRs and functioned as a negative regulator of TLR-associated signaling events in murine macrophages. We first demonstrated that the NMES1 transcript was a functional primary miR-147. miR-147 was induced in LPS-stimulated mouse macrophages and under in vivo conditions in the lungs of LPS-treated mice. Expression of miR-147 was greater after cellular activation by TLR4 than after engagement of either TLR2 or TLR3, suggesting that maximal induction of miR-147 required activation of both NF-κB and IRF3. TLR4-induced miR-147 expression was both MyD88- and TRIF-dependent. The miR-147 promoter was responsive to TLR4 stimulation and both NF-κB and STAT1α bound to the miR-147 promoter. miR-147 mimics or induced expression of miR-147 decreased, whereas miR-147 knockdown increased inflammatory cytokine expression in macrophages stimulated with ligands to TLR2, TLR3, and TLR4. These data demonstrate a negative-feedback loop in which TLR stimulation induces miR-147 to prevent excessive inflammatory responses.
Inflammatory cells, including macrophages and neutrophils, recognize invading microbial pathogens primarily through Toll-like receptors (TLRs) (1). To date, 13 TLRs have been discovered, each of which recognizes a unique PAMP (pathogen-associated molecule pattern) (2, 3). For example, TLR2 recognizes lipopeptides while TLR4 interacts with lipopolysaccharide (LPS) and TLR3, 7, 8, and 9 recognize microbial nucleic acids. Binding of ligands to TLRs activates the transcriptional factors NF-κB and IFN regulatory factor 3/7 (IRF3/7) (1–3). Based on the adaptor molecules recruited to the TLR intracellular domain after ligand engagement, the TLR-activated signaling events are largely defined as myeloid differentiation primary response gene 88 (MyD88)-dependent or TIR-domain-containing adapter-inducing IFN-β (TRIF)-dependent (4). For example, NF-κB activation upon TLR2 stimulation is mediated by MyD88, while activation of IRF3 in response to TLR3 engagement is TRIF-dependent (1). However, the TLR4 intracellular domain recruits both MyD88 and TRIF, leading to activation of both NF-κB and IRF3 after LPS stimulation (4). NF-κB activation results in the expression of numerous pro-inflammatory mediators, including the cytokines TNF-α and IL-6 (5), while activation of IRF3 leads to enhanced expression of type I IFN, which activates the signal transducers and activator (STAT) family of transcriptional factors and results in STAT-dependent amplification of IFN-associated responses (6).
MicroRNAs are 21–22-nucleotide (nt), non-coding small RNAs (7, 8) produced from actual genes within the genomic loci of cells. A majority of microRNA genes are intergenic or intronic. However, a small number of microRNA transcripts are derived from protein-coding mRNAs (7–9). The expression of microRNAs themselves is often subject to regulation by transcriptional factors in response to cellular stimuli (10). The primary transcripts of microRNAs (pri-miRs), which are similar to regular mRNAs, are processed by Drosha to form 70–80 nt precursor microRNAs (pre-miRs). The pre-miRs are then exported to the cytoplasm and processed by Dicer into mature microRNAs. The mature microRNA, together with Dicer and Argonaute proteins, forms the microRNA induced silencing complex (miRISC) (8). Guided by the microRNA through base pairing, the miRISC binds to the 3′ UTR of target genes, thereby repressing translation and/or inducing degradation of target gene mRNAs (8).
MicroRNAs have been shown to be centrally involved in immune system development and function (11). For example, miR-155, 181a, 223, and 150 have been demonstrated to directly participate in the differentiation and development of T and B lymphocytes (12–15). Emerging evidence also shows that microRNAs play an important role in both adaptive and innate immunity (11). For example, miR-155 modulates the formation and response of germinal-center B cells (12). miR-181a participates in regulating the strength and threshold of T cell receptor (TCR) signaling (13). Very recently, miR-146 expression was found to be increased in LPS stimulated monocytic cell line THP-1 (16). miR-146 may target IRAK-1 and TRAF6, two essential components of the TLR signaling pathways, thereby acting as a negative regulator of the inflammatory response (16).
In this study, we found that miR-147 is induced after cellular activation through multiple TLRs. miR-147 induction was dependent on both NF-κB and IRF3. We also demonstrated that miR-147 attenuates TLR stimulation induced-inflammatory response in macrophages. Our studies thus identified a previously unrecognized negative feedback loop in which TLR-induced miR-147 expression is capable of down-regulating excessive inflammatory responses.
