Summary
Type-I interferons (IFNs) are important for antiviral and autoimmune responses. Retinoic acid-induced gene I (RIG-I) and mitochondrial antiviral signaling (MAVS) proteins mediate IFN production in response to cytosolic double-stranded RNA or single-stranded RNA containing 5′-triphosphate (5′-ppp). Cytosolic B-form double-stranded DNA, such as poly(dA-dT)·poly(dA-dT) [poly(dA-dT)], can also induce IFN-β, but the underlying mechanism is unknown. Here we show that the cytosolic poly(dA-dT) DNA is converted into 5′-ppp RNA to induce IFN-β through the RIG-I pathway. Biochemical purification led to the identification of DNA-dependent RNA polymerase III (Pol-III) as the enzyme responsible for synthesizing 5′-ppp RNA from the poly(dA-dT) template. Inhibition of RNA Pol-III prevents IFN-β induction by transfection of DNA or infection with DNA viruses. Furthermore, Pol-III inhibition abrogates IFN-β induction by the intracellular bacterium Legionella pneumophila and promotes the bacterial growth. These results suggest that RNA Pol-III is a cytosolic DNA sensor involved in innate immune responses.
Introduction
Innate immunity is the first line of host defense against microbial pathogens. Host cells utilize their pattern recognition receptors (PRRs) to detect pathogen-associated molecular patterns (PAMPs). The Toll-like receptor family (TLRs) is one class of PRRs that recognize PAMPs including lipoproteins, lipopolysaccharides (LPS), double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), and unmethylated CpG DNA (reviewed by Akira et al., 2006). Ligand-engaged TLRs recruit the adaptor proteins MyD88 or TRIF to activate downstream kinases including IκB kinase complex (IKK) and IKK-related kinases (TBK1 and IKKε), which activate the transcription factors nuclear factor-kappa B (NF-κB) and interferon regulatory factors (IRFs), respectively. NF-κB and IRFs function together in the nucleus to induce type-I interferons (IFNs; e.g., IFN-α and IFN-β) and other cytokines.
In another PRR pathway, cytosolic RNAs are recognized by RIG-I-like receptors (RLRs), which include RIG-I, MDA5 and LGP2 (Yoneyama et al., 2004). RLRs contain RNA helicase domains that recognize viral double-stranded RNA. In addition, RIG-I and LGP2 contain a C-terminal regulatory domain that recognizes single-stranded RNA containing 5′-triphosphate, which distinguishes foreign (e.g, viral) RNAs from self-RNAs that normally contain 5′-modification (e.g, capped mRNA). RIG-I and MDA5 also contain N-terminal tandem caspase activation and recruitment domains (CARD), which interact with the CARD domain of mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, VISA and CARDIF) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS localizes on the mitochondrial outer membrane through its C-terminal transmembrane domain, and this localization is important for MAVS to activate the cytosolic kinases IKK and TBK1 to induce IFNs (Seth et al., 2005).
Like RNA, accumulation of foreign or self DNA in the cytosol also triggers potent innate immune responses. DNA can be introduced into the cytosol of mammalian cells following infection with DNA viruses or bacteria, and the detection of cytosolic DNA is important for mounting an immune response against these pathogens. Under certain conditions, self DNA is inappropriately delivered to the cytosol, resulting in autoimmune responses. For example, DNase II-deficient macrophages lack the ability to digest self-DNA from engulfed apoptotic cells, leading to IFN-β production (Okabe et al., 2005; Yoshida et al., 2005). However, the mechanism by which cytosolic DNA induces IFNs is not well understood. In particular, the sensor that detects cytosolic DNA and triggers IFN production has remained largely unknown. Although DNA-dependent activator of IFN-regulatory factors (DAI) has been proposed to be a potential cytosolic DNA sensor (Takaoka et al., 2007), DAI-deficient mice still produce interferons in response to B-form DNA and have similar innate and adaptive immune responses to those of wild-type mice (Ishii et al., 2008). Recent studies identify AIM2 as a cytosolic DNA sensor that activates the inflammasome and caspase-1 (reviewed by Schroder et al., 2009). However, AIM2 is not involved in type-I interferon induction by cytosolic DNA.
Genetic studies have shown that cytosolic DNA can induce IFN production in mouse cells lacking RIG-I or MAVS, suggesting that the DNA signaling pathway is distinct from the RIG-I pathway (Ishii et al., 2006; Sun et al., 2006). Nevertheless, there is evidence that in certain human cell lines the induction of IFN-β by transfected double-stranded DNA depends on RIG-I and MAVS (Cheng et al., 2007; Ishii et al., 2006). However, RIG-I binds to RNA but not DNA, raising the question of how DNA might activate the RIG-I pathway.
