Abstract
To cause rice blast disease, the fungus Magnaporthe oryzae elaborates specialized infection structures called appressoria, which use enormous turgor to rupture the tough outer cuticle of a rice leaf. Here, we report the generation of a set of 22 isogenic M. oryzae mutants each differing by a single component of the predicted autophagic machinery of the fungus. Analysis of this set of targeted deletion mutants demonstrated that loss of any of the 16 genes necessary for nonselective macroautophagy renders the fungus unable to cause rice blast disease, due to impairment of both conidial programmed cell death and appressorium maturation. In contrast, genes necessary only for selective forms of autophagy, such as pexophagy and mitophagy, are dispensable for appressorium-mediated plant infection. A genome-wide analysis therefore demonstrates the importance of infection-associated, nonselective autophagy for the establishment of rice blast disease.
Keywords: appressorium, fungus, plant pathogen
To cause plant disease, many plant pathogenic fungi elaborate specialized infection structures that are used to breach the plant cuticle and gain entry to internal tissue (1). These structures, known as appressoria, are a feature of some of the most important cereal pathogens, including the devastating rice blast disease-causing fungus, Magnaporthe oryzae (2, 3). Appressoria of the rice blast fungus are dome-shaped, single-celled structures that generate enormous turgor pressure through accumulation of very high concentrations of glycerol (4). Hydrostatic turgor is generated by rapid influx of water into the appressorium, where a layer of melanin in the appressorium cell wall prevents the efflux of glycerol, allowing turgor to increase to a level sufficient to rupture the plant surface (2, 4). A narrow penetration hypha enters the rice epidermis and differentiates into bulbous, branched invasive hyphae, which are bounded by the invaginated plant cell membrane, allowing the fungus to proliferate within living plant cells (2, 3, 4).
Appressoria are formed following germination of a 3-celled fungal spore, called a conidium, which attaches tightly to the hydrophobic rice leaf surface (2). The conidium germinates and develops a short cylindrical germ tube, which differentiates at its tip to form the appressorium. Development of these cells requires activation of the Pmk1 mitogen-activated protein kinase pathway (5, 6) and is regulated genetically by control of the cell cycle (7). During germination of the M. oryzae conidium, a single nucleus migrates into the germ tube and undergoes mitosis. After this, one of the resulting daughter nuclei migrates into the incipient appressorium, while the other migrates back into the conidium (7). Mitosis and the subsequent movement of nuclei are necessary for appressoria to develop and for plant infection to occur. Completion of mitosis also, however, leads to collapse and death of the fungal conidium, the contents of which are delivered to the maturing appressorium. Functional analysis of the M. oryzae (Mo) ATG8 gene, has suggested that type II autophagic cell death is necessary for appressorium maturation and plant infection (7, 8).
In this study, we set out to determine whether infection-related autophagy is necessary for rice blast disease solely as a result of its role in conidial cell death or whether appressoria also undergo autophagy during their maturation. We also aimed to define whether autophagy carried out by M. oryzae during plant infection is a selective or a nonselective form of autophagy (9, 10, 11). Genetic analysis in the budding yeast Saccharomyces cerevisiae has identified a family of 30 ATG genes, which encode proteins necessary for autophagy (11, 12). TOR kinase regulates initiation of autophagy (13, 14) leading to formation of a single membrane structure, the phagophore, which surrounds and engulfs cytoplasm, organelles, and other cellular components, developing into a spherical, double-membrane autophagosome. The autophagosome expands and then fuses with a vacuole, the lytic compartment (lysosome equivalent) of fungal cells, sequestering its contents and inner membrane for degradation by hydrolases (10, 11). Selective forms of autophagy degrade peroxisomes (pexophagy), mitochondria (mitophagy), and endoplasmic reticulum (reticulophagy) or can occur during the biosynthetic cytoplasm-to-vacuole-targeting (Cvt) pathway, described in S. cerevisiae, which is used to transport the inactive precursor of the vacuolar hydrolase aminopeptidase I to the vacuole (15). Selective forms of autophagy require a distinct set of proteins, such as Atg11, which encodes a peripheral membrane protein that is the adaptor required for cargo loading in pexophagy and for delivery of aminopeptidase I to the vacuole in the Cvt pathway (15–18).
To determine why fungal autophagy is necessary for rice blast disease and to define which type of autophagy takes place during plant infection, we decided to adopt a genome-wide approach in which we would systematically analyze the autophagic machinery of M. oryzae and define the role of each of the associated gene products. To do this, we first developed a rapid method for gene functional analysis in M. oryzae and deployed this method to characterize the 22 fungal genes involved in autophagy. Here, we provide comprehensive evidence that infection-related autophagy is nonselective and takes place in both conidia and appressoria of M. oryzae leading to death of the conidium and development of a functional appressorium essential for plant disease.
Results
Infection-Associated Autophagy Occurs in both Conidia and Appressoria of M. oryzae.