Results
miR-147 Is Derived from the NMES1 Gene Transcripts.
To identify microRNAs that are potentially involved in the regulation of TLR4 induced inflammatory responses, we performed a microRNA array analysis (analyzed by Exiqon) and found that the expression of miR-147 was increased in LPS treated mouse peritoneal macrophages. After sequence alignment, we found that mouse precursor miR-147 overlaps with mRNA transcripts for the mouse normal mucosa esophagus specific 1 (NMES1) gene. The sequence of pre-miR-147 is located within exon 4 of the NMES1 gene, but is downstream of the NMES1-coding sequence (CDS) (Fig. 1A).
Fig. 1.
miR-147 is derived from the NMES1 gene transcripts. (A) Schematic representation of the miR-147/NMES1 gene. (B) Full-length NMES1 cDNA gives rise to mature miR-147. HEK-293 cells were transiently transfected with pBabe-H1, pBabe-H1-miR-147, pcDNA4, or pcDNA-4-NMES1. Twenty-four hours after transfection, the cells were collected and RNA was purified. Ten micrograms RNA from each sample was resolved on denatured PAGE gels. As a positive control, the indicated amounts of synthetic miR-147 mimics were loaded onto the same gel. Northern blotting was performed with specific miR-147 probes. The PAGE gel was prestained with ethidium bromide to determine an equal loading of RNA. (C) RAW264.7-miR-147 cells were un-induced or induced to express miR-147 for 1 day. The cells were collected and RNA purified. Northern blotting was performed as in (B).
To determine if mature miR-147 is derived directly from the NMES1 transcript, we subcloned full length cDNA of NMES1 into an expressing vector, pcDNA4, and transfected the construct into HEK-293 cells. As shown in Fig. 1B, transfection of pcDNA4-NMES1 gave rise to mature miR-147. Of note, transfection of the empty vector did not result in expression of miR-147. To confirm that the bands on the northern blot are mature miR-147, we generated a construct that included only the mature miR-147 sequence under the control of the PolIII promoter H1 and found that transfection of this construct into HEK 293 cells resulted in a mature microRNA that was recognized by the miR-147 probes. As expected, the miR-147 probes recognized synthetic miR-147 mimics.
As should be noted, miR-147 expressed in the pcDNA4-NMES1 transfected cells does not have to be derived from the full length NMES1 transcript, but rather from a transcript produced by an embedded cryptic promoter within the NMES1 cDNA sequence. To address this possibility, we used a tet-on system in which the expression of full-length NMES1 cDNA was under the control of a tet operator. In this system, if the NMES1 cDNA contains no cryptic promoter, the expression of miR-147 can only be induced by addition of doxycycline. Using this system, we generated a stable mouse macrophage cell line in RAW264.7 cells. As shown in Fig. 1C, the RAW264.7 cells inducibly expressed mature miR-147 upon doxycycline addition. Of note, the minor expression of miR-147 in the uninduced cells was likely due to leakage. These data therefore indicate that miR-147 is derived from the NMES1 gene transcript and that the NMES1 gene transcript functions as primary miR-147.
miR-147 Is Up-Regulated in LPS-Treated Macrophages.
To delineate the kinetics of miR-147 induction, peritoneal macrophages were treated with LPS for different lengths of time. As shown in Fig. 2A, mature miR-147 was up-regulated as early as 2 h after LPS stimulation and levels continued to increase for 24 h. As a positive control, we also examined the expression of miR-146a and miR-155, two microRNAs that were previously shown to be induced in LPS stimulated human monocytes (16, 17), and found that both microRNAs were up-regulated in LPS-treated mouse macrophages (Fig. S1 A and B). Similar to the findings for miR-147, pri-mir-147 was also up-regulated 2 h after LPS stimulation (Fig. 2B). However, in contrast to the findings with miR-147 where expression continued to increase for 24 h after macrophage exposure to LPS, levels of pri-mir-147 peaked at 6h and gradually decreased thereafter (Fig. 2B). These data suggest that even though miR-147 is processed from pri-mir-147, miR-147 demonstrates greater stability than does pri-mir-147. Of note, levels of pri-mir-147 induced after LPS stimulation were much greater than those found for miR-147 (Fig. 2B), suggesting that only a portion of pri-mir-147 is processed into miR-147. As shown in Fig. 2 C and D, pri-mir-147 and miR-147 were dose-dependently induced by LPS stimulation. Northern blotting assays further demonstrated that both pre-mir-147 and miR-147 were up-regulated after LPS stimulation (Fig. 2E).