In this report, we show that the double-stranded DNA poly(dA-dT)·poly(dA-dT), herein referred to as poly(dA-dT), is converted to an RNA species in the cytosol to trigger the RIG-I pathway in human and mouse cells. This RNA species contains 5′-triphosphate and forms a double-stranded RNA. The conversion of DNA to RNA can be recapitulated in vitro using cytosolic extracts. Biochemical purification led to the identification of DNA-dependent RNA polymerase III (Pol-III) as the enzyme responsible for transcribing the DNA template into an RNA ligand that activates RIG-I. RNAi-mediated knock down of Pol-III expression or inhibition of its enzymatic activity impedes interferon induction by transfection of DNA or infection with several DNA viruses, including adenovirus, herpes simplex virus 1 (HSV-1), and Epstein-Barr virus (EBV). Moreover, Pol-III inhibition blocks interferon induction by the intracellular bacterium Legionella pneumophila. These results strongly suggest that Pol-III is a cytosolic DNA sensor that triggers type-I interferon production through the RIG-I pathway.
Results
Cytosolic DNA triggers the RIG-I pathway through an intermediary RNA
In the course of attempting to identify a cytosolic DNA sensor involved in IFN-β production, HEK293 cells were transfected with various DNA together with a luciferase reporter driven by the IFN-β promoter. The cell lysates were analyzed by luciferase activity assays as well as native gel electrophoresis to detect IRF3 dimerization (Fig. 1A). As controls, the cells were also transfected with the synthetic RNA poly(I:C) or infected with Sendai virus (SeV), an RNA virus known to induce IFN-β. Among the DNA tested, including poly(dA-dT), poly(dI-dC), calf thymus DNA, PCR fragments of different lengths, and a linearized plasmid (pcDNA3), only poly(dA-dT) could activate IRF3 and induce IFN-β. Silencing the expression of RIG-I or MAVS with two different pairs of siRNA oligos strongly inhibited IRF3 and IFN-β induction by poly(dA-dT) (Fig. 1B). Pretreatment of poly(dA-dT) with DNase-I completely abrogated its ability to induce IFN-β, whereas RNase A treatment had no effect (Fig. 1C), indicating that RIG-I - dependent induction of IFN-β was not due to some RNA contaminants in poly(dA-dT). Synthetic DNA oligos containing 50 or 100 nucleotides of AT sequence were potent inducers of IFN-β, whereas GCAT or GC sequences of comparable lengths had little activity (Supplementary Fig. S1). Interestingly, insertion of 10 GC nucleotides in the middle of AT50 abolished its IFN-β inducing activity. Poly-T, poly-A or the duplex DNA formed between poly-T and poly-A did not induce IFN-β. A duplex DNA containing 80% AT sequence was capable of inducing IFN-β, whereas a DNA containing 50% AT sequence was inactive (Fig. S1). Taken together, these results suggest that IFN induction by DNA in HEK293 cells requires AT-rich sequence and certain length (30-50 nucleotides, see below). Poly(dA-dT), but not other DNA tested, also activated IRF3 and induced IFN-β in HeLa cells; the IRF3 activation was inhibited by RNAi of RIG-I or MAVS (Supplementary Fig. S2), indicating that RIG-I - dependent induction of IFN-β by poly(dA-dT) is not restricted to HEK293 cells.
Figure 1. Poly(dA-dT) activates RIG-I-dependent IFN-β production through intermediary RNA.