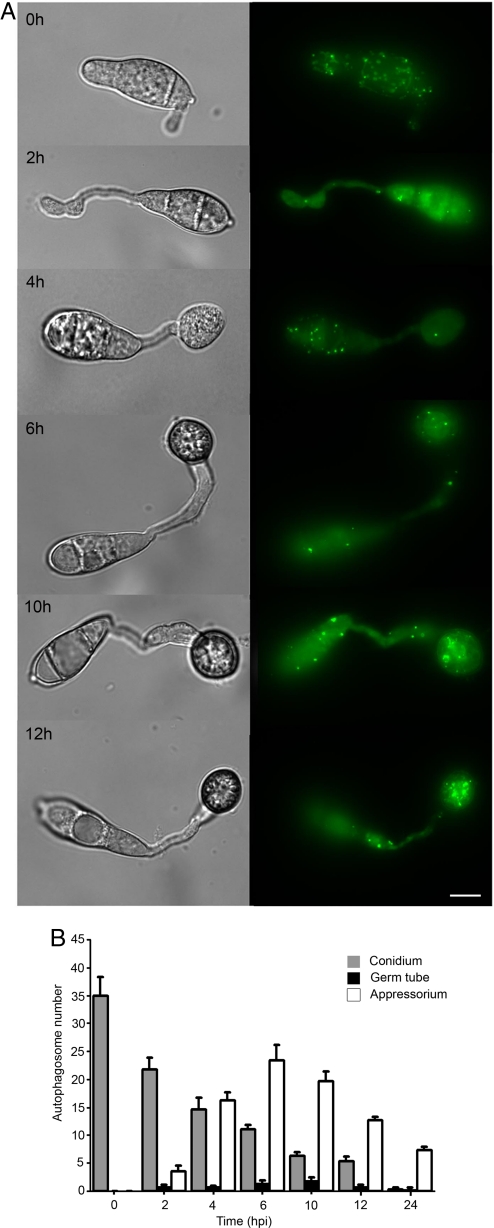
We set out first to visualize infection-associated autophagy in M. oryzae and determine the spatial and temporal dynamics of autophagosomes during appressorium development. To do this, we constructed a GFP-MoATG8 gene fusion, which was introduced into a wild-type strain of M. oryzae Guy11 and also the ΔMoatg8 mutant. Analysis of the cellular localization pattern and flux of Atg8 has been shown to be a reliable marker for autophagy (19). In yeast, ATG8 encodes an ubiquitin-like protein that can be modified at its C terminus by addition of phosphatidylethanolamine, tethering it to the autophagosome membrane where it is necessary for phagophore expansion during autophagosome formation (20). Expression of the GFP-MoATG8 fusion was sufficient to complement the ΔMoatg8 mutant phenotypes (7; supporting information (SI) Fig. S1), providing evidence that it was functional within M. oryzae and therefore a reliable marker for analysis of the cellular pattern of autophagy (19). Laser excitation epifluorescence microscopy showed that GFP-MoATG8-labeled autophagosomes accumulated in conidia during germination and then steadily decreased in number during the onset of conidial cell death and appressorium maturation (Fig. 1). Autophagosome number within developing appressoria increased during appressorium maturation and intense autophagic activity and vacuole expansion was associated with mature appressoria (Fig. 1). To investigate whether autophagy is specifically associated with appressorium development in M. oryzae, we expressed GFP-MoATG8 in a Δpmk1 MAP kinase mutant that does not elaborate appressoria and is consequently nonpathogenic (5, 6). In the Δpmk1 mutant, GFP-MoATG8-labeled autophagosomes were present in the conidium during germination but in significantly smaller numbers (see Fig. S2; t test, P < 0.05). Conidia of the Δpmk1 mutant remained intact throughout germination and germ tube elongation, indicating that conidial programmed cell death does not occur in the absence of appressorium formation. We conclude that infection-associated autophagy requires the Pmk1 MAP kinase and occurs during the onset of appressorium-mediated plant infection, initially within conidia, allowing the recycling of some of their contents to the developing appressorium where further autophagic activity occurs.
Fig. 1.
Cellular localization of autophagosomes during infection-related development of M. oryzae. (A) Conidia were harvested from a Guy-11 transformant expressing a GFP:MoATG8 gene fusion, inoculated onto glass coverslips, and observed by epifluorescence microscopy at the times indicated (Scale bar, 10 μm.). (B) Bar chart showing mean autophagosome numbers present in conidium, germ tube and appressorium at 0 h, 2 h, 4 h, 6 h, 10 h, and 12 h after inoculation (error bars indicate ± 2 SE).
Development of a Rapid Method for Gene Functional Analysis in M. oryzae.
