Summary
The self-incompatibility response of crucifers is a barrier to fertilization in which arrest of pollen tube development is mediated by allele-specific interactions between polymorphic receptors and ligands encoded by the S-locus haplotype. The binding of the stigma-expressed S-locus receptor kinase (SRK) [1] to the pollen coat-localized S-locus cysteine-rich (SCR) ligand [2-5], activation of the receptor, and pollen rejection occurs only if receptor and ligand are encoded by the same S haplotype [4, 6-8]. To identify residues within the extracellular domain of SRK (eSRK) that are required for its ligand-selective activation, we assayed chimeric receptors and receptor variants containing substitutions at polymorphic sites in transgenic Arabidopsis thaliana [9, 10]. We show that only a small number of the ∼100 polymorphic residues in eSRK are required for ligand-specific activation of self-incompatibility in vivo. These essential residues occur in two non-contiguous clusters located at equivalent positions in the two variants tested. They also correspond to sites showing elevated levels of substitutions in other SRKs, suggesting that these amino acids could define SI specificity in most SRKs. The results demonstrate that the majority of eSRK residues that show signals of positive selection and previously surmised to function as specificity determinants are not essential for specificity in the SRK-SCR interaction.
Results and Discussion
To understand the basis of ligand-selective activation of SRK, we focused on polymorphic residues in the eSRK, a region subject to strong diversifying selection, with extraordinarily high levels of polymorphisms that have persisted for long periods of time [11-16]. Comparison of SRK alleles, which can number over 50 in one species, has shown that their amino-acid sequences can diverge by as much as 35% in Brassica species [1, 17] and 51% in A. lyrata [18]. Sequence alignments have demonstrated that polymorphic residues, although scattered over the length of the eSRK, are particularly prevalent in several “hypervariable regions” [17, 19], in which non-synonymous to synonymous substitution (Ka/Ks) ratios are significantly greater than 1 [20] or elevated relative to the rest of the protein [21]. These “hypervariable regions” also contain many of the residues having a high posterior probability of being under selective pressure to change in physicochemical property [22]. These features have suggested that hypervariability in these regions is not due to relaxed constraint but rather to diversifying selection, and that the variable residues within these regions function as SI specificity determinants.
Swapping the hypervariable regions of eSRK
To test the hypothesis that SI specificity resides in the hypervariable (hv) regions, specifically hvI, hvII, and hvIII (Figure 1A), we assayed eSRK chimeras in which a segment encompassing these regions was swapped between different SRK variants in transgenic plants of the Arabidopsis thaliana C24 accession [9]. We first generated AtS1pr∷eSRKx:AlSRKb fusions (Figure 1A; Boggs et al. submitted; Supplemental Data) in which the stigma-specific AtS1 promoter [23] drives expression of an SRK consisting of the eSRK (minus the last 23 amino acids) from one SRK variant (eSRKx) fused to the last 23 amino acids of AleSRKb, followed by the AlSRKb transmembrane and kinase domains. The hvI-hvIII region of an AtS1pr∷eSRKx:AlSRKb fusion was then replaced with the corresponding region from other SRK variants to generate eSRKx(y)x chimeras (Figure 1B; Supplemental Data), where “x” and “y” correspond to the number or letter of the SRK allele from which the various regions were derived, and the central swapped hvI-hvIII region is indicated in parentheses (Figure 1B). For each chimera, several independent transgenic plants were assayed by pollinating stigmas with transgenic A. thaliana pollen expressing the SCRs that correspond to the parental SRKs used in chimera construction (Supplemental Data; Figure S1). In this expression system, AtS1pr∷AleSRKa:AlSRKb and AtS1pr∷AleSRK16:AlSRKb (Figure 1A) confer Sa and S16 specificity, respectively: transgenic stigmas expressing these chimeras inhibit transgenic A. thaliana AlSCRa- and AlSCR16-expressing pollen, respectively, but not wild type or AlSCRb-expressing pollen (Boggs et al., submitted).
Figure 1. The structure and function of chimeric SRK genes.
