Abstract
To what extent genotypic differences translate to phenotypic variation remains a poorly understood issue of paramount importance for several cornerstone concepts of microbiology including the species definition. Here, we take advantage of the completed genomic sequences, expressed proteomic profiles, and physiological studies of 10 closely related Shewanella strains and species to provide quantitative insights into this issue. Our analyses revealed that, despite extensive horizontal gene transfer within these genomes, the genotypic and phenotypic similarities among the organisms were generally predictable from their evolutionary relatedness. The power of the predictions depended on the degree of ecological specialization of the organisms evaluated. Using the gradient of evolutionary relatedness formed by these genomes, we were able to partly isolate the effect of ecology from that of evolutionary divergence and to rank the different cellular functions in terms of their rates of evolution. Our ranking also revealed that whole-cell protein expression differences among these organisms, when the organisms were grown under identical conditions, were relatively larger than differences at the genome level, suggesting that similarity in gene regulation and expression should constitute another important parameter for (new) species description. Collectively, our results provide important new information toward beginning a systems-level understanding of bacterial species and genera.
Keywords: comparative genomics, evolution, proteomics, speciation, phenotype
Predicting the phenotype of newly isolated organisms based upon the existing knowledge of previously characterized organisms constitutes one of the most fundamental goals of microbiology. Organisms isolated from diverse environments and habitats often have their phenotypic and physiological properties inferred from their evolutionary relatedness, measured by (mainly) the 16S rRNA gene sequence identity or other means (1, 2), to the type strains of known species. Although this practice has been broadly applied in studies of microbial communities, contributing greatly toward advancing microbiology knowledge, its use in this manner is rooted in rather low-resolution experimental methods and procedures (1, 3). The powerful genomic tools now available provide the opportunity for a much more detailed and informative evaluation of the relationship between genetic and phenotypic similarity. Simple questions that remain unanswered or only partially explored, such as to what degree do microorganisms encode and express the same metabolic pathways when grown under identical conditions, and to what extent are the similarities in expressed pathways determined by the genetic relatedness and/or the (distinct) ecological adaptations of the microorganisms, can now be answered accurately and quantitatively. Addressing such questions will provide long-needed information to better understand and to model the enormous microbial biodiversity that exists on the planet.
To this end, we have analyzed and compared, both at the whole-genome and the whole-proteome levels, 10 isolates belonging to the genus Shewanella, an important genus in cycling of organic and inorganic materials in the environment (4). These isolates originated from diverse geographic locations and habitats, including fresh and marine water columns, sediments, and subsurface environments (Fig. 1A and Table S1), and carry out a diverse range of metabolic processes (4). Although precise ecological information, e.g., in situ abundance and persistence in time, about each isolate is typically not available, the procedure used to isolate these strains, i.e., enrichment cultures from a variety of environmental samples for the phenotype or genotype of interest, is similar to common microbiology practice. Accordingly, our analyses with the Shewanella strains should be relevant for the questions described above and for broadening our understanding of the interrelationship between genotype, phenotype, environment, and evolution. Our results represent the first thorough and system-level assessment of an environmental representative of Proteobacteria, an enormously diverse and important group, that can be compared and contrasted to previous assessments of the heavily sampled human pathogens or the ecologically specialized organisms such as the photosynthetic Prochlorococcus (5). Such comparisons identified several trends that may apply to other environmentally versatile bacteria besides Shewanella.
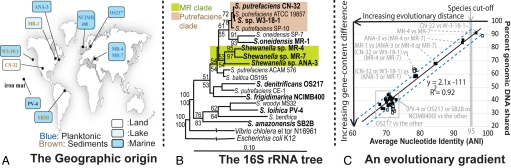
Fig. 1.
The 10 Shewanella genomes used in this study and their evolutionary gradient. The geographic origin (A) and the 16S rRNA-based phylogenetic tree (B) of the 10 genomes (in boldface type) are shown. The scale represents the number of substitutions per position, and the numbers above and below the nodes represent the bootstrap support from 1000 resamplings using parsimony and maximum likelihood methods, respectively. Bootstrap values <50 were omitted. A continuous genetic gradient was formed (C) when the fraction of the total genes in the genome shared between two genomes (y axis) was plotted against the ANI of the shared genes between the two genomes (45 comparisons in total are shown). Dashed blue lines represent the 90% prediction intervals of the regression line; open squares identify the outlier pairs of genomes observed (discussed in the text).
