Abstract
TRAF6 is a ubiquitin ligase essential for the activation of NF-κB and MAP kinases in multiple signaling pathways including those emanating from the interleukin-1 and Toll-like receptors (IL-1R/TLR)1-3. TRAF6 functions together with a ubiquitin-conjugating enzyme complex consisting of Ubc13 and Uev1A to catalyze Lys-63 (K63)-linked polyubiquitination, which activates the TAK1 kinase complex4,5. TAK1 in turn phosphorylates and activates IκB kinase (IKK), leading to activation of NF-κB. Although several proteins are known to be polyubiquitinated in the IL-1R/TLR pathways, it is not clear whether ubiquitination of any of these proteins is important for TAK1 or IKK activation. Herein, we reconstituted TAK1 activation in vitro using purified proteins and found that free K63 polyubiquitin chains, which are not conjugated to any target protein, directly activated TAK1 through binding to the ubiquitin receptor TAB2. This binding leads to autophosphorylation and activation of TAK1. We also found that unanchored polyubiquitin chains synthesized by TRAF6 and Ubc5 activated the IKK complex. Disassembly of the polyubiquitin chains by deubiquitination enzymes prevented TAK1 and IKK activation. These results indicate that unanchored polyubiquitin chains directly activate TAK1 and IKK, suggesting a novel mechanism of protein kinase regulation.
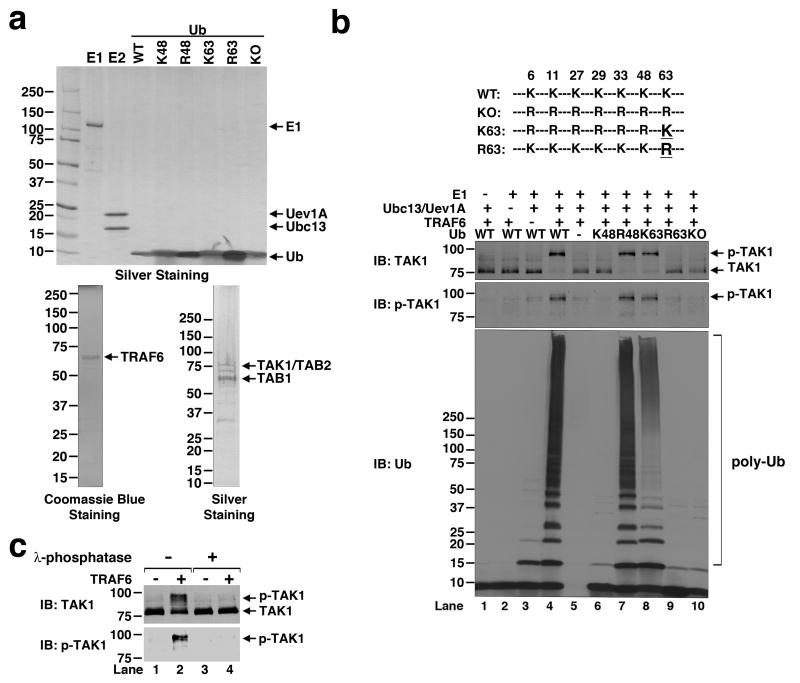
To investigate the biochemical mechanism of TAK1 regulation, we reconstituted TAK1 activation in vitro using purified proteins including E1, ubiquitin (Ub), Ubc13/Uev1A (E2), TRAF6 (E3) and the TAK1/TAB1/TAB2 kinase complex (Fig. 1a; see Supplementary Results for the source and purification of the proteins). These proteins were incubated together with an ATP buffer, and the activation of TAK1 was analyzed by immunoblotting with an antibody specific for TAK1 or TAK1 phosphorylated at Thr-1876. As shown in Figure 1b, in the presence of E1, Ubc13/Uev1A, TRAF6 and Ub, a slower migrating form of TAK1, which could be detected with the phospho-TAK1 antibody, was apparent (lane 4). Phosphorylation of TAK1 was not detectable in the absence of E1, Ubc13/Uev1A (E2) or TRAF6 (E3), or in the presence of Ub mutants harboring a mutation at K63, which prevented polyubiquitin (polyUb) chain synthesis. The phosphorylated form of TAK1 was converted to unphosphorylated TAK1 by lambda protein phosphatase (Fig. 1c), confirming that the large mobility shift of TAK1 was indeed caused by phosphorylation. These results demonstrate that TRAF6-catalyzed K63 polyubiquitination is both necessary and sufficient to activate TAK1 in vitro.