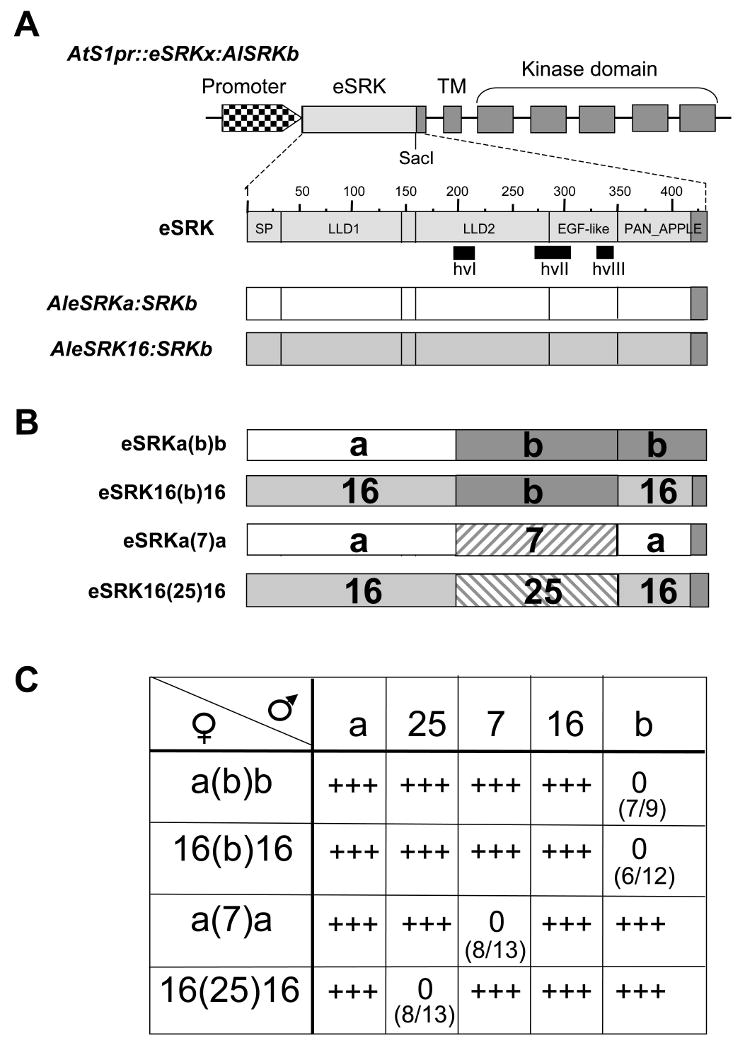
A. The AtS1pr∷eSRKx:AlSRKb genes used for construction of eSRK chimeras. The top diagram shows the structure of a generic AtS1pr∷eSRKx:AlSRKb gene, in which the AtS1 promoter (checkered arrowhead) drives an SRK transcriptional unit with its seven exons and native 3′ untranslated sequences. Exon 1 encodes the AlSRK extracellular domain (eSRK), exon 2 encodes the transmembrane domain (TM), and exons 3-7 encode the kinase domain. The unique SacI restriction site used for construction of chimeras is shown towards the 3′ end of the eSRK. AlSRKb sequences are shown in dark grey and eSRK sequences (from the initiating methionine codon to the unique SacI site) derived from other variants are shown in light grey. Due to the use of the SacI site, all constructs used in this study contain a common 23-amino-acid region derived from AlSRKb (shown in dark grey, spanning the last 23 amino acids of eSRK, i.e. residues 411-434 in SRKb). The middle diagram is a magnified view of the AleSRKb (with numbers indicating amino acids). The vertical lines delineate predicted structural subdomains in the eSRK [29]: SP, signal peptide; LLD1 and LLD2, lectin-like domains 1 and 2; EGF-like, epidermal growth factor-like domain; and PAN_APPLE domain. The locations of hypervariable regions are indicated below the diagrams and correspond to the following amino-acid segments in AlSRKb: 204-219 (hvI), 269-304 (hvII), 326-340 (hvIII). The lower diagrams show the eSRKs of AtS1pr∷AleSRKa:AlSRKb and AtS1pr∷AleSRK16:AlSRKb, two of the constructs used for domain swaps.
B. eSRK chimeras. The structures of four functional chimeric eSRKs are shown. The derivation of various segments is shown by different colors or patterns: AleSRKb: grey; AleSRKa: white; AleSRK16: hatched; AleSRK25: stippled; CgeSRK7: slanted bricks. The limits of the swapped region in these and other chimeras analyzed are indicated in Table 1 and their sequences are shown in Figure S2.
C. Pollination phenotypes of A. thaliana plants transformed with eSRK chimeras. First- and second-generation transgenic plants expressing each chimera were pollinated using plants expressing the cognate SCR, other SCRs, and wild-type pollen. eSRK chimeras are indicated in the column below the female symbol: a(b)b, AleSRKa(b)b:AlSRKb; 16(b)16, AleSRK16(b)16:AlSRKb; a(7)a, CgeSRKa(7)a:AlSRKb; 16(25)16, AleSRK16(25)16:AlSRKb. The SCR variants expressed in pollen used for pollination assays are indicated in the row to the right of the male symbol and correspond to the constructs shown in Figure S1: a, native AlSCRa; 25, native AlSCR25; 7, AlSCRb:CgSCR7; 16, AlSCRb:AlSCR16; b, native AlSCRb. The numbers in parentheses show the number of T1 plants that expressed an incompatibility response towards pollen expressing cognate SCR over the total number of primary transformants analyzed. 0 = an incompatible response (typically <5 pollen tubes per pollinated stigma); +++ = a compatible response (typically >50 pollen tubes per pollinated stigma). For each construct, although the majority of transformants exhibiting SI expressed a strong SI response (<5 pollen tubes per pollinated stigma), typically 1 or 2 transformants exhibited a weaker SI response (5-10 pollen tubes per pollinated stigma).