Fig. 2.
miR-147 is up-regulated in LPS-treated macrophages. (A and B) Time course of mature (A) and primary miR-147 (B) induction after LPS stimulation. Peritoneal macrophages were treated with 1 μg/ml LPS for the indicated lengths of time. Real-time PCR was performed as described in Materials and Methods. The non-treated cells were used as controls and values from these cells were regarded as 1. Values are expressed as mean ± SD from triplicate wells. Data shown are representative of three experiments. (C and D) LPS dose-dependently induced miR-147 expression. Peritoneal macrophages were treated with LPS at the indicated concentrations for 24 h. Real-time PCR was performed to determine the expression of mature (C) and primary miR-147 (D). (E) Peritoneal macrophages were treated with LPS at the indicated concentrations for 24 h. RNA was purified and Northern blotting was performed with the miR-147 probes. The same membrane was re-blotted with U6 probes to ensure equal loading.
miR-147 Is Induced in Multiple Macrophage Populations After LPS Treatment and in the Lungs of LPS-Exposed Mice.
We examined miR-147 expression in LPS treated alveolar macrophages and in mouse macrophage cell line RAW264.7. As shown in Fig. S1 C and D, expression of miR-147 was increased in both macrophage populations after LPS exposure.
The human homologue to mouse miR-147 is miR-147b (hsa-miR-147b) (18) and also overlaps with the human NMES1 transcript. To determine if hsa-miR-147b is induced by TLR4 stimulation, we treated the human monocytic cell line THP-1 with LPS for 4 and 24 h. As shown in Fig. S1 E and F, LPS stimulation up-regulated the expression of both primary and mature miR-147b, although to a lesser extent than that of mouse miR-147. These data suggest that TLR4 induced miR-147b expression in inflammatory cells is conserved across species.
To examine if miR-147 is induced under in vivo conditions, we determined the expression of miR-147 in the lungs of mice treated with intra-tracheal LPS. miR-147 levels were increased 4 h after LPS instillation and continued to rise for 72 h after LPS administration (Fig. S1 G and H). We also found that miR-155 was up-regulated in LPS-exposed lungs, with a pattern similar to that of miR-147 (Fig. S1 G and H).
miR-147 Is Induced by Stimulation of Multiple TLRs.
We examined miR-147 levels in peritoneal macrophages stimulated with TLR2 or TLR3 ligands. As shown in Fig. S2A, culture with the TLR2 ligand, PAM3CSK4 as well as with the TLR3 ligand, poly(I:C), induced miR-147 expression. However, although TLR2 and TLR4 stimulation induced comparable levels of TNF-α mRNA, the increase in miR-147 found after TLR2 stimulation was much less than that present after TLR4 stimulation (Fig. S2 A and B). As shown in Fig. 3A, poly(I:C), the TLR3 ligand, and LPS, the TLR4 ligand, induced miR-147 expression in a dose-dependent manner. However, the increase in miR-147 expression after exposure to PAMC3SK4, the TLR2 ligand, or poly(I:C) was less than that found after culture with LPS. Given that TLR2 stimulation only activates NF-κB and TLR3 stimulation primarily activates IRF3, these data suggest that maximal miR-147 induction may require the participation of both NF-κB and IRF3.
Fig. 3.