Figure 1. In vitro reconstitution of TAK1 activation by TRAF6.
a, Silver or Coomassie blue staining of purified proteins. b, Top: diagram of ubiquitin lysine mutants used in the assays. Bottom: purified TAK1 kinase complex was incubated with E1, Ubc13/Uev1A, TRAF6 and ubiquitin or its mutants in the presence of ATP for 1 hour at 30°C. Aliquots of the reaction products were analyzed by immunoblotting with an antibody against TAK1, p-TAK1 or ubiquitin. c, TAK1 activated by TRAF6-catalyzed ubiquitination as shown in (b) (lanes 3 &4) was treated with lamda protein phosphatase and then analyzed by immunoblotting.
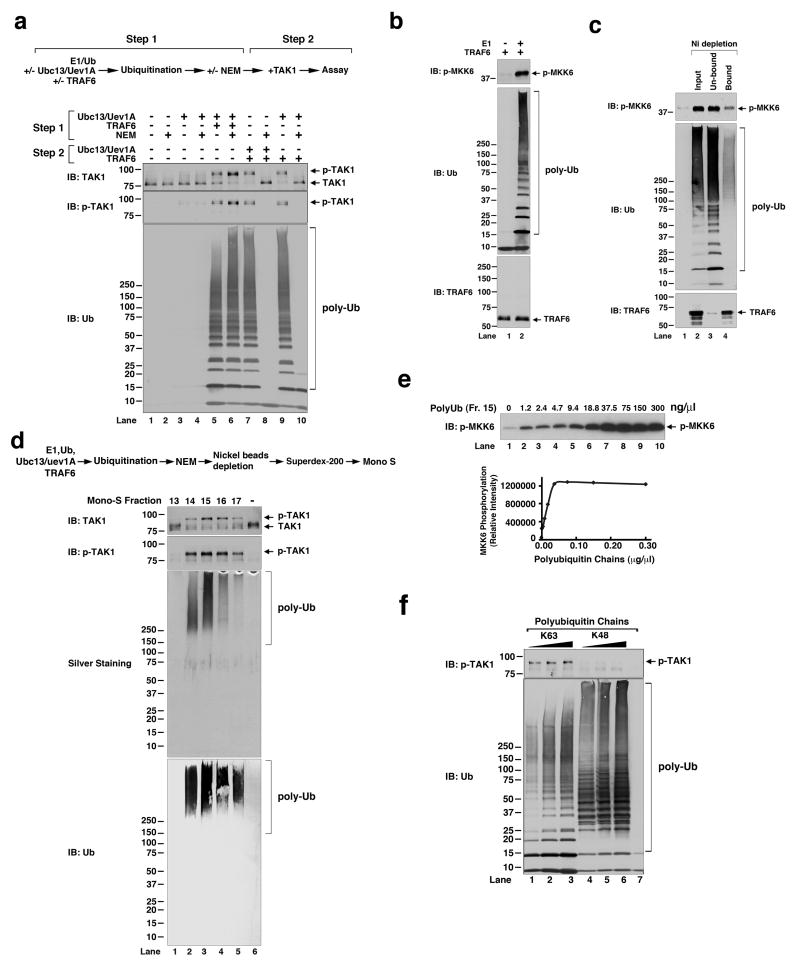
Two reactions occurred in the reconstitution system: ubiquitination and kinase activation. To uncouple these two steps, we first carried out the ubiquitination reaction in the presence of E1, Ub, Ubc13/Uev1A and TRAF6, but in the absence of the TAK1 kinase complex (Fig. 2a). The reaction mixtures were treated with N-ethylmaleimide (NEM), an alkylating agent that inactivates E1 and E2. After NEM was quenched by DTT and removed by buffer exchange, the ubiquitination mixtures were incubated with the TAK1 kinase complex in the presence of ATP. Remarkably, the ubiquitination product(s) synthesized in the first step was sufficient to activate the TAK1 kinase (Fig. 2a; lane 6). This activation was not caused by ubiquitination of the TAK1 kinase complex, because E1 and E2 had been inactivated by NEM in step 1 before they encountered the TAK1 complex in step 2. Inactivation of E1 and E2 by NEM was complete, because after E1 and Ubc13/Uev1A were incubated with NEM, they could no longer function together with TRAF6 to activate TAK1 in the second step (lane 7-10). The activation of TAK1 correlated perfectly with the formation of polyUb chains, whose synthesis required both Ubc13/Uev1A and TRAF6. These results indicate that the polyubiquitination reaction in the first step produces an “activator” that directly activates the TAK1 kinase complex.
Figure 2. Direct activation of TAK1 by unanchored polyubiquitin chains.
a, Ubiquitination reaction was carried out in the presence of E1, Ubc13/Uev1A, TRAF6 and ubiquitin, then E1 and E2 were inactivated with NEM (step 1) before incubation with the TAK1 complex in the presence of ATP to measure TAK1 activation (step 2). b, Ubiquitination reactions were carried out as in step 1 in (a), then incubated with TAK1 and its substrate MKK6(K82A). c, Ubiquitination mixtures were incubated with nickel-NTA beads, then the bound and unbound fractions were tested for stimulation of TAK1. d, Top: procedure for purification of TAK1 activator; bottom: the Mono S fractions were analyzed for TAK1 activation and by silver staining and immunoblotting. e, Different concentrations of purified polyubiquitin chains (Mono S fraction 15 in panel d) were tested for TAK1 activation. f, K63- and K48-linked polyubiquitin chains were tested for TAK1 activation.