We constructed 11 AtS1pr∷eSRKx(y)x:AlSRKb chimeras (hereafter “eSRK chimeras”) (Table 1; Figure S2) using AleSRKa, AleSRKb, and AleSRK16 (which are ∼62% similar), AleSRK25 (which is 82% similar to AleSRK16; Boggs et al. submitted), and Capsella grandiflora CgeSRK7 (which is 77% similar to AleSRKa [25]). Pollination assays of stigmas from several independent primary (T1) transformants using pollen that expresses SCR corresponding to the hvI-hvIII region of eSRK chimeras revealed that seven chimeras (Table 1; Figure S2B) failed to confer an incompatibility response, possibly due to the disruptive steric effect of combining diverged eSRK segments. However, the eSRK16(b)16:SRKb, eSRK16(25)16:SRKb, eSRKa(7)a:SRKb, and eSRKa(b)b:SRKb chimeras (Figure 1B; Figure S2A) were functional and each conferred the SI specificity of the SRK allele from which the hvI-hvIII region was derived (Table 1), as determined by pollination assays in T1 and T2 transgenic progenies (Figure 1C). Thus, residues within the ∼160-aa segment spanning the hvI-hvIII region of eSRK are sufficient for determining SI specificity in transgenic Arabidopsis.
Table 1. eSRK chimeras assayed in planta.
| Functional Chimeras | Observed Specificity a | Swapped region (SRKa #s) b | No. of polymorphic sites within the swapped region c | No. of polymorphic sites outside the swapped region c |
|---|---|---|---|---|
| eSRKa(b)b | b | 198-403 (197-403) | 91 | 67 |
| eSRK16(b)16 d | b | 198-357 (197-357) | 70 | 91 |
| eSRKa(7)a | 7 | 196-351 (200-356) | 42 | 58 |
| eSRK16(25)16 | 25 | 197-355 (198-357) | 46 | 28 |
| Nonfunctional Chimeras | Expected Specificity | Swapped region (SRKa #s) b | No. of transgenic plants analyzed | |
| eSRKa(b)a | b | 198-357 (197-357) | 7 | |
| eSRK25(16)25 | 16 | 197-355 (198-357) | 16 | |
| eSRK25(16)16 | 16 | 197-401 (198-403) | 17 | |
| eSRKa(16)a | 16 | 197-354 (198-356) | 8 | |
| eSRKb(16)b | 16 | 196-355 (197-357) | 12 | |
| eSRK25(a)25 | a | 198-357 (198-357) | 13 | |
| eSRK16(a)16 | a | 198-356 (198-356) | 8 | |
SI specificity was determined by pollinating the stigmas of transgenic plants expressing each chimera with pollen expressing the SCR variant corresponding to the swapped hvI-hvIII region. The source of pollen was A. thaliana plants transformed with one of the SCR constructs diagrammed in Figure S1. Functional chimeras expressed the expected specificities. For nonfunctional chimeras, the indicated specificity was expected but not conferred in transgenic stigmas.
The numbers show the limits of the swapped region in each eSRK chimera (see alignments in Supplementary Figure 2), with the corresponding numbers in SRKa shown in parentheses for reference.
The eSRK16(b)16 chimera contains the smallest swapped region of all chimeras tested.
The numbers indicate the number of amino-acid differences between the pair of SRK variants used to generate each chimera.