miR-147 is induced by stimulation of multiple TLRs. (A) Dose-dependent induction of miR-147 by poly(I:C) and LPS. Peritoneal macrophages were treated with PAM3CSK4, poly(I:C) or LPS at the indicated concentrations for 6 h (PAM3CSK4 and LPS) or 9 h (poly(I:C)). Real-time PCR was performed. Values are presented as mean ± SD from triplicate wells. ###, P < 0.001 compared to cells treated with 10 μg/mL PAM3CSK4. ***, P < 0.001 when compared to cells treated with 50 μg/mL poly(I:C). (B) LPS treatment rapidly induces IFN β expression. Peritoneal macrophages were treated with 1 μg/mL LPS for 0, 30, 60, and 120 min. The expression of IFN β was determined by real-time PCR analysis. (C and D) LPS-induced miR-147 expression requires de novo protein synthesis. Peritoneal macrophages were pretreated with or without 5 μg/mL cycloheximide (CHX) for 1 h. CHX was then removed and the cells were cultured with or without 1 μg/mL LPS for 3 h. Real-time PCR was performed to determine the levels of pri-mir-147 (C) and IL-1β (D). Values are presented as mean ± SD from four wells. *, P < 0.05 compared to cells treated with LPS alone. (E) Poly(I:C), but not PAM3CSK4-induced miR-147 expression requires de novo protein synthesis. Peritoneal macrophages were treated as in (C), except replacing LPS with 1 μg/mL PAM3CSK4 or 5 μg/mL poly(I:C).
LPS stimulation induces the expression of type I IFN mediated by IRF3 activation. Type I IFN subsequently activates STATs and STAT-dependent gene expression. To determine if such autocrine or paracrine events are involved in LPS-induced miR-147 expression, we first demonstrated that IFN β, but not IFN α, is rapidly induced upon LPS stimulation (Fig. 3B and Fig. S2C). Next, we pretreated macrophages with cycloheximide (CHX), a protein synthesis inhibitor, to block new protein syntheses and the resultant autocrine or paracrine effects. We found that LPS-induced pri-mir-147 expression was diminished by CHX pretreatment (Fig. 3C). However, the expression of IL-1β, which requires no new protein synthesis, was not affected by CHX pretreatment (Fig. 3D). These data suggest that LPS-induced miR-147 expression requires de novo protein synthesis. To further demonstrate the dependency of miR-147 induction on both NF-κB and IRF3, macrophages were pretreated with CHX before being stimulated with PAMC3SK4 or poly(I:C). As shown in Fig. 3E, CHX pretreatment diminished poly(I:C)-induced, but not PAMC3SK4-induced pri-mir-147 expression. These data suggest that miR-147 induced by TLR3 stimulation is mediated by IRF-dependent autocrine and/or paracrine activities and that TLR2 induced miR-147 expression occurs directly through NF-κB activation.
TLR4-Induced miR-147 Expression Requires Both MyD88 and TRIF.
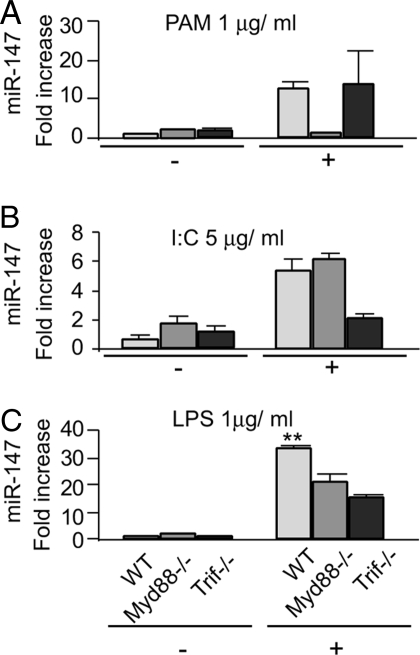
TLR4 stimulation activates both MyD88 and TRIF mediated signaling events. To determine whether LPS-induced miR-147 expression is MyD88 or TRIF dependent, we examined miR-147 expression in peritoneal macrophages isolated from wild-type (WT), MyD88−/−, and TRIF−/− mice. As shown in Fig. 4 A and B, PAM3CSK4 induced comparable levels of miR-147 expression in WT and TRIF−/− macrophages, but not in MyD88−/− cells whereas poly(I:C) induced comparable levels of miR-147 expression in WT and MyD88−/− macrophages, but not in TRIF−/− cells. These data are consistent with previous results showing the requirement of MyD88 for TLR2 mediated signaling events and of TRIF for cellular activation following TLR3 stimulation (1, 19, 20). In contrast, LPS-induced miR-147 expression was significantly decreased in both MyD88−/− and TRIF−/− macrophages, suggesting that the induction of miR-147 is both MyD88−/−- and TRIF−/−-dependent upon TLR4 engagement (Fig. 4C).
Fig. 4.