A likely candidate that serves as the TAK1 activator is ubiquitinated TRAF6 4,7. However, consistent with a previous report 8, ubiquitinated TRAF6 was undetectable in the in vitro ubiquitination reactions containing varying concentrations of Ub (Fig. 2b and Supplementary Fig. S2). Despite the absence of detectable TRAF6 ubiquitination, the ubiquitination reaction stimulated TAK1 to phosphorylate MKK6, a physiological substrate (Fig. 2b, lane 2)4. Furthermore, when the ubiquitination reaction mixtures were incubated with nickel-NTA agarose to separate His-tagged proteins, including E1, Ubc13/Uev1A and TRAF6, from the untagged protein (ubiquitin), the majority of TAK1 stimulatory activity was present in the unbound fraction, which was largely depleted of TRAF6 (Fig. 2c, lane 3). Therefore, the TAK1 stimulatory activity correlated with the polyUb chains, but not TRAF6.
Next, we tried to identify the TAK1 activator in an unbiased manner through biochemical fractionation of TRAF6-catalyzed ubiquitination mixtures (see Methods and the diagram in Fig. 2d). The reaction mixtures were treated with NEM and then incubated with nickel-NTA agarose to deplete His-tagged proteins (E1, Ubc13/Uev1A and TRAF6). The unbound material, which contained the majority of TAK1 stimulatory activity, was further fractionated by gel filtration (Superdex 200) followed by cation exchange chromatography (Mono S). The fractions from the Mono S column were assayed for their ability to activate TAK1 in the presence of ATP. The same fractions were also analyzed by silver staining and immunoblotting with a Ub antibody (Fig. 2d). Strikingly, the TAK1-stimulatory activity co-fractionated with high molecular weight polyubiquitinated species. Immunoblotting experiments with an antibody against TRAF6 or the His6-tag revealed no detectable signal (Supplementary Fig. S3), suggesting that the polyUb chains were not conjugated to any of these proteins. These “unanchored” polyUb chains stimulated the TAK1 kinase to phosphorylate MKK6 in a dose-dependent and saturable manner (Fig. 2e). The concentration of polyUb chains required to achieve half maximal activation of TAK1 was approximately 16 ng/μl. In contrast to K63 polyUb chains, K48-linked polyUb chains failed to activate TAK1 (Fig. 2f).