A Continuous Genetic Gradient Within a Genus.
Phylogenetic analysis of the 16S rRNA gene sequences revealed that the 10 Shewanella isolates formed a tight cluster, with the intra-cluster sequence identity ranging from 92 to ≈100% (Fig. 1B). Hence, these genomes belong justifiably to the same genus according to the most frequently used standards of bacterial taxonomy (2, 6). To gain further insight into the diversity of this group, the average nucleotide identity (ANI) of all pairwise conserved genes between (any) two genomes, a more sensitive parameter for measuring evolutionary relatedness among closely related genomes than the 16S rRNA gene (7), was used. The ANI analysis revealed that these genomes form a continuing gradient of genetic relatedness, which was not readily apparent from the 16S rRNA gene analysis (Fig. 1C). In particular, S. putrefaciens strains W3–18-1 and CN-32 as well as Shewanella sp. MR-4 and MR-7 are the most closely related pairs, showing ANI values of ≈96.5% and ≈98.4%, respectively. These values are well above the 95% ANI that corresponds to the 70% DNA-DNA hybridization (DDH) standard frequently used for species demarcation, which is consistent with the experimentally derived DDH values for these organisms (6). Hence, these pairs of genomes sample the subspecies level. The MR-4 and MR-7 genomes show ≈92%, ≈85%, and ≈79% ANI to Shewanella sp. ANA-3, S. putrefaciens CN-32, and S. oneidensis MR-1 genomes, respectively. Thus, these genome pairs represent varied levels of genetic relatedness within the Shewanella genus. Finally, all of the previously mentioned genomes show ≈69.7–72% ANI to S. frigidimarina NCIMB400, S. denitrificans OS217, S. loihica PV-4, and S. amazonensis SB2B strains, which represent the four most divergent species sampled within the genus. This gradient provided the opportunity to precisely estimate the number of changes in the genes, pathways, and subsystems of the cell over time and as a result of environmental adaptations and selection pressures.
Gene Content Variation as a Function of Evolutionary Time and Ecology.
The 10 Shewanella isolates have similar genome sizes, varying from 4.3 to 5.3 Mbp (Table S1). Comparative analysis revealed extensive gene content diversity among the genomes. From the 9782 predicted nonredundant (orthologous genes removed) protein-coding sequences (CDS) annotated in the 10 genomic sequences (the pangenome), only ≈2128 (22%, constituting ≈54% of the total genes in the genome, on average) were present in all genomes (core CDS set); ≈2965 (30%) were found in at least two genomes (variable CDSs), whereas the remaining CDS (4689 or 48%) were strain specific (Fig. 2B, Fig. S1, and Table S2). Nonetheless, the majority of the variable CDSs were found to be specific to clades, i.e., the MR or S. putrefaciens clades (Fig. 1B), whereas a smaller fraction had a more sporadic distribution among the strains (note the similarity between the gene content tree and the phylogeny of the genomes in Fig. 3). Accordingly, the overall extent of CDS content similarity showed a very strong linear decrease with increasing evolutionary distance between the genomes compared (R2 > 0.9; Fig. 1C), which is consistent with results reported previously based on other bacterial groups (7). The strong linear trend suggests that, despite the extensive gene diversity and apparent genome fluidity, the genotypic similarity of bacteria may be generally predictable from their evolutionary relatedness.
Fig. 2.
The Shewanella pangenome. (A) Contribution of different categories of genes to the pangenome as a function of ANI. The genes that differed in all pairwise whole-genome comparisons among the 10 Shewanella genomes (45 comparisons in total) were assigned to five major functional categories (graph legend). The number of genes in each category, expressed as a fraction of the total genes that differed between the two genomes (y axis), is plotted against the genomic ANI value of the two genomes compared. Individual data points representing each comparison have been removed for clarity; trendlines representing the mean, and bars representing one standard deviation from the mean, are shown instead. (B) Comparisons with the enterics pangenome. The number of genes that remained conserved (y axis) with the inclusion of more genomes in the analysis is plotted against the number of genomes (x axis) used (light colors). The total number of nonredundant unique genes in all genomes used is also shown (dark colors). Bars represent one standard deviation based on 10 random combinations in adding the genomes to the analysis.