TLR4 induced miR-147 expression requires both MyD88 and TRIF. (A–C) Peritoneal macrophages were isolated from WT, MyD88−/−, and TRIF−/− mice. The cells were treated with 1 μg/mL PAM3CSK4 (A), 5 μg/mL poly(I:C) (B), or 1 μg/mL LPS (C) for 0 or 6 h. Real-time PCR was performed to determine the levels of miR-147. Values are presented as mean ± SD from triplicate wells. **, P < 0.01 compared to MyD88−/− or TRIF−/− macrophages.
The miR-147 Promoter Is Responsive to LPS Stimulation.
To determine if miR-147 is directly induced by NF-κB and/or IRF3, we analyzed the promoter region of the miR-147 gene. We found three potential NF-κB consensus binding sequences within approximately 2 kb upstream of the transcriptional start site (Fig. 5A). We found no IRF consensus sequence, but there is a GAS element between the two NF-κB sites within the proximal promoter region (Fig. 5A). To determine if the miR-147 promoter is responsive to LPS stimulation, we cloned the 2 kb promoter region into a luciferase reporter. We also generated mutants with deletion of various consensus sequences (Fig. 5A). As shown in Fig. 5B, the 2-kb promoter was responsive to LPS stimulation in RAW264.7 cells. Deletion of the two distal NF-κB binding sites had no effect on luciferase activity, suggesting that they are not necessary for the promoter activation. However, when the GAS element and the NF-κB binding site closer to the transcriptional start site were deleted, the promoter became significantly less responsive to LPS stimulation (Fig. 5B). These data suggest that the activation of the miR-147 promoter by LPS stimulation requires both the GAS element and the NF-κB binding site. Mutant promoter with an internal deletion of the GAS element and the proximal NF-κB binding site was still responsive to LPS stimulation, although to a lesser extent than the full-length promoter (Fig. 5C), suggesting that the two distal NF-κB binding sites alone can be activated in the luciferase assay.
Fig. 5.
The miR-147 promoter is responsive to LPS stimulation. (A) Schematic representation of the miR-147 promoter, the locations of various transcriptional factor binding sites, the luciferase reporters containing the 2-kb miR-147 promoter and reporters with various deletions. (B) The miR-147 promoter is responsive to LPS stimulation. The luciferase reporters (0.2 μg) were transfected into RAW264.7 cells, and 24 h after transfection, the cells were cultured without or with 1 μg/ml LPS for an additional 16 h. Luciferase activities were determined and normalized to the protein concentration of the lysates. Luciferase activity with the empty vector pGL2 was regarded as 1. Values are presented as mean ± SD from 3–4 wells. *, P < 0.05 compared to cells transfected with pGL2. (C) Mutant promoter with an internal deletion of the GAS element and the NF-κB binding site proximal to the transcriptional start site is responsive to LPS stimulation. Experiments were performed as in (B). *, P < 0.05 compared to cells transfected with FL. (D) NF-κB and STAT1α bind to the miR-147 promoter. RAW264.7 cells were treated with 1 μg/ml LPS for 0, 30, 60, and 120 min. The cell extracts were immunoprecipitated with anti-p65 or STAT1α Abs. Specific primers were used to amplify the promoter region (−789 to −596) containing the potential NF-κB binding site and the GAS element in the miR-147 promoter and the NF-κB binding site in the IκB-α promoter.
GAS elements bind to STAT family members, which are activated in LPS stimulated cells through IRF3 induced type I IFN (21). To further examine the role that NF-κB and STATs may play in LPS induced miR-147 expression, we performed ChIP assays and found binding of both the NF-κB p65 subunit and STAT1α to the promoter region containing the NF-κB-binding site and the GAS element (Fig. 5D). As expected, the NF-κB p65 subunit bound to the promoter of the NF-κB-dependent gene, IκB-α (Fig. 5D).
miR-147 Is a Negative Regulator of Macrophage Inflammatory Responses.
Given that miR-147 was induced in TLR2-, TLR3-, and TLR4-stimulated macrophages, we asked if this microRNA might participate in regulating inflammatory responses. As shown in Fig. 6A, transfection of miR-147 mimics or control microRNA mimics with scrambled sequences alone did not activate macrophages to produce pro-inflammatory cytokines, such as TNF-α or IL-6. However, transfection of miR-147 mimics into peritoneal macrophages significantly decreased LPS induced TNF-α and IL-6 production as well as the expression of several other LPS induced genes, such as PYHIN, UBCH8, ZBP1, and miR-147 itself (Fig. S3A). These data suggest that miR-147 can function as a negative regulator of inflammatory response. Given the ability of miR-147 to down-regulate TLR4 induced inflammatory responses, we next investigated if miR-147 can also modulate activation of TLR2 and TLR3 stimulated cells. As shown in Fig. 6 B and C, miR-147 attenuated TNF-α and IL-6 production induced by PAM3CSK4 and poly(I:C) in macrophages.