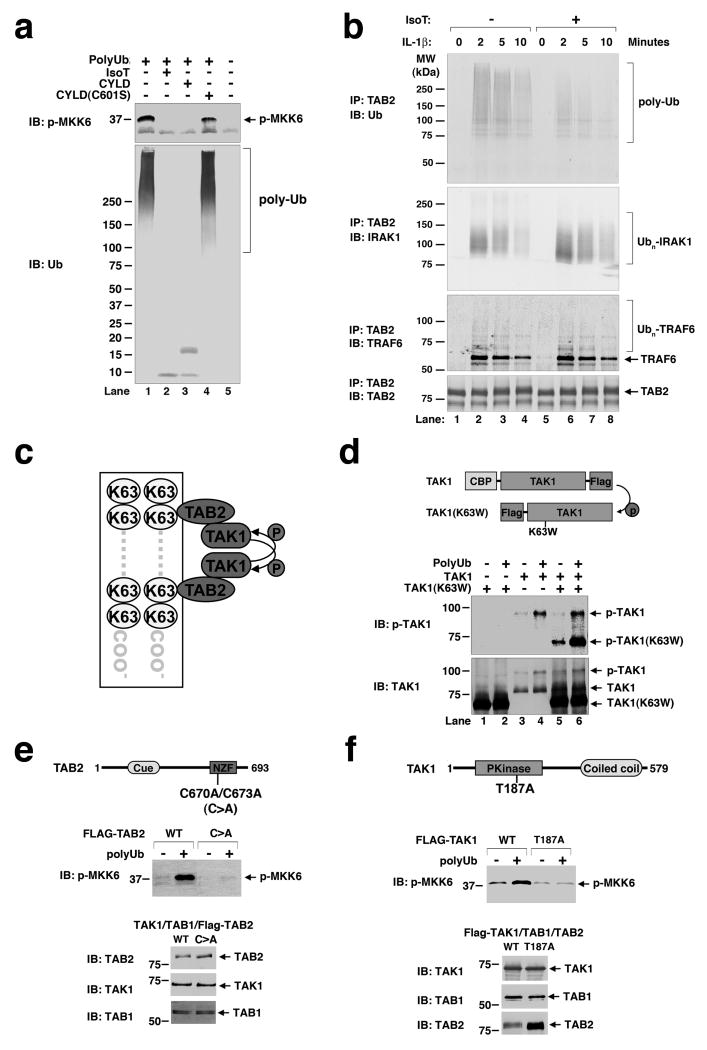
To further demonstrate that unanchored K63 polyUb chains were responsible for TAK1 activation, we incubated the purified polyUb with the deubiquitination enzymes CYLD or isopeptidase T (USP5 or IsoT). CYLD is a tumor suppressor protein exhibiting deubiquitination enzyme (DUB) activity that specifically cleaves K63-linked polyUb9,10, whereas IsoT specifically cleaves unanchored polyUb from the proximal end owing to its high-affinity binding to the C-terminal tail of ubiquitin11. As shown in Figure 3a, both CYLD and IsoT effectively converted K63 polyUb chains into mono-Ub and completely blocked the activation of TAK1. In contrast, a catalytically inactive CYLD mutant (C601S) was unable to cleave K63 polyUb chains or inhibit TAK1. These results demonstrate that unanchored K63 polyUb chains directly activate TAK1 in vitro.