To investigate the molecular control of autophagy in M. oryzae, we reasoned that it would first be necessary to develop a high throughput method for gene functional analysis. Recently, it has been shown that deletion of genes encoding components of the nonhomologous DNA end-joining pathway can be used to generate fungal strains with enhanced frequencies of homologous recombination (for review see ref. 21). We therefore deleted the ku70-encoding gene of M. oryzae and tested the resulting strain for the frequency of targeted gene replacement. We used deletion of the BUF1 gene, which encodes tri-hydroxy-naphthalene reductase, an enzyme required for melanin biosynthesis (22), as a visual test for the frequency of gene replacement (Fig. S3). Because Buf1 mutants have a buff color compared to the olive green/gray color of wild-type M. oryzae cultures (22), identifying mutants was straightforward. We found that homologous gene replacement occurred at a frequency of 80% (n = 100) in the Δku70 mutant background. This compares to the highly variable frequency of gene replacement in M. oryzae, which is locus-dependent and ranges from 1% to 25% (21). Growth rate, sporulation, and pathogenicity of the Δku70 mutant were found to be unaltered by the mutation and therefore we selected the mutant as an entry strain for rapid evaluation of the function of new genes in M. oryzae (Fig. S3).
Functional Analysis of Genes Necessary for Nonselective Autophagy in M. oryzae.
Analysis of the M. oryzae genome sequence (3) provided evidence for the presence of 23 autophagy-related genes, which are described in detail in Table S1. By reference to a series of molecular analyses in yeast (9–19), these genes could be functionally separated into those that putatively play a role in the initiation of autophagy (MoATG1, MoATG13, MoATG17), nucleation (MoATG6), phagophore, and autophagosome expansion (MoATG3, MoATG4, MoATG5, MoATG7, MoATG8, MoATG10, MoATG12, and MoATG16), and recycling (MoATG2, MoATG9, MoATG15, and MoATG18). The most significant differences compared to the S. cerevisiae autophagy gene family was the absence of clear orthologues of ATG20, ATG21, and ATG23 required for the selective Cvt and pexophagy pathways in yeast (10) but which are not found in the genome sequences of filamentous fungi or other multicellular eukaryotes, and ATG19, which is a Cvt pathway-specific receptor protein confined to S. cerevisiae (18). Other S. cerevisiae genes absent from the M. oryzae gene set were ATG14, which in yeast encodes the autophagy-specific subunit of phosphatidylinositol 3-kinase complex (23), and ATG31, which encodes a protein that interacts with Atg17p and Atg29p forming a complex involved in localizing other Atg proteins to the phagophore assembly site (24).
We classified the M. oryzae autophagy gene set into those predicted to be required for nonselective autophagy and those necessary for pexophagy, mitophagy, or the Cvt pathway (Table S2). To test the role of each gene in plant infection by M. oryzae, we carried out targeted gene replacements using the Δku70 mutant as recipient strain. In this way, we were able to generate a set of 22 isogenic mutants differing with respect to a single ATG gene. Two putative orthologues of ATG22 (25) were encoded in the M. oryzae genome, precluding analysis of this gene function by a single gene deletion. To check the efficacy of using the Δku70 mutant, we also deleted MoATG8, MoATG4, MoATG9, and MoATG12 in Guy11 so that comparative phenotype analysis could be carried out (Fig. S1). Phenotypic analysis of mutants generated in both genetic backgrounds gave identical results.