Fig. 6.
miR-147 is a negative regulator of macrophage inflammatory responses. (A–C) miR-147 mimics attenuate LPS, PAM3CSK4, and poly(I:C) induced inflammatory response in primary macrophages. Peritoneal macrophages were transfected with 40 nM control microRNA mimics or miR-147 mimics. At 48 h after transfection, the cells were cultured without or with 1 ng/mL LPS (A), 1 μg/mL PAM3CSK4 (B), or 5 μg/mL poly(I:C) (C) for 18 h. The levels of TNF-α and IL-6 in the supernatants were determined by ELISA. (D and E) miR-147 knockdown enhanced LPS and poly(I:C) induced IL-6 expression. Peritoneal macrophages were transfected with 40 nM control LNA knockdown probes or LNA miR-147 knockdown probes. At 48 h after transfection, the cells were cultured without or with 1 ng/mL LPS (D) or 5 μg/mL poly(I:C) (E) for 18 h. The levels of TNF-α and IL-6 in the supernatants were then determined. Values are presented as mean ± SD from triplicate wells. *, P < 0.05, **, P < 0.01, and ***, P < 0.001 compared to control groups.
To further address this question, we transfected microphages with specific miR-147 knockdown or control probes with scrambled sequences. The miR-147 knockdown probes or the control probes alone did not activate macrophages to produce TNF-α or IL-6. However, miR-147 knockdown markedly enhanced LPS and poly(I:C) induced TNF-α and IL-6 production (Fig. 6 D and E).
As shown in Fig. S3B, cytokine expression is similar in LPS-stimulated cells that were not transfected with probes (i.e., in the absence of control microRNA probes) and in cells transfected with control microRNA probes, but was decreased in cells transfected with miR-147 probes. This suggests that transfection of control microRNA probes does not affect macrophage inflammatory response. To further rule out any possibility that the observed anti-inflammatory effects associated with transfection of miR-147 mimics or knockdown probes may be artifactual and caused by transient transfection of short RNA oligos, we used the RAW264.7 stable cell line that can inducibly express mature miR-147. As shown in Fig. S3 C–E, the production of TNF-α and IL-6 after culture of RAW264.7 cells with LPS, PAM3CSK4, or poly(I:C) was decreased after miR-147 induction. These data demonstrate that miR-147 attenuates the inflammatory response of macrophages activated through TLR2, TLR3, or TLR4.
Since mature miR-147 is located in the 3′ UTR of the NMES1 transcript, the miR-147 knockdown probes may suppress NMES1 expression. To determine if NMES1 is involved in the effects of LNA probes on macrophage inflammatory responses, we used siRNA that targets the coding region of the NMES1 transcripts. As shown in Fig. S4A, LPS stimulation significantly increased NMES1 expression in the non-targeting control siRNA transfected macrophages. However, LPS induced NMES1 expression was markedly reduced in the NMES1 siRNA transfected cells. As noted, NMES1 siRNA did not decrease LPS-induced mature miR-147 expression (Fig. S4B). These data suggest that the NMES1-coding region targeting siRNA only knocks down NMES1, but not mature miR-147 expression. As further demonstrated in Fig. S4 C and D, down-regulation of NMES1 had no effects on LPS-induced TNF-α and IL-6 expression, suggesting that the effects of the LNA probes are independent of NMES1.
Discussion
In this study, we found that the microRNA, miR-147, is induced in murine macrophages after stimulation with TLR2, TLR3, and TLR4 ligands and in the lungs of mice after LPS exposure. miR-147 appears to have potent anti-inflammatory properties as it attenuates the expression of proinflammatory cytokines in TLR2-, TLR3-, and TLR4-stimulated macrophages. These results therefore demonstrate the participation of miR-147 in a negative feedback loop that is able to inhibit the proinflammatory response of macrophages to multiple TLR ligands.