Figure 3. IL-1β induces the synthesis of unanchored polyubiquitin chains to activate TAK1.
a, Purified polyUb chains were pre-incubated with the indicated enzymes and then tested for TAK1 activation. b, HEK293T/IL-1R cells were stimulated with IL-1β before cell lysates were immunoprecipitated with a TAB2 antibody. The precipitated materials were incubated with isopeptidase T and then analyzed by immunoblotting. c, A model of polyUb-induced TAK1 autophosphorylation. d, Purified polyUb was incubated with the wild-type TAK1 and/or TAK1(K63W) complexes in the presence of ATP. e, TAK1 complexes containing wild-type (WT) or C670A/C673A (C>A) mutant of TAB2 were incubated with polyUb and MKK6(K82A) to measure TAK1 activation. f, Similar to e, except that the TAK1 complexes contained wild-type (WT) or T187A mutant of TAK1.
To determine whether unanchored polyUb chains are synthesized in IL-1β-stimulated cells, we used a TAB2 antibody to immunoprecipitate the TAK1 complex and associated polyUb chains from HEK293 cells stably expressing the IL-1 receptor (IL-1R), which were stimulated with IL-1β for different lengths of time (Fig. 3b)12. The TAB2-associated proteins were treated with IsoT, then immunoblotted with an antibody against Ub, IRAK1 or TRAF6. Consistent with previous reports4,7,13,14, IRAK1 and TRAF6 were rapidly polyubiquitinated following stimulation of cells with IL-1β, but the polyUb chains on these proteins were resistant to disassembly by IsoT, which only cleaves polyUb with an unanchored C-terminal tail. In contrast, immunoblotting with the Ub antibody showed that the majority of polyUb chains associated with TAB2 were cleaved by IsoT. Similarly, immunoprecipitation experiments with an antibody against NEMO, which is known to bind to polyUb chains to mediate IKK activation15,16, revealed that the polyUb chains associated with NEMO in the IL-1β-stimulated cells were sensitive to IsoT treatment (Supplementary Fig. S4). These results indicate that IL-1β induced rapid formation of polyUb chains that associated with TAB2 and NEMO but were not conjugated to any target protein. Additional experiments further suggest that ubiquitination of IRAK1 or TRAF6 is dispensable for IKK activation (see Supplementary Results and Figure S5), consistent with a recent report showing that a TRAF6 mutant lacking all lysines fully rescued IL-1β-induced TAK1 and NF-κB activation in TRAF6-deficient cells17.
How might unanchored polyUb chains activate the TAK1 kinase complex? One possibility is that the polyUb chains bind to the TAB2 subunit of the TAK1 complex, facilitating autophosphorylation of TAK1 at Thr-187, which is known to be important for TAK1 activation (Fig. 3c)6. To test this possibility, we expressed and purified kinase complexes containing wild type or a kinase-dead (K63W) mutant of TAK1. The two forms of TAK1 differ in size due to a calmodulin-binding peptide (CBP) appended to the N-terminus of the wild-type TAK1, allowing these proteins to be distinguished when probed with a TAK1 antibody. We incubated the wild type and catalytically inactive TAK1 complex, both of which contained endogenous TAB2, in the presence of polyUb and ATP, then measured the phosphorylation of TAK1 by immunoblotting. As shown in Figure 3d, both wild type and mutant TAK1 were phosphorylated at Thr-187 in the presence of polyUb when they were incubated together. Since TAK1 (K63W) cannot phosphorylate itself, its phosphorylation at Thr-187 must have been carried out by the wild type TAK1, suggesting that the TAK1 complexes are brought into close proximity through binding to polyUb. Alternatively, polyUb binding may allosterically activate the TAK1 complex.
The model proposed in Figure 3c predicts that TAK1 activation by polyUb requires an intact Ub-binding domain of TAB2 as well as Thr-187 of TAK1. Indeed, mutations of two conserved cysteine residues (C>A) within the NZF-type ubiquitin-binding domain of TAB212 severely impaired the activation of TAK1 by polyUb (Fig. 3e). Similarly, the TAK1 (T187A) mutant failed to phosphorylate MKK6 in the presence of polyUb (Fig. 3f).