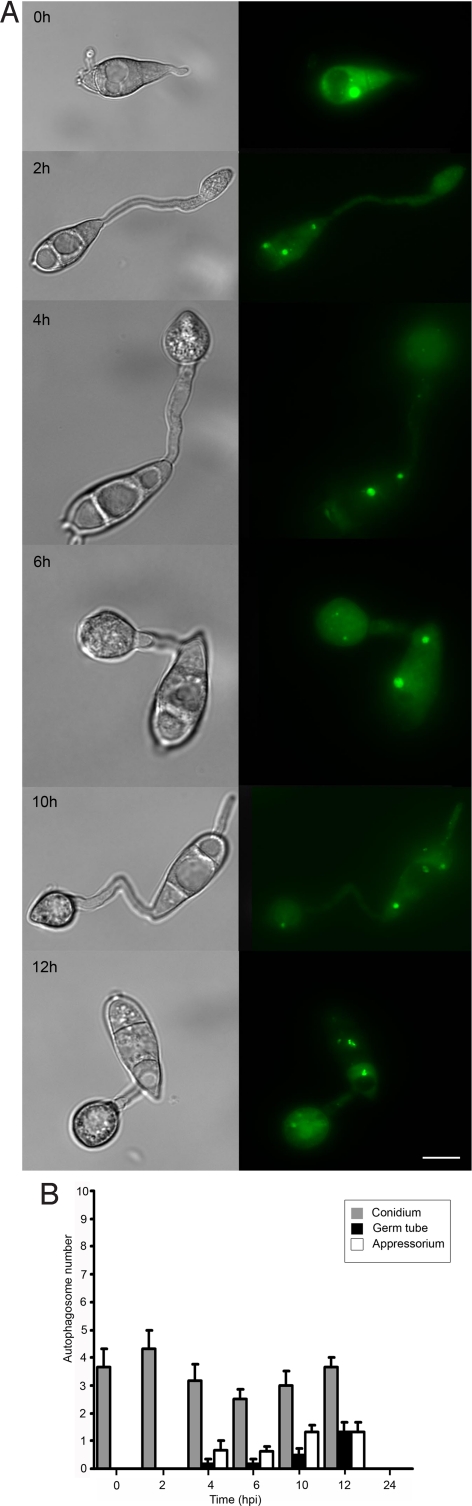
To confirm the role of these genes in autophagy, the GFP-MoATG8 gene fusion was introduced into a subset of the ATG gene deletion set and the distribution of autophagosomes was assessed (19). In a M. oryzae ΔMoatg4 mutant expressing GFP-MoATG8, we found that autophagosomes did not accumulate in fungal spores or appressoria and were very significantly reduced in number (Fig. 2; t test P < 0.0001). Large aggregates of GFP-MoAtg8 were observed and were excluded from vacuoles. MoATG4 encodes a cysteine protease necessary for processing of Atg8 (26) and therefore required for phagophore initiation, autophagosome formation, and vacuole fusion (10, 11, 26). Consistent with this predicted role, GFP-MoAtg8 puncta were significantly reduced in number (P < 0.0001) in both appressoria and conidia of ΔMoatg4 when compared to Guy11 and excluded from vacuoles. Furthermore, conidia did not collapse and die during appressorium formation, and there was no intense burst of autophagic activity in appressoria. When considered together, these results indicate that autophagy is arrested by the absence of MoATG4-encoded cysteine protease, disrupting appressorium maturation.
Fig. 2.
Cellular localization of autophagosomes during infection-related development of a ΔMoatg4 mutant of M. oryzae. (A) Conidia were harvested from a ΔMoatg4 transformant expressing a GFP:MoATG8 gene fusion, inoculated onto glass coverslips and observed by epifluorescence microscopy at the times indicated (Scale bars, 10 μm.). (B) Bar chart showing mean autophagosome numbers present in conidium, germ tube, and appressorium 0 h, 2 h, 4 h, 6 h, 10 h and 12 h after inoculation with ΔMoatg4 mutant expressing a GFP:MoATG8 (error bars indicate ± 2 SE).
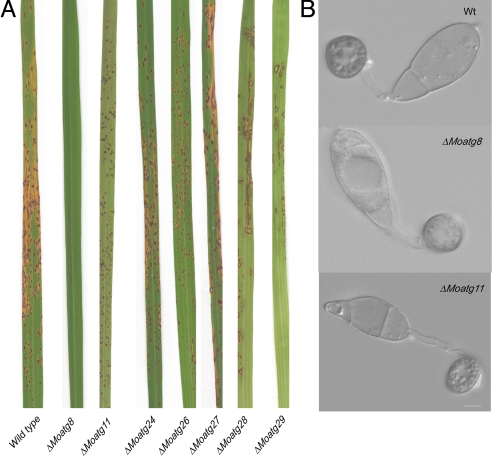
To test whether nonselective autophagy is necessary for rice blast disease, spores were collected from ΔMoatg1, ΔMoatg2, ΔMoatg3, ΔMoatg4, ΔMoatg5, ΔMoatg6, ΔMoatg7, ΔMoatg8, ΔMoatg9, ΔMoatg10, ΔMoatg12, ΔMoatg13, ΔMoatg15, ΔMoatg16, ΔMoatg17, and ΔMoatg18 mutants and used to inoculate 21-day-old seedlings of the blast-susceptible rice cultivar CO-39 (Fig. 3). The isogenic Δku70 mutant and the wild-type Guy11 strain were both able to cause severe rice blast symptoms. In contrast, Atg mutants impaired in nonselective autophagy were nonpathogenic or highly reduced in virulence. Only the ΔMoatg13 and ΔMoatg18 mutants were able to cause any disease symptoms, and lesion numbers were significantly reduced when compared to the wild type (t-tests, P < 0.001 and P < 0.01 respectively). Furthermore, cytological analysis of prepenetration structures revealed that conidial cell collapse was prevented in all cases by inhibition of nonselective autophagy (Fig. 3). The loss of rice blast symptoms was found to be due to impaired appressorium function, although inoculation of wounded seedlings with either conidia or hyphae also failed to produce rice blast symptoms, indicating a role for autophagy in proliferation of the fungus in plant tissue, in addition to its requirement for cuticle penetration. The ΔMoatg6 mutant showed reduced viability upon storage and a severe impairment in sporulation. Complementation analysis was performed on a subset of the Δatg mutants and in all cases tested led to restoration of the wild-type phenotype (Fig. S1). We conclude that each individual gene product that is necessary for nonselective fungal autophagy in M. oryzae is also required for rice blast disease.
Fig. 3.