Fig. 3.
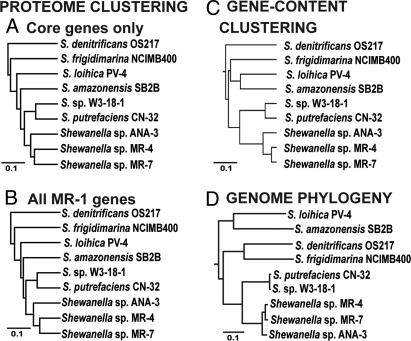
Genome vs. proteome comparisons among nine Shewanella strains. The protein profiles of nine Shewanella strains were compared based on the 2128 core genes (A) and the 4300 genes found in the genome of strain MR-1 (B) for gene expression, and the nine strains were subsequently clustered based on their overall similarity in the expression patterns of these two gene sets as follows. For each gene set, a full (all genes by all genomes) 0/1 matrix was built, with “1” denoting expression (defined as the detection of at least two unique peptides per protein) and “0” denoting no expression of the corresponding protein; the derived matrices were clustered as described in the SI Materials and Methods and the resulting cladograms are shown. Similarly, the nine strains were also clustered based on the presence/absence of the 4300 MR-1 gene orthologs in their genome (sequence comparisons, C). A maximum-likelihood phylogenetic tree of the concatenated alignment of 1507 single-copy core genes that had no detectable signal for recombination by Phi test analysis (22) is also shown (D). Scale bars represent percent similarity in the derived matrices for (A), (B), and (C); and number of substitution per site for (D).
Although a tight relationship between shared CDS content and evolutionary relatedness was observed, several significant departures (outliers) from this main trend were also noted and were most likely attributable to ecological adaptations. For instance, the two most closely related genomes based on ANI, CN-32, and W3–18-1 (98.4% ANI) showed substantially more CDS content differences compared with what was expected based on their small evolutionary divergence (see regression trendline in Fig. 1C) or compared with the more distantly related (96.4% ANI) pair of MR-7 and MR-4 (≈530 vs. ≈430 CDSs, respectively, not counting CDS on mobile elements; Table S2). CN-32 and W3–18-1 were isolated from more diverse environments (deep-subsurface sandstone vs. marine sediment, respectively) compared with MR-4 and MR-7 (5-m vs. 60-m depth in the Black Sea, respectively). Hence, it is likely that genetic adaptations specific to these environments account for the larger gene content differences observed in the former strains relative to the latter ones. In agreement with the latter interpretation, CN-32-specific genes included several genes that might be important for survival in the subsurface environment, such as an arsenate reductase, copper resistance system, heavy metal efflux pump, and a polysaccharide biosynthesis cluster.
Similarly, S. denitrificans strain OS217 is as divergent as three other isolates (strains PV-4, NCIMB400, and SB2B) are from the remaining six Shewanella isolates in our collection (e.g., Fig. 3D). Yet, the OS217 genome contained substantially more strain-specific genes and showed the greatest loss of “core-like” CDSs (i.e., CDSs present in all other Shewanella genomes) compared with the genomes of PV-4, NCIMB400, or SB2B (Table S2). For instance, the core set increased by 265 genes when OS217 was removed from the analysis compared with <60 genes when PV-4, NCIMB400 or and SB2B were individually removed. Our genomic, physiological (e.g., Table S3), and proteomic data collectively suggest that strain OS217 has undertaken a unique evolutionary path, possibly driven by the loss of the three menaquinone biosynthetic gene clusters (menDHCE, menF, menB) common to the other Shewanella strains and resulting in an inability to exploit strictly anaerobic habitats. These results are consistent with previous findings suggesting that strain OS217 is a specialized denitrifier (4) and are also consistent with the longstanding observation that respiratory denitrification is not found in organisms that are strong fermentors (8). These findings may indicate that more extensive genetic changes are involved for an organism to diverge to the opposite physiology. Finally, the (outlier) pairs of genomes with a higher percentage of shared genes than the average, i.e., CN-32 or W3–18-1 vs. MR-4 or MR-7 (Fig. 1C), are attributable to the substantially smaller size of these genomes (i.e., 4.6–4.7 Mbp) relative to that of the rest of the genomes (i.e., ≈5.2 Mbp; Table S1) rather than to more similar ecological adaptations (the number of shared orthologs and mobile gene content in these pairs is comparable to that of other pairs).