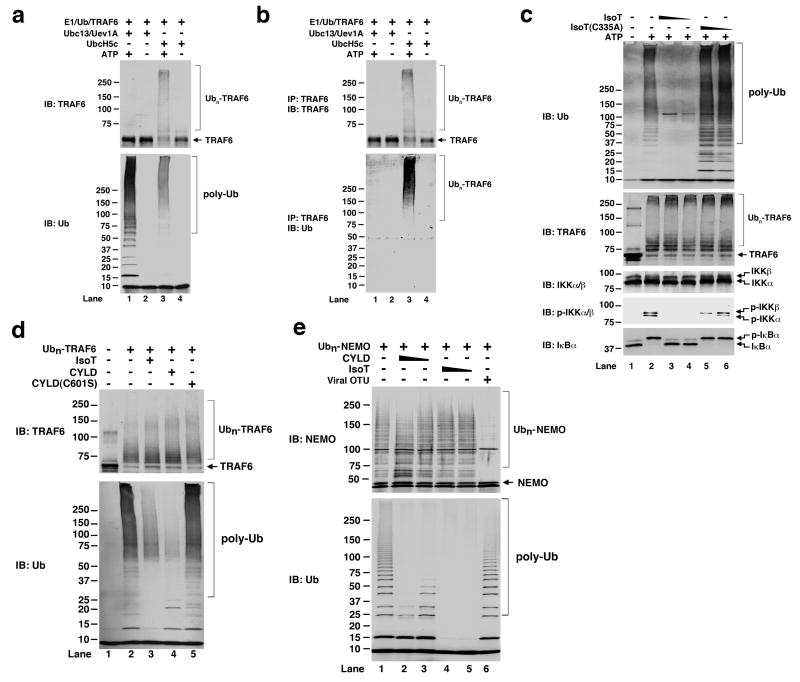
Whereas TRAF6 did not undergo auto-polyubiquitination in the presence of Ubc13/Uev1A in vitro, it was strongly polyubiquitinated in the presence of Ubc5 (Fig. 4a). Immunoprecipitation of TRAF6 under a denaturing condition confirmed that the polyUb chains were covalently attached to TRAF6 (Fig. 4b). Interestingly, the polyubiquitination products synthesized in the presence of Ubc5 preferentially activated IKK, whereas those synthesized by Ubc13 activated TAK1 (Supplementary Figure S6a & S6b). Treatments of polyUb chains with IsoT effectively cleaved the unanchored polyUb chains, but not polyubiquitinated TRAF6 (Fig. 4c). Importantly, the IsoT treatment abrogated IKK activation, indicating that the Ubc5-synthesized unanchored polyUb chains, but not ubiquitinated TRAF6, directly activated the kinase. A point mutation within a Ub-binding domain of NEMO (Y308S) prevented IKK activation by unanchored polyUb chains in vitro (Supplementary Fig. S7), suggesting that the polyUb chains activate IKK by direct binding to NEMO. The direct activation of IKK by TRAF6 and Ubc5 may explain the observations that ablation of Ubc13 in some mouse cell types prevents the activation of TAK1 and MAP kinases, but not IKK18, and that TAK1-deficient cells retain residual IKK activity19.
Figure 4. Regulation of IKK by unanchored polyUb chains and CYLD.
a. Ubiquitination reactions containing TRAF6 and Ubc13/Uev1A or Ubc5 were analyzed by immunoblotting with the indicated antibodies. b. Aliquots of reaction mixtures from (a) were denatured by heating in 1% SDS, then diluted to 0.1% SDS before immunoprecipitation with a TRAF6 antibody. c, PolyUb synthesized in the presence of TRAF6 and Ubc5 were pre-incubated with isopeptidase T or its mutant C335A, then incubated with the IKK complex and IκBα to measure IKK activation. d. Polyubiquitinated TRAF6 (Ubn-TRAF6) was synthesized in the presence of Ubc5, then incubated with the indicated DUBs before immunoblotting. e, Linear polyubiquitination of NEMO (Ubn-NEMO) was carried out in the presence of Ubc5 and HOIL-1L/HOIP (E3), then Ubn-NEMO was incubated with the indicated DUBs followed by immunoblotting.