Deletion of any of the 16 genes required for nonselective autophagy renders M. oryzae unable to cause rice blast disease. (A) Seedlings of rice cultivar CO-39 were inoculated with uniform conidial suspensions (1 × 105 ml−1) of Δku70, Guy11 (wt) and autophagy mutants ΔMoatg1, ΔMoatg2, ΔMoatg3, ΔMoatg4, ΔMoatg5, ΔMoatg6, ΔMoatg7 ΔMoatg8, ΔMoatg9, ΔMoatg10, ΔMoatg11, ΔMoatg12, ΔMoatg13, ΔMoatg15, ΔMoatg16, ΔMoatg17, and ΔMoatg18. Seedlings were incubated for 5 days to allow development of disease symptoms. Very reduced symptom development was observed on some plants sprayed with ΔMoatg18 conidia, but lesion density was significantly reduced compared to Guy-11 (P < 0.001). All other macroautophagy mutants were completely nonpathogenic. (B) Conidia were germinated on hydrophobic glass coverslips and incubated for 24 h to form appressoria. Micrograph shows conidial collapse in Guy11. Conidia of macroautophagy mutants such as ΔMgatg4 and ΔMgatg8 did not show conidial cell death (Scale bars, 10 μm.)
Functional Analysis of Genes Associated with Selective Autophagy.
Our analysis of the M. oryzae genome sequence revealed the presence of 6 genes specifically associated with pexophagy or mitophagy (16, 17). There was, however, no evidence of a functional Cvt pathway in M. oryzae based on the absence of orthologues of ATG19, ATG20, ATG21 and ATG23 genes (Tables S1 and S2). We therefore carried out targeted gene deletions to generate ΔMoatg11, ΔMoatg24, ΔMoatg26, ΔMoatg27, ΔMoatg28 and ΔMoatg29 mutants. These were used to inoculate rice seedlings and disease symptoms were evaluated and quantified. We found that ΔMoatg11, ΔMoatg24, ΔMoatg26, ΔMoatg27, ΔMoatg28 and ΔMoatg29 mutants were each able to cause rice blast disease and did not affect conidial or appressorial autophagy (Fig. 4). We conclude that selective autophagy (16, 17) is dispensable for appressorium-mediated plant infection.
Fig. 4.
Genes involved in selective autophagy in M. oryzae are not required for rice blast disease. (A) Seedlings of rice cultivar CO-39 were inoculated with uniform conidial suspensions of Guy-11, Δku70, and autophagy mutants ΔMoatg8, ΔMoatg11, ΔMoatg24, ΔMoatg26, ΔMoatg27, ΔMoatg28, and ΔMoatg29. Seedlings were incubated for 5 days to allow development of disease symptoms. ΔMoatg24, ΔMgatg26, ΔMoatg27, ΔMoatg28, ΔMgatg11, and ΔMoatg29 produced similar disease lesion density on rice seedlings to Guy11 or Δku70. (B) Conidia were germinated on hydrophobic glass coverslips and incubated for 24 h to form appressoria. Micrograph shows conidial collapse in Guy11. Conidia of selective autophagy mutants such as ΔMgatg11 showed conidial cell death in contrast to ΔMgatg8 (Scale bars, 10 μm.).
Discussion
Autophagy is a cell survival response that is triggered normally by starvation stress and used to recycle cytoplasm, organelles, and proteins within cells (9–11). It is becoming increasingly clear, however, that in addition to its homeostatic functions, autophagy may be necessary for cellular differentiation, defense from infection, and many aspects of development in multicellular organisms. In fungi, there are relatively few reports of autophagy (27), but the osmotrophic, mycelial growth habit of fungi and their extraordinary capacity to invade heterogeneous, nutrient poor substrates strongly suggests that autophagy may be fundamental to the fungal lifestyle. Autophagy is, for example, known to be important in heterokaryon incompatibility in Podospora anserina (28) and for metal ion homeostasis in Aspergillus fumigatus (29).