SRK residues required for ligand-specific activation of the incompatibility response
To identify residues within the hvI-hvIII region that determine SI specificity, we focused on the S7- and S25-determining regions of eSRKa(7)a and eSRK16(25)16 (Table 1). The eSRKa(7)a and eSRK16(25)16 chimeras were modified by site-directed mutagenesis (Supplemental Data) to generate a series of mutants containing single-site substitutions at each of the 42 polymorphic sites that differ between the hvI-hvIII regions of eSRKa and eSRK7, and 44 out of the 46 polymorphic sites that differ between the hvI-hvIII regions of eSRK16 and eSRK25 (Figure 2A). Each of the resulting eSRKa(7)a and eSRK16(25)16 mutants had the chimeric eSRK sequence except for one amino-acid residue within the hvI-hvIII region that was replaced with the residue found at the corresponding position in eSRKa or eSRK16, respectively (Figure 2A). The stigmas of T1 plants transformed with each of the 42 eSRKa(7)a and 44 eSRK16(25)16 mutants were pollinated with A. thaliana SCR7- and SCR25-expressing pollen (Figure S3), respectively. In all, 35 eSRKa(7)a mutants and 38 eSRK16(25)16 mutants (Figure 2A) conferred a strong incompatibility response in at least some of the T1 transformants analyzed, indicating that the mutant chimeras retained the specificity of the “wild type” eSRka(7)a or eSRK16(25)16 chimeras, and that residues that were replaced in these mutants are not required for S7 or S25 specificity.
Figure 2. Structure and immunoblot analysis of eSRKa(7)a:SRKb and eSRK16(25)16:SRKb substitution mutants.
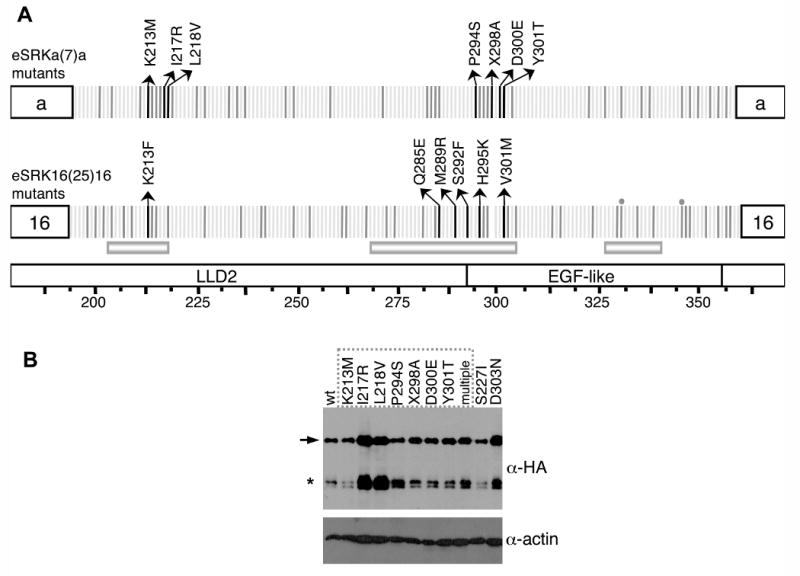
A. Single-codon substitutions in eSRK chimeras. The diagrams show the specificity-determining hvI-hvIII regions of the eSRKa(7)a and eSRK16(25)16 chimeras, with amino-acid residues depicted by vertical bars. The bottom diagram shows the location of the LLD2 and EGF-like domains and hypervariable regions hvI, hvII, and hvIII. Residues that do not differ between eSRKa and eSRKa(7)a or between eSRK16 and eSRK16(25)16 are shown by light grey bars. Polymorphic residues that were modified by substitution mutagenesis are shown by dark grey and black bars. Each of these variable residues, with the exception of 2 residues in eSRK16(25)16 (marked by gray circles, mutants of which failed to generate transgenic plants), were individually replaced in eSRKa(7)a with residues found at the equivalent positions in eSRKa, and in eSRK16(25)16 with residues found at the equivalent positions in eSRK16. Transgenic stigmas expressing each of the single-codon substitution eSRKa(7)a or eSRK16(25)16 derivatives were tested by pollination with SCR7- or SCR15-expressing pollen, respectively. For most substitution mutants (substituted residues shown as dark grey bars), the stigmas of at least some transformants [13-88% of transformants for eSRKa(7)a mutants and 12-71% of transformants for eSRK16(25)16 mutants] exhibited an incompatible response. For each of the substitution mutants that failed to confer an incompatibility response, between 10 and 18 independent transformants were assayed. Amino-acid residues found to be required for the function of eSRK chimeras are shown by black bars with arrows indicating the amino-acid substitution that caused loss of chimera function. The L218V substitution in eSRKa(7)a and the Q285E substitution in eSRK16(25)16 conferred a weakened incompatibility response (Figure S3). The X298A change in eSRKa(7)a involved inserting alanine at amino-acid site 298. Note that substitutions at two sites, 213 and 301, disrupted the function of both eSRKa(7) and eSRK16(25)16 chimeras: site 213 was sensitive to a change from the polar and charged lysine to a non-polar and uncharged methionine or phenylalanine, while site 301 was sensitive to changes in volume.