These experiments present the initial identification of primary miR-147 and suggest that the NMES1 transcripts function as primary miR-147. The evidence for this conclusion is two-fold: first, not only the expression of mature miR-147, but also that of NMES1 was induced by the same set of TLR ligands and, under these conditions, the expression of mature miR-147 and NMES1 shared similar kinetics; second, transfection of full-length NMES1 cDNA produced mature miR-147. However, as shown in Fig. 2, only a small portion of NMES1 transcripts are processed into mature miR-147. The functional consequences of NMES1 induction by TLR stimulation in macrophages are presently unknown. A previous study suggested that NMES1 may have a role as a tumor suppressor (22). Given that inflammation has been closely associated with carcinogenesis, elevation of NMES1 in inflammatory diseases may be a protective mechanism to prevent cellular transformation. The human homologue to miR-147, hsa-miR-147b, also overlaps with the human NMES1 transcript. However, the basal level of hsa-miR-147b in human monocytic cells is very low. Furthermore, compared to the up-regulation of miR-147 expression in LPS stimulated mouse macrophages, the increase of hsa-miR-147b in LPS treated human monocytic cells seems marginal, suggesting that the mechanisms leading to the induction of miR-147 and hsa-miR-147b may be different and that the function of hsa-miR-147b may be distinct from that of miR-147.
In contrast to early responsive genes in inflammation, such as TNF-α and IL-1β, which show diminished transcription within hours after TLR engagement, mature miR-147 appears to be highly stable and levels were elevated for prolonged periods in LPS-stimulated macrophages. Since microRNAs repress gene expression through binding to the 3′ UTR of target mRNAs, miR-147 is unlikely to regulate the early inflammatory response, which normally has peaked within hours after TLR engagement. It is more likely that miR-147 targets an inducible positive modulator in the TLR signaling pathways. Consistent with this hypothesis, we found that enhanced expression of miR-147 attenuated inflammatory cytokine production only at later time points, such as 18 h after TLR2, TLR3, or TLR4 stimulation, but not at earlier time points.
miR-147 was induced in the lungs of LPS-treated mice. Because the lung contains a mixture of cell populations, it is currently unknown which cell population contributed to the elevation of pulmonary miR-147 levels. The numbers of inflammatory cells, which are primarily neutrophils and accumulate in the lungs after intratracheal LPS exposure, had diminished to near normal levels by three days after instillation of LPS (23). However, the levels of mature miR-147 in the lungs were at their highest levels at this time point, suggesting that pulmonary cells other than neutrophils and macrophages also had increased miR-147 expression. Given that miR-147 appears to have anti-inflammatory effects, its late increase in the lungs after TLR4 engagement may play a role in attenuating pulmonary inflammatory responses.
Both TLR2 and TLR3 engagement induced miR-147, but to a more limited extent than did TLR4 stimulation. TLR2 stimulation activates NF-κB and TLR3 engagement primarily activates IRF3, whereas TLR4 stimulation activates both (1). These results suggested that maximal miR-147 induction requires both NF-κB and IRF3 activation. This conclusion is supported by our findings that significantly lower levels of miR-147 were found in LPS-treated MyD88−/− or TRIF−/− macrophages than in WT cells. Of note, MyD88 participates in LPS induced NF-κB activation, whereas TRIF mediates LPS-induced IRF3 activation (1). Furthermore, functional assays with the miR-147 promoter demonstrated that the GAS element and the NF-κB-binding site closest to the transcriptional start site were essential for LPS-induced activation. In addition, results with ChIP assays provided direct evidence that NF-κB and STAT1α bound to the miR-147 promoter region.
miR-147 was found to regulate inflammatory responses to TLR2, TLR3, and TLR4 stimulation. Given that these TLRs share common pathways that lead to activation of NF-κB and/or IRF3, it would seem likely that miR-147 targets upstream mediators, such as MyD88 and TRIF. However, we found no change in the degradation or phosphorylation of IκB-α or in the phosphorylation of p65, indicating that miR-147 expression does not modulate signaling upstream of these events. Targets for miR-147 identified by computational prediction programs, such as TargetScan, have not previously been shown to be directly involved in TLR signaling events. Thus, the mechanism by which miR-147 regulates TLR-induced inflammatory responses requires further investigation.