Our previous work in M. oryzae demonstrated a role for MoATG8 in autophagic cell death of conidia during appressorium development (7). ΔMoatg8 mutants were unable to undergo conidial cell collapse, and although they could form appressoria, these were nonfunctional and unable to cause plant disease. MoATG8 has also been shown to be involved in regulation of glycogen metabolism during conidiogenesis (30), and ΔMoatg8 mutants produce less conidia than an isogenic wild-type strain of M. oryzae (7, 30). The MoATG1 gene was independently identified as a differentially expressed gene during appressorium formation and shown to be necessary for pathogenicity (31). These previous studies did not, however, investigate the precise onset or spatial pattern of autophagy during infection-related development. In this report, we were able to demonstrate that autophagosomes are enriched in conidia as soon as they begin to germinate, less than an hour after landing on a rice leaf surface. The large burst of autophagic activity continues in the conidium until it collapses and undergoes cell death. Significantly, we also observed that autophagy also occurs in the appressorium during its development and maturation. Autophagosomes and are highly enriched in appressoria and a large central autophagic vacuole acts as the lytic compartment in maturing appressoria, consistent with previous studies of lipolysis during appressorium maturation (32). During infection-related morphogenesis in M. oryzae, autophagy is therefore necessary for programmed cell death in the conidium and for differentiation and active growth in the appressorium. How autophagy occurs in cells with such different fates in M. oryzae is an intriguing question. Autophagy is normally considered a pro-survival response that is essential for cells to contend with nutrient shortage in the extracellular environment. Autophagy is therefore up-regulated when the nutrient supply is insufficient to meet cellular energy demands and when cells are exposed to different forms of stress (8–10). Under these conditions, several studies have shown that autophagy acts to protect cells from death in a variety of eukaryotic organisms (8, 33). However, autophagy has also been shown to be a contributing factor in cell death (34–36), indicating that a nonapoptotic programmed cell death pathway exists in eukaryotes that is dependent on autophagy genes (8, 36). It is likely that these dual roles for autophagy are highly context-dependent in most cases, dependent on the prevailing extracellular environmental conditions. It is therefore striking that M. oryzae can spatially regulate autophagy in such a way that it is necessary both for conidial cell death and also for maturation and differentiation of functional appressoria. Induction of autophagy during infection-related development is developmentally regulated and requires the Pmk1 MAP kinase pathway (6), but is also a consequence of starvation stress. The purpose of autophagy is therefore to fuel infection-related development in the absence of exogenous nutrients before entry into the plant; for this reason it has to be tightly coupled to genetic control of development of the appressorium, which occurs via the cell cycle since completion of mitosis is a necessary prerequisite both for appressorium maturation and conidial cell death.
We cannot, however, preclude the possibility that conidial cell death also involves apoptosis and M. oryzae possesses 2 metacapsase-encoding genes that require functional analysis. It is clear, however, that the absence of any component of the autophagic machinery is sufficient to prevent both conidial collapse and appressorium-mediated plant infection. Understanding the role of TOR kinase in the initiation of infection-associated autophagy and the potential interplay with cAMP-dependent protein kinase A signaling, which has been shown to be necessary for appressorium morphogenesis in M. oryzae, will therefore be highly informative. In S. cerevisiae, protein kinase A and the Sch9 kinase, for instance, cooperatively regulate induction of autophagy (14).
Our other major aim in this study was to test whether infection-associated autophagy is a selective or nonselective process, which could not be determined by analysis of MoATG8 (7). We decided that it would be necessary to adopt a genome level analysis to verify this correctly. The analysis of gene function in plant pathogenic fungi has generally proceeded by the analysis of single genes using targeted gene replacement normally to validate the role of a gene product in pathogenesis (31). This process has been a powerful means of identifying new virulence factors but has been less successful in defining cellular processes critical for plant diseases to occur. Furthermore, gene replacement is time-consuming, has rather poor levels of efficiency, and has been carried out on an ad hoc basis, with little further validation of predictions made from initial studies. This means that while a large number of discrete fungal genes are known to be necessary for plant disease, there has been less associated new insight into the molecular basis of plant disease (37). To develop a rapid method for gene functional analysis in M. oryzae, we therefore generated a Δku70 mutant and showed that it significantly enhanced the frequency of homologous recombination to 80%. A recent study has demonstrated that deletion of the Ku80-encoding gene in M. oryzae has a similar effect (38), and we were able to validate use of the Δku70 mutant in this systematic analysis of autophagy in M. oryzae.
Our results comprehensively demonstrated that nonselective autophagy is necessary for rice blast appressoria to form. Deletion of any of the 16 gene products necessary for macroautophagy rendered the fungus unable to cause blast disease, because of an impairment in appressorium function. The only mutants that were able to cause any disease symptoms, ΔMoatg13 and ΔMoatg18, were severely reduced in virulence. The phosphoprotein Atg13 is part of a multiprotein regulatory protein complex with Atg17, Vac8, and other proteins, which activate the Atg1 protein kinase in S. cerevisiae and is necessary for induction of autophagy (9). Atg13 mutants, however, show a reduction, but not elimination of autophagy, and this may also be the case for M. oryzae ΔMoatg13 mutants, explaining the phenotype. Similarly, Atg18 is involved in recycling of Atg9, together with a number of other proteins (9) and may have a partially redundant function in M. oryzae. Conidial collapse was impaired in all macroautophagy mutants and cytological analysis of ΔMoatg4 carrying the GFP-MoATG8 reporter confirmed disruption of autophagosome generation and autophagy in both cellular compartments—conidia or appressoria. In contrast, deletion of any of the 6 specific genes associated with pexophagy or mitophagy did not affect fungal pathogenicity. We can conclude that these selective forms of autophagy are therefore not necessary for plant infection. The significance of peroxisome biogenesis to appressorium function and fatty acid β-oxidation to appressorium physiology (39–41) predicts that pexophagy may play a role in subsequent stages of plant tissue colonization following breach of the cuticle. A recent study in the pathogenic fungus Colletotrichum orbiculare, for example, suggests that pexophagy is important in plant infection (42). Investigating the efficiency of growth in plant tissue of ΔMoatg11, ΔMoatg24, ΔMoatg26, ΔMoatg27, ΔMoatg28, and ΔMoatg29 will therefore be important, although they are clearly not severely impaired due to the severe disease symptoms observed at 72–96 h. Our study also demonstrates that the Cvt pathway, as defined in S. cerevisiae (10, 11) is absent from M. oryzae, providing further evidence that it is restricted to yeast (10, 11) and not present in multicellular filamentous fungi (27).