B. Immunoblot analysis of eSRK chimeras. For immunoblot analysis of HA-tagged eSRKa(7)a chimeras, proteins were extracted from the stigmas of plants transformed with AtS1pr∷eSRKa(7)a:AlSRKb (wt) and its single- and multiple-codon substitution derivatives, and subjected to protein immunoblot analysis (Supplemental Data). The “wt” lane shows the level of non-mutated “wild type” eSRKa(7)a protein found in stigmas exhibiting an incompatibility response toward SCR7-expressing pollen. The remaining lanes show representative patterns for eSRKa(7)a substitution derivatives: nine single-codon substitution derivatives labeled according to the amino-acid substitution introduced into each chimera (numbering as in panel A), and a multiple-codon substitution derivative (multiple). The dashed box indicates the substitution derivatives that did not confer an incompatible response towards SCR7-expressing pollen. The blot was probed sequentially with an anti-HA monoclonal antibody (top panel) and an anti-actin antibody as loading control (bottom panel). The arrow indicates the full-length eSRKa(7)a:SRKb receptor and the asterisk indicates the alternative smaller SRK products typically produced in stigmas [39].
For the remaining mutants (Figure 2A), none of the 10-18 independent T1 transformants analyzed per construct conferred a strong incompatibility response towards pollen expressing the SCR corresponding to the swapped specificity-determining region, indicating that the substituted residues are important for the S7 or S25 specificity to be manifested. The majority of these mutant eSRKa(7)a and eSRK16(25)16 chimeras produced only plants whose stigmas were fully compatible with SCR7- and SCR25-expressing pollen, respectively. However, chimeras carrying the L218V substitution in eSRKa(7)a and the Q285E substitution in eSRK16(25)16 conferred a weakened incompatibility phenotype (Figure S3), characterized by variable expressivity of the response or a degree of leakiness that allowed the germination of some SCR7- or SCR25-expressing pollen grains. This partial disruption of SRK function might be due to the partial insensitivity of the sites to volume- (L218V) or charge- (Q285E) changing substitutions.
To determine if the identified essential sites are not only necessary for eSRKa(7)a:SRKb and eSRK16(25)16:SRKb function, but are sufficient for S7 or S25 specificity, we generated a construct that combined substitutions at the seven sites essential for eSRKa(7)a function into the eSRKa backbone, and another construct that combined substitutions at the six sites essential for eSRK16(25)16 function into the eSRK16 backbone. However, neither of these two multiple-substitution mutants conferred an incompatibility response in transgenic stigmas pollinated with SCR7- or SCR25-expressing pollen.
To exclude the possibility that the non-functionality of mutant eSRK chimeras is due to their sub-optimal accumulation in stigmas, the original “wild type” eSRKa(7)a and eSRK16(25)16 chimeras, as well as their mutant derivatives, were tagged by inserting a hemagglutinin (HA) tag at the N-terminus of mature eSRK (Supplemental Data). The HA-eSRK16(25)16 chimeras, including the “wild type” chimera, were not functional, likely due to disruptive effects of the HA tag, and were not investigated further. In contrast, the HA-eSRKa(7)a chimeras recapitulated the pollination phenotypes observed with their non-tagged counterparts. Importantly, for all non-functional HA-eSRKa(7)a chimeras, T1 transformants were obtained whose stigmas accumulated SRK to levels equivalent to, or higher than, those of “wild type” HA-eSRKa(7)a in stigmas that expressed S7 specificity (Figure 2B). Thus, the non-functionality of chimeras containing substitutions at essential sites cannot be explained by reduced SRK levels in transgenic stigmas. Therefore, amino acids at essential sites most likely function as specificity-determining residues. However, the failure of multiple-substitution eSRKa(7)a and eSRK16(25)16 mutants to confer an incompatibility response suggests that additional residues in the hI-hvIII region might also be required for SRK specificity. Such residues might have escaped detection because a single amino-acid substitution at these sites might not produce a detectable effect on SRK function.
Nevertheless, the observation that the majority of substitutions (even those that changed charge, volume, or polarity) were not disruptive, indicates that the specific amino acids occupying most polymorphic sites in the hvI-hvIII region are not critical for function. Interestingly, the few sites found to be essential for eSRKa(7)a and eSRK16(25)16 function are clustered in two non-contiguous regions located at equivalent positions in the hvI and hvII regions of the two chimeras, and two of these sites were found to be essential for the function of both chimeras.