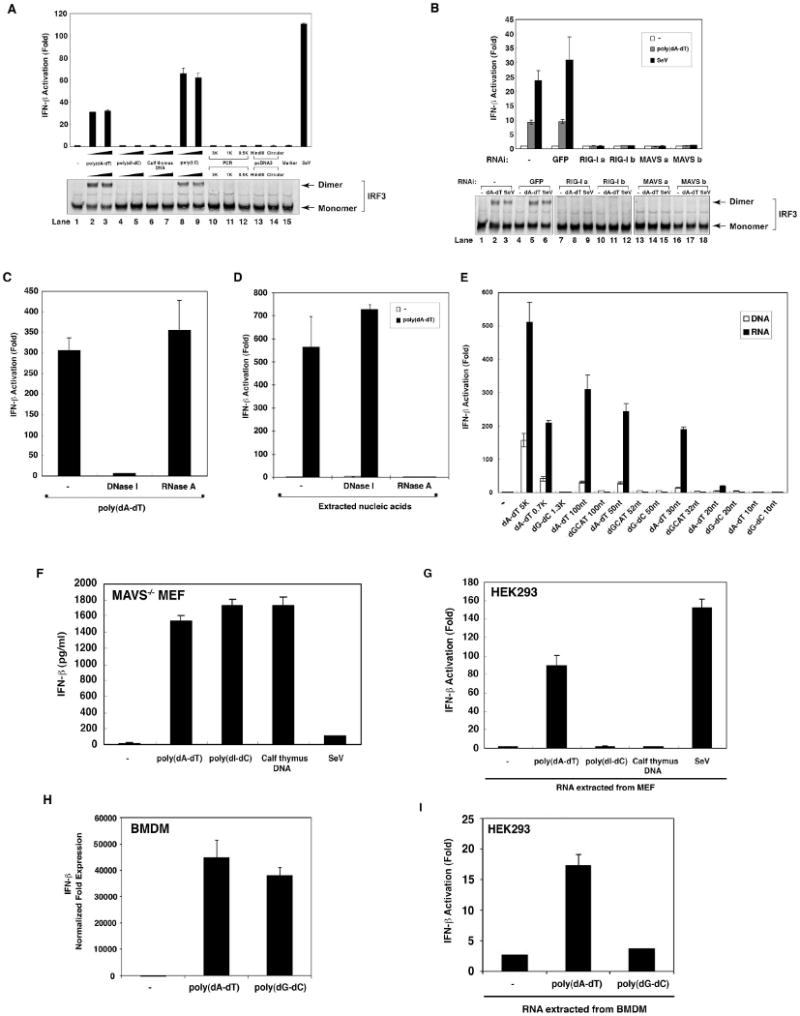
(A) HEK293 cells were transfected with the reporter plasmid IFN-β-Luc (25 ng/ml), pCMV-LacZ (50 ng/ml) and various types of DNA or RNA [poly(I:C)] as indicated (1 μg/ml). Cells were lysed 16 hours after transfection, followed by luciferase reporter assay (upper panel) or IRF3 dimerization assay (lower panel). PCR: linear PCR fragments of the indicated size; HindIII: HindIII-linearized fragment of pcDNA3; marker: DNA marker (0.1-10 kB; NEB); SeV: Sendai virus infection. (B) HEK293 cells were transfected with control siRNA oligos against GFP, two different pairs of siRNA oligos (a & b) against RIG-I, or MAVS. Subsequently, cells were cotransfected with IFN-β-Luc, pCMV-LacZ and pcDNA3. After 24 hours, cells were transfected with poly(dA-dT) (1 μg/ml) or infected with SeV. Luciferase reporter assay (upper panel) or IRF3 dimerization assay (lower panel) were performed 16 hours after transfection or infection. (C) HEK293 cells were transfected with poly(dA-dT) which had been pre-digested with DNase I (0.2 U/μl) or RNaseA (0.1 mg/ml), and then IFN-β luciferase reporter assay was carried out as described in (A). (D) HEK293 cells were transfected with or without poly(dA-dT) for 16 hours, and then nucleic acids were prepared from the cell lysates by phenol/chloroform extraction. After digestion with DNase I (0.2 U/μl) or RNase A (0.1 mg/ml), the nucleic acids were transfected into HEK293-IFNβ-luciferase reporter cells to measure IFN-β induction. (E) HEK293 cells were transfected with DNA of different sizes and compositions as indicated (1 μg/ml). Total RNA was extracted with TRIzol and then transfected into HEK293-IFNβ-luciferase reporter cells. In parallel experiments, the DNA was directly transfected into HEK293-IFNβ-luciferase reporter cells. (F) MAVS-deficient MEF cells were transfected with various DNA or infected with SeV. Culture supernatants were collected 16 hours after treatment for measurement of IFN-β by ELISA. (G) MAVS-deficient MEF cells were treated as described in (F) and then total RNAs were extracted by TRIzol and transfected into HEK293-IFNβ-luciferase reporter cells. (H) Mouse BMDM were transfected with poly(dA-dT) or poly(dG-dC) for 8 hours before total RNAs were extracted for qPCR. The expression level of IFN-β gene was normalized with that of β-actin gene. (I) BMDM cells were transfected as described in (H) and then total RNAs were extracted by TRIzol and transfected into HEK293-IFNβ-luciferase reporter cells.