In conclusion, we have provided the first genome-wide analysis of autophagy in a filamentous fungus and have validated the importance of nonselective autophagy in the establishment of plant disease by M. oryzae. Controlling the initiation of fungal autophagy may provide an effective target for development of new and novel antifungal strategies, given the fact that the plant infection process is so sensitive to perturbation of this process.
Materials and Methods
Fungal Strains, Growth Conditions, and DNA Analysis.
Growth, maintenance, and storage of M. oryzae isolate, media composition, nucleic acid extraction, and transformation were all as previously described (43). Gel electrophoresis, restriction enzyme digestion, gel blots, and sequencing were performed by using standard procedures (44).
Generation of the Δku70 Mutant of M. oryzae.
The MoKu70 gene was identified from the published genome sequence (3) and primers designed to amplify the 2 regions flanking the gene. All primers are listed in Table S3. The primers used were Ku70Ff and Ku70LFr to amplify a 1.0 kb region upstream from the start codon and introduce a NdeI site at the 3′ end and primers Ku7-RFf and Ku70RFr to amplify a 1.0 kb region downstream of the gene and introducing a NdeI site at the 5′ end. The 2 flanking DNA fragments were cloned into pGEM-T (Promega) and then the left flank excised with NdeI/NotI and cloned into the vector with the right flank, giving pMG12.1. The ILV1 allele conferring resistance to sulfonylurea (44) was amplified as a 2.8 kb fragment using primers SurF and SurR introducing an NdeI site to both ends of the amplicon. This fragment was cloned into the NdeI site of pMG1 to create pMJG2. NotI and ApaI restriction sites within the pGEM-T polylinker were used to liberate the gene disruption cassette from pMJG2. Transformants were selected in the presence of chlorimuron ethyl (100 μgml−1). Two independent ΔMoku70 deletion mutants were obtained as assessed by Southern blot.
Generation of ΔMoatg Mutants.
Targeted gene replacement of the M. oryzae MoATG genes was performed using the split marker strategy (45). Vectors were constructed using a hygromycinB resistance selectable marker, hph (46), for transformation of ΔMoku70. The hph gene cassette was cloned into pBluescript (Stratagene) as a 1.4 kb EcoRI-XbaI fragment. To amplify the split hph templates the primers used were M13 F with HY and M13R with YG, as described in ref. 45 and shown in Fig. S4. M. oryzae autophagy genes were identified by homology to S. cerevisiae ATG genes obtained from the Saccharomyces genome database (Department of Genetics at the School of Medicine, Stanford) (Table S1). The sequence data for each M. oryzae autophagy gene was retrieved from the M. oryzae genome database at the Broad Institute (Massachusetts Institute of Technology, Cambridge, MA) (www.broad.mit.edu/annotation/fungi/magnaporthe) and used to design specific primers (see Table S4). The M. oryzae Δku70 mutant was transformed with each Mgatg:hph deletion cassette (2 μg of DNA of each flank). Transformants were selected in the presence of hygromycin B (200 μg. mL−1). Two independent deletion mutants were obtained for each autophagy gene as assessed by Southern blot. Complementation analysis was performed as described in Fig. S1.
GFP:MoATG8 Gene Fusion Construction.
The MoATG8 gene was amplified as a 1.6 kb fragment using primers ATG8.5 and ATG8.3. creating ClaI and XhoI sites at the ends of the fragment (See Table S2). The amplicon was digested and cloned into pCB1532 (5), which carries a selectable marker bestowing resistance to sulfonyurea. The promoter region of the MoATG8 gene was amplified as a 1.4 kb fragment using primers ATG8p5 and Atg8p3 creating SpeI and NcoI sites. The fragment was sub cloned into pMJK142.2 in frame with sGFP gene as a SpeI - NcoI fragment. MoATG8p:GFP was obtained by PCR using primers ATG8p5 and GFPrev, and the 2.15bp amplicon was digested with SpeI-ClaI and subcloned in-frame with the MoATG8 ORF. The resulting vector pCB GFP-MoATG8 was transformed into Guy-11, Δpmk1 and ΔMoatg4. Transformants were selected in the presence of chlorimuron ethyl (100 μg mL-1).
Light and Epifluorescence Microscopy.