Unlike Ubc13/Uev1A, which catalyzes specific K63 polyUb chain synthesis5,20,21, Ubc5 promotes the synthesis of polyUb chains of various linkages, including K48 and K63. Indeed, neither K48R nor K63R mutation prevented polyUb chain synthesis or IKK activation by TRAF6 and Ubc5, whereas ubiquitin mutants containing a single lysine at position 48 or 63 failed to support polyUb chain synthesis or IKK activation (Supplementary Figure S8). These results suggest that alternative ubiquitin linkages in the polyUb chains synthesized by TRAF6 and Ubc5 support IKK activation. A recent study suggests that linear Ub chains in which the N-terminus of one Ub is linked to the C-terminus of the preceding Ub may mediate IKK activation 22. However, we found no evidence that linear Ub chains were efficient activator of TAK1 or IKK in vitro (see Supplementary Results and Fig. S9-S10).
Surprisingly, like IsoT, CYLD specifically removed unanchored polyUb chains, but not polyUb chains conjugated to TRAF6 (Fig. 4d). To test if CYLD could remove polyUb chains from another substrate, we carried out a ubiquitination reaction using Ubc5 and the HOIP/HOIL-1L complex, which have been shown to function as E2 and E3, respectively, to catalyze linear polyubiquitination of NEMO22. Interestingly, CYLD and IsoT efficiently cleaved unanchored linear polyUb chains, but not the chains conjugated to NEMO (Fig. 4e). In contrast, a deubiquitination enzyme containing the OTU domain of the Crimean Congo hemorrhagic fever virus (CCHFV) large (L) protein removed polyUb chains from NEMO, but not unanchored linear polyUb chains23 (Fig. 4e, lane 6). Additional experiments indicated that the viral OTU, but not CYLD, was capable of cleaving K63 polyubiquitin chains from RIG-I, a viral RNA sensor24 (see Supplementary Results and Fig. S11). Taken together, these results suggest that CYLD preferentially cleaves unanchored polyUb chains in vitro. However, our data do not exclude the possibility that other proteins could assist CYLD in cleaving polyUb chains from protein targets or interfering with ubiquitination of these targets in vivo. Nevertheless, since the physiological function of CYLD in inhibiting TAK1 and IKK activation has been validated by abundant biochemical and genetic evidence9, our finding that CYLD preferentially cleaves unanchored polyUb chains further supports the role of these chains in protein kinase activation in the NF-κB pathway.
In much the same way that second messengers such as cyclic AMP (cAMP) are generated to activate protein kinases following stimulation of cells, unanchored polyUb chains are rapidly synthesized in cells stimulated with IL-1β. Also like cAMP, which can be hydrolyzed by phosphodiesterases, polyUb chains can be disassembled by specific deubiquitination enzymes, such as CYLD. The mechanism by which ubiquitin polymers activate protein kinases may be analogous to that employed by polymeric nucleic acids, such as DNA and RNA, which bind to protein kinases containing nucleic acid-binding domains or subunits (e.g, DNA-PK or protein kinase R). It is known that binding of double-stranded RNA polymers to protein kinase R leads to its dimerization and autophosphorylation25,26. Similarly, we propose that binding of unanchored polyUb chains to TAB2 or TAB3 leads to the dimerization or oligomerization of the TAK1 kinase complex and subsequent autophosphorylation of TAK1 at Thr-187, resulting in TAK1 activation. We also suggest that binding of unanchored polyUb chains to NEMO leads to autophosphorylation and activation of the IKK complex.