Processes Contributing to Gene Content Variation.
To provide further quantitative insights into the processes contributing to gene content variation, the genes that differed in pairwise whole-genome comparisons were assigned to five major functional categories and the percentage of genes in each category was evaluated against the genetic relatedness of the two genomes compared. The five categories were as follow: (i) pseudogenes, denoting genes predicted to encode insertions, deletions, or sequence alterations that would result in premature termination of the encoded protein; (ii) IS/Tn, denoting insertion sequences or transposons; (iii) mobile islands, denoting runs of neighboring genes (genomic islands) that included integrase genes; (iv) other, denoting all other unique genes, including genomic islands that do not contain clear evidence of being mobile; and (v) hypothetical or conserved hypothetical, denoting the fraction of the genes in category iv that had no detectable homolog in any of the fully sequenced genomes except in other Shewanella genomes (Table S4). Our results revealed that mobile islands and insertion elements dominated the gene content differences among genomes of the same species, but their contribution gradually decreased in comparisons among genomes of increasing evolutionary divergence at the expense of genes in the “other” category (Fig. 2A). These findings are consistent with rapid turnover of mobile islands over short evolutionary scales. Furthermore, the majority (>75%) of the genes in the “other” category were typically found in clusters of ≈5–40 genes, reflecting presumably their “mobile island” origin. These findings are consistent with preferential deletion of the mobility/transposition genes (presumably because of negative selection) in the course of evolution and retention of only the potentially ecologically important genes of mobile islands. Therefore, the Shewanella organisms evaluated here appear to acquire most of their new functions as follows: acquisition of mobile islands, followed by selection for the islands carrying ecologically important genes, and finally loss of the mobile and ecologically unimportant genes.
The Shewanella Pangenome and Conserved Gene Core.
Comparative analysis of the 10 Shewanella genomic sequences revealed that sampling of the genus pangenome remained unsaturated (Fig. 2B, blue bars); this result was attributable to the large number (468, on average) of strain-specific genes. Only 10–25% of the latter genes, depending on the genome evaluated, found a homolog in a genome outside the Shewanella genus when queried against all bacterial genomes available at the end of 2008, indicating the great potential for discovering novel genes with more Shewanella strains sequenced. The number of new genes per genome is an order of magnitude higher than those calculated for highly specialized human pathogens (9) but significantly lower than that of the opportunistic pathogen Escherichia coli (7). It must be pointed out, however, that these pan-genome calculations are not directly comparable and should be interpreted with caution. For instance, the average ANI value among all pairs of Shewanella genomes is ≈76%, which is significantly lower than that within the E. coli group (≈96%), and there appears to be a strong positive correlation between the number of novel genes carried in a genome and the (higher) degree of evolutionary divergence of the genome, regardless of the effect of ecology or environmental adaptation (Fig. 1C) (7). On the other hand, the prophage content of the E. coli genomes is substantially higher than that of the Shewanella genomes (10–20% vs. 0–5%, respectively), and this accounts for much of the difference observed. When the groups were adjusted for comparable intragroup diversity, by including selected Salmonella (≈82% ANI to E. coli) and Yersinia (≈72% ANI to E. coli) genomes together with E. coli genomes and with prophage genomes removed from the analysis, the gene diversity observed within the enterics was comparable to that of the Shewanella (Fig. 2B). Therefore, the evaluation of these two important groups suggests that sequencing of any new organism, as long as the organism belongs to a versatile genus and has a different ecological history relative to the previously sequenced members of the genus, should be expected to expand substantially the pangenome of the genus.