Prediction of functionally-important amino-acid residues in other SRK variants
To determine if the amino-acid sites we identified as likely specificity determinants in CgSRK7 or AlSRK25 might also determine specificity in other SRK variants, we performed pairwise alignments of the hvI-hvIII region for 34 intra- or inter-specific pairs of the most closely-related eSRK variants that are known or assumed to encode different SI specificities (Figure S4). We reasoned that this strategy is preferable to comparing highly-diverged SRK sequences, which can impede the prediction of specificity-determining residues due to overall high variability in eSRKs [21]. Alignment of pairwise consensus sequences (Figure 3, Figure S4) showed that none of the 11 essential sites identified in planta was polymorphic in all sequences. However, seven of these sites were polymorphic in >50% of the comparisons, and the two sites found to be essential for both CgSRK7 and AlSRK25 were polymorphic in >60% of the comparisons (Figure 3). Additionally, several of the 15 sites in hvI and hvII that exhibited polymorphisms in >50% of pairwise comparisons coincided with, or clustered near, the functionally-important residues identified in planta (Figure 3).
Figure 3. Extent of variability at individual sites within the hvI-hvIII region observed in pairwise alignments of closely-related eSRK pairs.
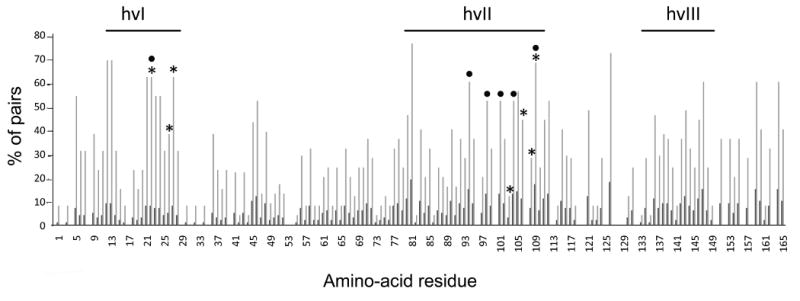
eSRK sequences from Arabidopsis lyrata, A. halleri, Capsella grandiflora, Brassica oleracea, B. rapa, and Raphanus sativus were analyzed by pairwise alignment of the most closely-related sequences that are either known or likely to encode different SI specificities (Figure S4). Pairwise consensus sequences were generated and aligned (Figure S4), and the percentage of consensus sequences that differed at a particular site was calculated. Each site was assigned a “substitution score” between 0 and 100, as shown on the y-axis: a score of “0” indicates that 0% of the variant pairs differ at that site, and a score of “100” indicates that 100% of the pairs differ at that site. The x-axis indicates amino-acid site number along the hvI-hvIII region after removal of indels (Figure S4); this numbering was used to highlight the overlap and clustering of highly variable residues relative to the essential residues identified in planta. The short dark bars indicate the number of sequence pairs with substitutions for each site. Asterisks and circles indicate the residues found to be essential for the function of the eSRK16(25)16 and eSRKa(7)a chimeras, respectively. The locations of the essential sites using eSRKa as a reference sequence (Figure 2) are: K213, I217, L218, P294, X298, D300, and Y301 in eSRK7, and K213, Q285, M289, S292, H295, and V301 in eSRK25. Note that the hvI and hvII regions, and in particular clusters of sites in the vicinity of the essential residues identified in planta, are enriched for residues showing elevated variability relative to other segments, as indicated by the number of residues that are polymorphic in more than 50% of the pairwise alignments (indicated by the horizontal dashed line). Substitution scores for the essential sites identified in vivo are significantly different from the non-essential sites (see Figure S4).
Conclusions
Our results provide the first empirical support for the hypothesis suggested by previous comparisons of SRK sequences [24, 27], that SCR-specific activation of SRK is a function of hvI and hvII (which are conserved in functionally-equivalent SRKs) rather than hvIII (which differs by many substitutions between functionally-equivalent SRKs). Although theory predicts high diversity at amino acids that are closely linked to sites subject to balancing selection [28], the finding that so very few of the residues previously shown to bear signals of positive selection (and therefore presumed to function as specificity determinants) are essential for SRK function (Figure 3) was unexpected. While some “positively-selected” sites that lie outside the hvI, hvII, and hvIII regions [22, 24] might be important for receptor functions unrelated to ligand recognition [e.g. see 29], polymorphisms at many “positively-selected” sites within the hvI-hvIII region of eSRK may have little functional importance. Rather, they might exhibit false signals of positive selection, as shown for “rapidly-evolving” regions in the human genome, in which “positively-selected” polymorphisms were found to result, not from selection, but from biased gene conversion [30-32]. Our finding that 17 out of 24 “positively-selected” sites in eSRK (Figure S4) can be replaced with other amino acids with no consequence for receptor function and ligand selectivity, together with the documented occurrence of gene conversion at the SRK gene [20, 33], suggests that a non-selective process might similarly drive accumulation of polymorphisms in eSRK.