We found that miR-147 attenuated the inflammatory responses of macrophages after stimulation through TLR2, TLR3, and TLR4. The specificity of these observations was shown because miR-147 mimics and miR-147 knockdown had opposite effects. Furthermore, a RAW264.7 cell line that inducibly expressed mature miR-147 also demonstrated the inhibitory effects of this microRNA on TLR induced inflammatory responses. However, it should be noted that the RAW264.7 cell line expresses full-length NMES1 cDNA. The effects of NMES1 over-expression on TLR-induced macrophage activation are presently unknown. However, given our results with transient transfection of mature miR-147 mimics as well as with the stable miR-147-expressing cell line, a role for miR-147 as a negative regulator of inflammatory response in macrophages appears to be well-established.
Materials and Methods
Reagents.
Thioglycolate and LPS from Escherichia coli 0111:B4 were from Sigma-Aldrich. PAM3CSK4 and poly(I:C) were from Invivogene. Rabbit anti-p65 and anti- STAT1α antibodies were from Santa Cruz.
Mice.
C57BL/6 mice were purchased from NCI-Frederick. TRIF−/− mice were purchased from Jackson Labs. MyD88−/− mice were previously described in (24). All animal protocols were approved by the UAB Institutional Animal Care and Use Committee (IACUC).
Cell Culture.
Culture of HEK-293, RAW 264.7, THP-1, and mouse peritoneal and alveolar macrophages were described in SI Text.
Plasmids.
pcDNA4-miR-147 and pBabe-H1-miR-147 were described in SI Text.
Generation of a Stable Cell Line Inducibly Expressing miR-147.
A RAW264.7 cell line that expressed the tet repressor was first established. The cells were then transfected with pcDNA4-miR-147 and cultured under zeocin selection. Two individual clones that inducibly expressed miR-147 (RAW264.7-miR-147#5 & #8) were used for experiments.
Transfection of MicroRNA Mimics and Inhibitors.
Transfection of microRNA mimics and inhibitors was described in SI Text.
Real-Time RT-PCR.
Real-time PCR was performed using a Lightcycler 480 SYBR Green I Master system (Roche). The expression of mmu-miR-147 and hsa-miR-147b was determined by Real-time PCR using the Taqman Probe Master (Roche) system with Taqman hsa-miR-147b probes (Applied Biosystems).
Luciferase Reporters.
Approximately 2 kb of miR-147 promoter region upstream of the transcriptional starting site and the promoter region with deletion of the two distal NF-κB binding sites, deletion of the two distal NF-κB binding sites and the GAS site, deletion of all three NF-κB-binding sites and the GAS site, or internal deletion of the GAS site and the NF-κB-binding site proximal to the transcriptional starting site were obtained by PCR amplification and subcloned into pGL2. The resulting constructs were termed FL, Δ1, Δ2, Δ3, and Δ4 respectively.
Statistical Analysis.
One-way ANOVA and Student t test were used. P < 0.05 was regarded statistically significant.
Supplementary Material
Acknowledgments.
This work was supported in part by a pilot grant to G.L. under National Institutes of Health (NIH) grant # 5P30DK072482. This work was also supported in part by NIH grants HL62221, HL76206, and GM87748 to E.A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.A.J.O. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901216106/DCSupplemental.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 3.Bell JK, et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 4.O'Neill LA. How Toll-like receptors signal: What we know and what we don't know. Curr Opin Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Moynagh PN. The NF-kappaB pathway. J Cell Sci. 2005;118:4589–4592. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 6.Honda K, Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 10.Kulshreshtha R, et al. Regulation of microRNA expression: The hypoxic component. Cell Cycle. 2007;6:1426–1431. [PubMed] [Google Scholar]
- 11.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 13.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 16.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell RM, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akira S, Takeda K, Kaisho T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 21.Qin H, et al. LPS induces CD40 gene expression through the activation of NF-kappaB and STAT-1alpha in macrophages and microglia. Blood. 2005;106:3114–3122. doi: 10.1182/blood-2005-02-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, et al. A novel gene, NMES1, downregulated in human esophageal squamous cell carcinoma. Int J Cancer. 2002;101:311–316. doi: 10.1002/ijc.10600. [DOI] [PubMed] [Google Scholar]
- 23.Abraham E. Neutrophils and acute lung injury. Crit Care Med. 2003;31:S195–199. doi: 10.1097/01.CCM.0000057843.47705.E8. [DOI] [PubMed] [Google Scholar]
- 24.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.