Both the Shewanella and the enterics core gene sets were highly enriched in translational, transcriptional, DNA replication, and central metabolism genes and overlapped extensively (≈50% of the genes were shared between the two cores). Shewanella-specific core functions were associated mainly with metabolic pathways as well as with chemotaxis and sensory-transduction processes. Using the BioCyc pathway schema (10), 104 pathways were identified as being common to all Shewanella genomes, including pathways for energy metabolism, synthesis of building blocks (amino acids, cofactors, fatty acids, and nucleotides), and degradation or interconversion of metabolites and all but two amino acids and metabolites (Fig. S2 and Table S5). A common trait of the Shewanella strains appears to be the use of the pentose phosphate and Entner-Doudoroff pathways for hexose degradation. This is based on the lack of the enzyme 6-phosphofructokinase (Pfk; the most important regulatory enzyme of the canonical glycolysis pathway), initially observed in previous gene expression studies of MR-1 cultures (11). Members of the Shewanella genus also have fewer phosphotransferase system (PTS) transporters than usually encountered in proteobacterial genomes. Whether there is a connection between the reduced PTS and lack of Pfk is not clear, but it is possible that the lower level of phosphoenolpyruvate (PEP) synthesized as a result of not using the glycolytic pathway may render the PEP-dependent PTS system inefficient.
When the core was defined as the genes present in all but one of the 10 genomes, the dataset increased by 411 protein coding genes (265 when OS217 was excluded from the analysis), corresponding to, on average, 12–14% of the Shewanella genome (Table S2). These findings suggest that gene loss, including loss of genes that are apparently indispensable for the majority of the strains of a species, might be a successful strategy for rapid evolution and environmental adaptation. A representative example of strain-specific adaptations related to a group core function, which involved considerable gene deletion and/or gene acquisition, is given below. All Shewanella strains except for S. denitrificans OS217, which shows limited anaerobic growth capabilities presumably because of gene loss during the process of ecological specialization (discussed above), were able to reduce several metals and metalloids (Table S3), a well-known characteristic of the genus (12). The main metal reductase locus, encoded by mtrCAB genes, is virtually identical for the nine strains but the adjacent loci vary, reflecting evolutionary history and possibly metal respiratory specialization (4). These dissimilarities explained some, but not all, of the variation in metal respiration among strains observed during our growth experiments. For example, although their mtr locus and flanking genes are identical, strain CN-32 was able to grow on lactate (20 mM) when six different metals or metalloids were used as electron acceptors, whereas strain W3–18-1 grew only with Fe, Mn, and Se, under the conditions tested. These results may reflect differences in the upstream pathways to metal reduction between the two strains and underscore the need for more research to understand better the details of the metal respiration cascade.
Gene Presence vs. Expression as a Function of Time and Ecology.
Transcriptome comparisons have shown that gene expression rather than gene content differences, occurring either at different times and/or in different tissues, are mainly responsible for the differential development of eukaryotic organisms, e.g., human and chimpanzees (13), and the adaptive evolution of natural populations (14). It follows that, in addition to the number of shared genes, gene expression constitutes an important factor determining phenotypic similarity (or dissimilarity). Although the latter applies presumably to bacteria as well, systematic assessments of the role of gene expression on the phenotypic differences observed among closely related organisms are lacking.