We propose that for many if not all SRK variants, SI specificity is determined primarily by small amino-acid regions located toward the C-terminal end of hvI and C-terminal half of hvII. We further suggest that it is the overall sequence or three-dimensional conformation of these small segments, rather than individual residues within them, which determines SI specificity. The fact that residues in two discrete regions underlie SRK specificity is not surprising, as similar results were obtained for other recognition molecules [34-38]. It is tempting to speculate that the essential eSRK residues identified in this study, or the two clusters that encompass them, might be surface-exposed regions that are brought into close proximity in a three-dimensional structure to form part of an SCR-binding pocket. A high-resolution three-dimensional structure of the eSRK in its ligand-bound and unbound forms is required to address this issue. Nevertheless, three-dimensional models of eSRK sub-domains predict with confidence that hvI is a solvent-exposed segment of the LLD2 domain [29]. Accordingly, it is not surprising that the charge-, polarity-, or volume-changing substitutions (Figure S3) that were introduced at essential residues in this region would be disruptive. In contrast, the resolution of the hvII region, which maps to the structurally distinct LLD2 and EGF-like domains, is less clear [29], and it is difficult to surmise how substitutions in this region might impact SRK structure and function. Future in vivo structure-function analyses of other SRK variants, together with high-resolution structural studies, will undoubtedly elucidate the contribution of essential eSRK residues to ligand binding, and help explain how large numbers of SRK and SCR variants have coevolved to maintain their highly-specific interaction.
Supplementary Material
Acknowledgments
We thank Tiffany Crispell and Lei Zhu for technical help. A. thaliana C24 seed was obtained from the Arabidopsis Biological Resource Center in Columbus, Ohio. This work was supported by US National Institutes of Health Grant GM057527 and National Science Foundation Grant IOS-0744579.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci U S A. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 3.Shiba H, Iwano M, Entani T, Ishimoto K, Shimosato H, Che FS, Satta Y, Ito A, Takada Y, Watanabe M, Isogai A, Takayama S. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell. 2002;14:491–504. doi: 10.1105/tpc.010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science. 2001;293:1824–1826. doi: 10.1126/science.1062509. [DOI] [PubMed] [Google Scholar]
- 5.Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci U S A. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, Iwano M, Isogai A. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature. 2001;413:534–538. doi: 10.1038/35097104. [DOI] [PubMed] [Google Scholar]
- 7.Chookajorn T, Kachroo A, Ripoll DR, Clark AG, Nasrallah JB. Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc Natl Acad Sci U S A. 2004;101:911–917. doi: 10.1073/pnas.2637116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimosato H, Yokota N, Shiba H, Iwano M, Entani T, Che FS, Watanabe M, Isogai A, Takayama S. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in brassica self-incompatibility. Plant Cell. 2007;19:107–117. doi: 10.1105/tpc.105.038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasrallah M, Liu P, Sherman-Broyles S, Boggs N, Nasrallah J. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: Implications for the evolution of selfing. Proc Natl Acad Sci U S A. 2004;101:16070–16074. doi: 10.1073/pnas.0406970101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasrallah ME, Liu P, Nasrallah JB. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science. 2002;297:247–249. doi: 10.1126/science.1072205. [DOI] [PubMed] [Google Scholar]
- 11.Edh K, Widen B, Ceplitis A. The Evolution and Diversification of S-locus Haplotypes in the Brassicaceae Family. Genetics. 2009 doi: 10.1534/genetics.108.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- 13.Dwyer KG, Balent MA, Nasrallah JB, Nasrallah ME. DNA sequences of self-incompatibility genes from Brassica campestris and B. oleracea: polymorphism predating speciation. Plant Mol Biol. 1991;16:481–486. doi: 10.1007/BF00024000. [DOI] [PubMed] [Google Scholar]
- 14.Kimura R, Sato K, Fujimoto R, Nishio T. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant Journal. 2002;29:215–223. doi: 10.1046/j.1365-313x.2002.01208.x. [DOI] [PubMed] [Google Scholar]
- 15.Sato Y, Fujimoto R, Toriyama K, Nishio T. Commonality of self-recognition specificity of S haplotypes between Brassica oleracea and Brassica rapa. Plant Mol Biol. 2003;52:617–626. doi: 10.1023/a:1024819129785. [DOI] [PubMed] [Google Scholar]