Epifluorescence microscopy to visualize eGFP and MDC-stained samples was routinely carried out using a Zeiss Axioskop 2 microscope (Zeiss) with differential interference microscopy (DIC) used for bright field images. For epifluorescence examination of the GFP:MoATG8 transformants, conidia were incubated onto coverslips and placed onto a 2% agar cushion, then observed using a IX81 motorized inverted microscope (Olympus) equipped with a UPlanSApo 100X/1.40 Oil objective (Olympus). Excitation of fluorescently-labeled proteins was carried out using a VS-LMS4 Laser-Merge-System with solid state lasers (488 nm/50mW). The laser intensity was controlled by a VS-AOTF100 System and coupled into the light path using a VS-20 Laser-Lens-System (Visitron System). Images were captured using a Charged-Coupled Device camera (Photometric CoolSNAP HQ2, Roper Scientific). All parts of the system were under the control of the software package MetaMorph (Molecular Devices).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901477106/DCSupplemental.
References
- 1.Tucker SL, Talbot NJ. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol. 2001;39:385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- 2.Talbot NJ. On the trail of a cereal killer: Investigating the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 3.Dean R, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 4.De Jong JC, McCormack BJ, Smirnoff N, Talbot NJ. Glycerol generates turgor in rice blast. Nature. 1997;389:244–245. [Google Scholar]
- 5.Xu JR, Hamer JE. MAP kinase and cAMP signalling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Kim Y, Park G, Xu JR. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 8.Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ. 2009;16:21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 11.Klionsky DJ. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Molec Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimori T. Autphagy: A regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, Strandberg L, Lenardo MJ. The selectivity of autophagy and its role in cell death and survival. Autophagy. 2008;4:567–573. doi: 10.4161/auto.5902. [DOI] [PubMed] [Google Scholar]
- 17.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:604–609. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, et al. Guidelines for the use and interpretations of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krappman S. Gene targeting in filamentous fungi: Benefits of impaired repair. Fungal Biol Rev. 2007;21:25–29. [Google Scholar]
- 22.Chumley FG, Valent B. Genetic analysis of melanin-deficient, nonpathogenic mutants of. Magnaporthe grisea. Molec Plant Microbe Interact. 1990;3:135–143. [Google Scholar]
- 23.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol-3-kinase comlexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollack JK, Harris SD, Marten MR. Autophagy in filamentous fungi. Fungal Genet Biol. 2009;46:1–8. doi: 10.1016/j.fgb.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Pinan-Lucarré B, Paoletti M, Dementhon K, Coulary-Salin B, Clavé C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol. 2003;47:321–333. doi: 10.1046/j.1365-2958.2003.03208.x. [DOI] [PubMed] [Google Scholar]
- 29.Richie DL, et al. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryotic Cell. 2007;6:2437–2447. doi: 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng YZ, Ramos-Pamploma M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy. 2009;5:33–43. doi: 10.4161/auto.5.1.7175. [DOI] [PubMed] [Google Scholar]
- 31.Liu XH, et al. Involvement of a Magnaporthe grisea serine/threonine kinase gene MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell. 2007;6:997–1005. doi: 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber RWS, Wakley GE, Thines E, Talbot NJ. The vacuole as central element of the lytic system and sink for lipid droplets in maturing appressoria of Magnaporthe grisea. Protoplasma. 2001;216:101–112. doi: 10.1007/BF02680137. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 34.Yu L, et al. Regulation of an ATG7-beclin1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 35.Yu L, Lenardo MJ, Baehrecke EH. Autophagy and caspases: A new cell death program. Cell Cycle. 2004;3:1124–1126. [PubMed] [Google Scholar]
- 36.Shimuzu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 37.Idnurm A, Howlett BJ. Pathogenicity genes of phytopathogenic fungi. Mol Plant Pathol. 2001;2:241–255. doi: 10.1046/j.1464-6722.2001.00070.x. [DOI] [PubMed] [Google Scholar]
- 38.Villalba F, et al. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol. 2008;45:68–75. doi: 10.1016/j.fgb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZY, Thornton CR, Kershaw MJ, Debao L, Talbot NJ. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol. 2003;47:1601. doi: 10.1046/j.1365-2958.2003.03412.x. [DOI] [PubMed] [Google Scholar]
- 40.Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol. 2007;61:61–75. doi: 10.1111/j.1365-2958.2006.05194.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. Functional analysis of lipid metabolism in the rice blast fungus Magnaporthe grisea reveals a role for peroxisomal b-oxidation in appressorium-mediated plant infection. Molec Plant–Microbe Interact. 2007;20:475–491. doi: 10.1094/MPMI-20-5-0475. [DOI] [PubMed] [Google Scholar]
- 42.Asakura M, et al. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell. 2009;21:1291–1304. doi: 10.1105/tpc.108.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a Gene Involved in Pathogenicity from the Rice Blast Fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 45.Sweigard J, Chumley F, Carroll A, Farrall L, Valent B. A series of vectors for fungal transformation. Fungal Genet Newsl. 1997;44:52–53. [Google Scholar]
- 46.Catlett NL, Lee B-N, Yoder OC, Turgeon BG. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genetics Newslett. 2003;50:9–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.