To begin exploring this issue, the 10 strains were grown under identical batch-culture conditions to obtain their whole-cell proteome profiles and to contrast the profiles against the evolutionary relatedness among the strains. Overall, the degree of similarity in proteome profiles was congruent with the evolutionary relatedness among the strains; i.e., the fraction of orthologous proteins detected to be expressed in the cultures was higher in closely related strains than in more divergent strains. However, the differences in expressed proteins among the strains were consistently larger than their differences at the gene content level when gene expression and gene content were assessed for the same 4300 (reference) genes found in the MR-1 genome (compare branch lengths in Fig. 3B and 3C), which minimized the effect of gene- or strain- specific variations in the measurements. More surprisingly, the same pattern was observed even when gene expression was assessed for the core genes only (Fig. 3A and Fig. S2), which circumvented the dependency of the proteome profiles on the underlying gene content differences in the previous comparisons. These results were attributable to a high number of proteins expressed by one or a few, but not all, of the strains possessing the corresponding gene, with proportions that varied from 1.9 to 2.6 times more than those proteins expressed by all strains possessing the corresponding gene (Table S6). For instance, although 20% of the core proteins (556 genes) were expressed by all strains, a substantially larger fraction of core proteins (36%, or 993 genes) were expressed by one or more (but not all) strains. Although some of these differences may be caused by higher noise in the proteomics data relative to the genomics data, we believe that many of these differences are biologically relevant because of the high reproducibility (>80%) of proteomics measurements on batch cultures such as those used in the present study (15), our high stringency in processing and analyzing the proteomics data (see Materials and Methods), and the fact that very similar results were found when a subset of five specific regions of traditional two-dimension protein gels were overlaid and compared for absence or presence of protein spots (Fig. S3). Finally, proteins characteristic of the stationary growth phase, such as the RpoS sigma factor (16), were not detected in the expressed proteomes, suggesting that all of our cultures were sampled at their exponential growth phase.
Our findings revealed that although strains CN-32 and W3–18-1 are significantly more closely related than are strains MR-4 and MR-7—e.g., a 2% higher ANI value translates to substantially higher gene content and evolutionary relatedness, as we and others have shown (7)—the former strains showed comparable differences in expressed proteins compared with the latter strains for the same genes analyzed (Fig. 3). These findings could therefore be attributable to a higher degree of environmental/ecological adaptations (which may have altered metabolic and regulatory networks) in the CN-32/W3–18-1 pair relative to the MR-4/MR-7 pair. Similarly, S. denitrificans OS217, which appeared to be the most ecologically specialized organism of the set, also showed the most unique proteomic profile (Fig. 3). The larger gene expression differences observed for OS217 and CN-32/W3–18-1 than anticipated based on their evolutionary divergence alone echoes the results described above based on the gene content analysis. Furthermore, the largest fraction (44%) of the proteins detected in the protein profiles was strain specific and included many nonhypothetical proteins such as outer membrane proteins, TonB-dependent receptors, proteases, restriction-modification enzymes, glycosylases, and polysaccharide biosynthesis enzymes. Most of these proteins can be linked to metabolic fitness or interaction with the environment, and hence could possibly underlie important physiological and/or regulatory differences among the strains. The extensive variability in core proteins and the high number of strain-specific proteins expressed under identical growth conditions indicates a multifaceted and highly dynamic control of whole genome expression. Collectively, our proteomics analyses suggest that changing this control appears to represent a particularly important mechanism, in addition to gene acquisition or loss, for rapid adaptation in changing and diverse environments. Consistent with these conclusions, the first mutations observed in experimentally evolved E. coli strains for 20,000 generations under laboratory conditions involved regulatory genes and networks (17).
Compartmentalized Microbial Evolution.
To characterize which cellular functions evolve faster in the Shewanellae, the percent conservation of selected functional gene categories (see Materials and Methods for details) was evaluated against the evolutionary relatedness among the strains compared (measured by percent ANI). As evolutionary distance increased, the percent conservation of all categories decreased, but the extent of decline (i.e., the slope) differed, presumably reflecting the varied selection pressures on the corresponding genes. The analysis revealed the following order: pathways were substantially more conserved than individual orthologs; orthologs more conserved than transcriptional regulators, sensing and respiration genes, and expressed proteins (Fig. 4). The most rapidly changing individual functions, both in terms of gene presence/absence and sequence conservation, were TonB-dependent outer membrane receptors followed by methyl-accepting chemotaxis proteins, transcription regulators and cytochromes. These results are consistent with our previous findings and suggest that genomic and regulatory changes in sensing mechanisms represent the first line of adaptive response to different redox conditions. Experimentally determined anaerobic growth characteristics such as biomass produced and electron acceptors used (Table S3) were also very different among the Shewanella strains and ranked among the fastest changing functional entities (Fig. 4). A growth phenotype encompasses the sensing of a substrate, expression of relevant regulators, transporters, and enzymes, in addition to physiological parameters related to the change in growth conditions. These potential sources for additional variation among the strains may explain why the growth phenotype is significantly less conserved compared with pathways, orthologs, and protein expression patterns.