Methods Summary
Expression plasmids, proteins, antibodies and cell lines are described in the Full Methods. The TAK1 kinase complex was affinity purified from a HEK293T cell line stably expressing a TAP-tagged TAK1. The IKK complex was purified from HeLa S100 through affinity and conventional chromatography. K63-linked polyUb chains were synthesized in a reaction mixture containing Ubc13/Uev1A and TRAF6, then purified by affinity and conventional chromatography. K48-linked polyUb chains were synthesized using Ubc3 and Skp1-Cul1-Roc1-βTrcP complex as the E2 and E3, respectively. The TAK1 and IKK activation assays were performed using polyUb chains as the activator. For deubiquitination experiments, polyUb chains were incubated with CYLD or isopeptidase T. PolyUb chains induced by IL-1β were isolated by immunoprecipitation with an antibody against TAB2 or NEMO, incubated with or without IsoT, then analyzed by immunoblotting. A detailed description of the experimental procedures is provided in the Full Methods.
Supplementary Material
Acknowledgments
We thank Dr. Chee-Kwee Ea for generating the HEK293 cell line stably expressing the TAP-tagged TAK1, Dr. Jonathan Ashwell (NIH) for the bacterial GST-Ub2 and –Ub3 expression plasmids, Dr. Adolfo Garcia-Sastre (Mount Sinai School of Medicine) for the expression vector encoding the viral OTU enzyme, CCHFV-L(1-169), and Dr. Xiaoxia Li (Cleveland Clinics) for the IRAK1-deficient HEK293 cells line. We also thank Mr. Brian Skaug for critically reading the manuscript. This work was supported by grants from National Institute of Health (RO1-AI09919 and RO1-GM63692) and the Robert Welch Foundation (I-1389). Z.J.C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Author Contribution: Z-P. X., L. S., and Z.J.C. designed the experiments, which were performed by Z-P. X. and L.S., with assistance from X. C. G.P., X.J., A.A., and W.Z. contributed reagents. The manuscript was written by Z.J.C and Z-P. X.
References
- 1.Inoue J, Gohda J, Akiyama T. Characteristics and biological functions of TRAF6. Adv Exp Med Biol. 2007;597:72–79. doi: 10.1007/978-0-387-70630-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krappmann D, Scheidereit C. A pervasive role of ubiquitin conjugation in activation and termination of IkappaB kinase pathways. EMBO Rep. 2005;6:321–326. doi: 10.1038/sj.embor.7400380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 5.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 6.Singhirunnusorn P, Suzuki S, Kawasaki N, Saiki I, Sakurai H. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-beta-activated kinase 1 (TAK1) in a signaling complex containing TAK1-binding protein TAB1 and TAB2. J Biol Chem. 2005;280:7359–7368. doi: 10.1074/jbc.M407537200. [DOI] [PubMed] [Google Scholar]
- 7.Lamothe B, et al. Site-specific Lys-63-linked Tumor Necrosis Factor Receptor-associated Factor 6 Auto-ubiquitination Is a Critical Determinant of I{kappa}B Kinase Activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petroski MD, et al. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. J Biol Chem. 2007;282:29936–29945. doi: 10.1074/jbc.M703911200. [DOI] [PubMed] [Google Scholar]
- 9.Courtois G. Tumor suppressor CYLD: negative regulation of NF-kappaB signaling and more. Cell Mol Life Sci. 2008;65:1123–1132. doi: 10.1007/s00018-007-7465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komander D, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Reyes-Turcu FE, et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 12.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008;28:1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 17.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS ONE. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto M, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 19.Chen ZJ, Bhoj V, Seth RB. Ubiquitin, TAK1 and IKK: is there a connection? Cell Death Differ. 2006;13:687–692. doi: 10.1038/sj.cdd.4401869. [DOI] [PubMed] [Google Scholar]
- 20.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 22.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 23.Frias-Staheli N, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 25.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, et al. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.