- 16.Schierup MH, Mable BK, Awadalla P, Charlesworth D. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics. 2001;158:387–399. doi: 10.1093/genetics/158.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusaba M, Nishio T, Satta Y, Hinata K, Ockendon D. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: Implications for the evolution and recognition mechanism. Proc Natl Acad Sci U S A. 1997;94:7673–7678. doi: 10.1073/pnas.94.14.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schierup MH, Mable BK, Awadalla P, Charlesworth D. Identificaiton and Characterization of a Polymorphic Receptor Kinase Gene Linked to the Self-Incompatibility Locus of Arabidopsis lyrata. Genetics. 2001;158:387–399. doi: 10.1093/genetics/158.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasrallah JB, Kao TH, Chen CH, Goldberg ML, Nasrallah ME. Amino-acid sequence of glycoproteins encoded by three alleles of the S locus of Brassica oleracea. Nature. 1987;326:617–619. [Google Scholar]
- 20.Sato K, Nishio T, Kimura R, Kusaba M, Suzuki T, Hatakeyama K, Ockendon D, Satta Y. Coevolution of the S-Locus genes, SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002;162:931–940. doi: 10.1093/genetics/162.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awadalla P, Charlesworth D. Recombination and selection at Brassica self-incompatibility loci. Genetics. 1999;152:413–425. doi: 10.1093/genetics/152.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sainudiin R, Wong WS, Yogeeswaran K, Nasrallah JB, Yang Z, Nielsen R. Detecting site-specific physicochemical selective pressures: applications to the Class I HLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J Mol Evol. 2005;60:315–326. doi: 10.1007/s00239-004-0153-1. [DOI] [PubMed] [Google Scholar]
- 23.Dwyer KG, Kandasamy MK, Mahosky DI, Acciai J, Kudish BI, Miller JE, Nasrallah ME, Nasrallah JB. A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell. 1994;6:1829–1843. doi: 10.1105/tpc.6.12.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasrallah JB, Liu P, Sherman-Broyles S, Schmidt R, Nasrallah ME. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics. 2007;175:1965–1973. doi: 10.1534/genetics.106.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castric V, Vekemans X. Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol Biol. 2007;7:132. doi: 10.1186/1471-2148-7-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miege C, Ruffio-Chable V, Schierup MH, Cabrillac D, Dumas C, Gaude T, Cock JM. Intrahaplotype polymorphism at the Brassica S Locus. Genetics. 2001;159:811–822. doi: 10.1093/genetics/159.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordborg M, Innan H. The genealogy of sequences containing multiple sites subject to strong selection in a subdivided population. Genetics. 2003;163:1201–1213. doi: 10.1093/genetics/163.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naithani S, Chookajorn T, Ripoll DR, Nasrallah JB. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc Natl Acad Sci U S A. 2007;104:12211–12216. doi: 10.1073/pnas.0705186104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurst LD. Evolutionary genomics: A positive becomes a negative. Nature. 2009;457:543–544. doi: 10.1038/457543a. [DOI] [PubMed] [Google Scholar]
- 30.Galtier N, Duret L, Glemin S, Ranwez V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 2009;25:1–5. doi: 10.1016/j.tig.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth D, Bartolome C, Schierup MH, Mable BK. Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol Biol Evol. 2003;20:1741–1753. doi: 10.1093/molbev/msg170. [DOI] [PubMed] [Google Scholar]
- 33.Fowler TJ, Mitton MF, Vaillancourt LJ, Raper CA. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics. 2001;158:1491–1503. doi: 10.1093/genetics/158.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gola S, Kothe E. The little difference: in vivo analysis of pheromone discrimination in Schizophyllum commune. Curr Genet. 2003;42:276–283. doi: 10.1007/s00294-002-0353-4. [DOI] [PubMed] [Google Scholar]
- 35.Gola S, Hegner J, Kothe E. Chimeric pheromone receptors in the basidiomycete Schizophyllum commune. Fungal Genet Biol. 2000;30:191–196. doi: 10.1006/fgbi.2000.1222. [DOI] [PubMed] [Google Scholar]
- 36.Bergwitz C, Jusseaume SA, Luck MD, Juppner H, Gardella TJ. Residues in the membrane-spanning and extracellular loop regions of the parathyroid hormone (PTH)-2 receptor determine signaling selectivity for PTH and PTH-related peptide. J Biol Chem. 1997;272:28861–28868. doi: 10.1074/jbc.272.46.28861. [DOI] [PubMed] [Google Scholar]
- 37.Garrett TP, Saper MA, Bjorkman PJ, Strominger JL, Wiley DC. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature. 1989;342:692–696. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- 38.Dixit R, Nasrallah ME, Nasrallah JB. Post-transcriptional maturation of the S receptor kinase of Brassica correlates with co-expression of the S-locus glycoprotein in the stigmas of two Brassica strains and in transgenic tobacco plants. Plant Physiol. 2000;124:297–311. doi: 10.1104/pp.124.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