Fig. 4.
Modeling bacterial genotypic and phenotypic conservation across an evolutionary gradient. The presence of orthologous proteins, TonB outer membrane receptors, cytochromes, methyl-accepting chemotaxis proteins (MCPs), transcriptional regulators, metabolic pathways, protein expression patterns, and reduction of metal or metalloids (anaerobic growth) was determined for the 10 Shewanella strains (see Materials and Methods). Each of the traits was compared among the Shewanella strains in a pairwise manner (45 comparisons in total). The fraction of shared traits was determined for each pair of strains and plotted against the average nucleotide identity (ANI) of the respective strain pair. Inset graph depicts the relationships between conservation of the traits and evolutionary distance using linear regression trendlines adjusted to intersect with the x and y axis at 100%. The r2 values of the regressions are also shown.
Summary and Perspectives for the Future.
Microbiologists have been primarily focused on comparisons among either very closely related strains of the same species or distantly related species to advance understanding of the microbial life on Earth. The 10 Shewanella genomes studied here were selected to represent a range of evolutionary distances, providing for a more unconstrained view of microbial diversity and evolution. Comparisons among these genomes revealed that the Shewanella genus is genomically and, more so proteomically, diverse. Although a high degree of variation in protein expression profiles was anticipated among distantly related species, the variation observed among strains of the same species was comparatively much larger than expected, given also the single growth condition used (Figs. 3 and 4). It also appears that, in some cases, the variation in expressed proteomes correlated positively with the extent of environmental adaptation (specialization). These findings have important implications for the correspondence between genotype and phenotype and, hence, for the bacterial species concept. The evolutionary and functional gradients reported here also suggested that specialization might occur over a very short time span, much shorter compared with that corresponding to the current species standards. Specialization appeared to take place primarily through changes at the regulatory level and through the high plasticity and fluidity of the Shewanella chromosomes (Fig. 4).
The power of “omics” compared with traditional approaches to unravel an organism's environmental and ecological adaptations and make robust predictions about the similarity (or difference) in phenotypic traits among organisms was also highlighted by our analyses. The published literature, as well as our experimentally derived physiological and growth data, could not easily distinguish between most of the strains used in this study or even define general properties for the major clades represented by these strains. This was also reflected in the very low correlation obtained between anaerobic growth characteristics (Table S3) and the evolutionary relatedness of the strains compared. In contrast, genomic and proteomic data correlated well with the phylogeny of the strains and identified congruently strain-specific adaptations that might be linked to speciation for several of the strains studied. These results further corroborate the notion that it is time to start replacing the traditional approaches for defining diagnostic phenotypes for new species or clades with omics-based procedures.
Distinguishing the effect of ecological adaptation from that of evolutionary divergence alone represents the most limiting factor in increasing the power of our predictions on phenotype based on the genotype. Toward this direction, studying the extent of variation among members of the same natural population, i.e., among organisms with very similar environmental adaptations, and contrasting it to the levels of variation detected in this study with diverse organisms will allow fruitful conclusions. The trendlines obtained in this study (Fig. 1C and Fig. 4) also provide a reference for comparing organisms of narrower (or broader) metabolic versatility than the Shewanellae. Furthermore, although the growth conditions used in this study were very limited, the conditions remain artificial compared with the environmentally relevant conditions and hence may represent different stresses for each isolate evaluated. Replicate experiments and experiments performed with continuous cultures (chemostats) are currently underway to provide further quantitative insights into the role of variation in gene expression. Finally, a major limitation remains in that, despite the dedicated efforts of numerous laboratories, many of the genes in the genome have not been experimentally characterized, and their physiological roles are unknown. Continuing the efforts to establish the functions of as many genes in the genome as possible is critical for a thorough understanding of a bacterium that could serve as a model for versatile environmental bacteria.
Regardless of these limitations, the results presented here constitute important information toward better modeling the correspondence between genotype and phenotype, and provide directions and testable hypotheses that will bring us one step closer to systems-level understanding of microbial species and populations.
Materials and Methods
The organisms used in this study, their genomic features, gene content, and accession numbers of the versions of the genomic sequences used in the study are provided in Table S1. Orthologs were identified for the 10 Shewanella genomes by a combination of three methods: (i) protein–protein pairwise reciprocal BLAST (blastp) (18); (ii) reciprocal protein-genomic sequence best match (tblastn); and (iii) Darwin pairwise best hit (19). Genes found in plasmids or mobile elements were excluded from ortholog and proteome comparisons among the strains. The degree of conservation of cellular functions or traits between two strains (Fig. 4) was determined as follows. (i) For orthologs, transcriptional regulators, TonB receptors, MCPs, and cytochromes: all genes in the genome assignable to each of these categories were determined based on the gene annotation and the number of orthologous genes shared between two strains for each category (according to Table S2) was divided by the total number of genes assignable to the category for each strain. The two values were averaged to provide the values used in Fig. 4. (ii) A total of 163 unique pathways were identified in the 10 Shewanella genomes according to the BioCyc pathway schema. The number of shared pathways between the strains, as a fraction of the total pathways carried by a strain, was determined based on the presence/absence of the corresponding pathway genes. (iii) For proteomes and anaerobic growth, the number of orthologous proteins expressed (Table S6) and metal/metalloids respired (Table S3) by both strains in a pair were divided by the total number of (nonredundant) proteins expressed and metal/metalloids respired by either strain, respectively. The use of “total traits counted for both strains” as the denominator (as opposed to “counts for one strain”) provided also for more direct comparisons with the sequence-based traits (i and ii, above) because otherwise the latter traits would have been penalized relatively higher because of the high number of “auxiliary” genes, which remained unexpressed under the simple growth conditions tested. For proteomics analysis, cultures were grown aerobically in Tryptic Soy Broth to a final optical density (OD) of 0.5. Cells were lysed, proteins extracted and digested with trypsin, and the resulting peptides analyzed by mass spectrometry as previously described (20), with the only exception that filtering of the data was performed as described in ref. 21. Two-dimensional proteomic gels were carried out as described previously (15). A detailed description of materials and methods is included in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors thank the numerous members of the Shewanella Federation for useful discussions during the course of their genomic investigations of Shewanella. The authors were supported by the U.S. Department of Energy through the Shewanella Federation consortium and the Proteomics Application project. The Michigan State University work relevant to speciation was also supported by the National Science Foundation (DEB 0516252). Portions of this research were performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy national scientific user facility located at the Pacific Northwest National Laboratory in Richland, Washington.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902000106/DCSupplemental.
References
- 1.Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 2.Brenner D, Staley J, Krieg N. Bergey's Manual of Systematic Bacteriology. 2nd Ed. New York: Springer-Verlag; 2001. pp. 27–31. [Google Scholar]
- 3.Vandamme P, et al. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fredrickson JK, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 5.Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 7.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiedje JM. Ecology of Denitrification and Dissimilatory Nitrate Reduction to Ammonium. New York: Wiley; 1988. pp. 179–244. [Google Scholar]
- 9.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Caspi R, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driscoll ME, et al. Identification of diverse carbon utilization pathways in Shewanella oneidensis MR-1 via expression profiling. Genome Inform. 2007;18:287–298. [PubMed] [Google Scholar]
- 12.Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 13.Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 14.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 15.Elias DA, et al. The influence of cultivation methods on Shewanella oneidensis physiology and proteome expression. Arch Microbiol. 2008;189:313–324. doi: 10.1007/s00203-007-0321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 17.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonnet GH, Hallett MT, Korostensky C, Bernardin L. Darwin v. 2.0: An interpreted computer language for the biosciences. Bioinformatics. 2000;16:101–103. doi: 10.1093/bioinformatics/16.2.101. [DOI] [PubMed] [Google Scholar]
- 20.Fang R, et al. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol Cell Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.