To identify the ligands responsible for RIG-I activation in cells transfected with poly(dA-dT), we extracted nucleic acids from the cell lysates with phenol and chloroform and tested the ability of these nucleic acids to induce IFN-β. The nucleic acids extracted from cells transfected with poly(dA-dT) potently induced IFN-β. Surprisingly, the activity of nucleic acids extracted from poly(dA-dT) transfected cells was sensitive to RNase A, but not DNase I, suggesting that some intermediary RNAs capable of inducing IFN-β were activated or produced after poly(dA-dT) transfection (Fig. 1D).
To test whether the production of IFN-inducing RNA depends on the length and/or composition of cytosolic DNA, we transfected synthetic DNA oligos, as well as longer poly(dA-dT) and poly(dG-dC) (purchased from GE Healthcare), into HEK293-IFNβ-Luc reporter cells to measure IFN-β induction by the DNA. RNA from the transfected cells was extracted, then transfected into the same IFN-β reporter cells to measure IFN-β induction by the RNA (Fig. 1E). Interestingly, poly(dA-dT) containing as few as 30 base pairs was capable of directing the production of IFN-inducing RNA in the transfected cells. By contrast, even long poly(dG-dC) and GCAT DNA sequences were unable to produce any IFN-inducing RNA.
Previous studies showed that mouse embryonic fibroblasts (MEFs) lacking RIG-I or MAVS were still capable of producing IFN-β following transfection of DNA, including poly(dA-dT) (Ishii et al., 2006; Sun et al., 2006). To determine if the transfection of DNA in MEF cells also leads to the generation of IFN-inducing RNA, we transfected MAVS-deficient primary MEF cells with various DNA and then measured IFN-β production by ELISA. Indeed, poly(dA-dT), poly(dI-dC)·poly(dI-dC) [poly(dI-dC)] and calf thymus DNA, but not Sendai virus infection, induced IFN-β production in MAVS-deficient MEF cells (Fig 1F). When RNA extracted from these cells was transfected into HEK293-IFNβ-Luc reporter cells, only the RNA extracted from poly(dA-dT)-transfected and Sendai virus - infected cells was capable of inducing IFN-β (Fig 1G). These results suggest that MEF cells possess two cytosolic DNA sensing pathways, one detecting DNA irrespective of sequence composition and inducing IFN-β through a MAVS-independent mechanism, the other recognizing the poly(dA-dT) sequence and producing RNA ligands that trigger the RIG-I – MAVS pathway. Both poly(dA-dT) and poly(dG-dC) DNA induced IFN-β in bone marrow-derived macrophages (BMDM), but only RNAs extracted from poly(dA-dT)-transfected BMDM were capable of inducing IFN-β when they were transfected into the HEK293-IFNβ-Luc reporter cells (Fig. 1H and 1I). Therefore, poly(dA-dT) leads to the generation of IFN-inducing RNA not only in transformed cells (HEK293 and HeLa), but also in primary fibroblasts (MEF) and macrophages (BMDM).
Identification of the intermediary RNA as double-stranded RNA containing 5′-triphosphate
RIG-I recognizes double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA) bearing 5′-triphosphate (Hornung et al., 2006; Pichlmair et al., 2006). To determine the features of the IFN-inducing RNA generated in cells transfected with poly(dA-dT), we treated the RNA isolated from HEK293 cells with shrimp alkaline phosphatase (SAP) or polynucleotide kinase (PNK) and then tested its ability to induce IFN-β (Fig. 2A). The activity of the RNA was destroyed by SAP, but not PNK treatment. Using PNK to add one phosphate to the SAP-treated RNA did not restore the activity, indicating that RNA containing 5′-monophosphate fails to induce IFN-β. Treatment of the RNA extracted from poly(dA-dT)-transfected cells with terminator exonuclease (Ter Ex), an enzyme that specifically digests RNA containing 5′-monophosphate, did not impair the IFN-inducing activity of the RNA (Fig. 2B; lower panel), whereas treatment with RNase If (an RNase sensitive to heat inactivation) destroyed IFN-β induction. Control experiments showed that Ter Ex efficiently degraded 5′-p RNA generated by the treatment of RNA with SAP followed by PNK (Fig. 2B; upper panel). These results show that the IFN-inducing RNA contained more than one phosphate at the 5′ end, most likely 5′-triphosphate (5′-ppp; see Discussion).
Figure 2. Properties of the IFN-inducing RNA derived from poly(dA-dT).
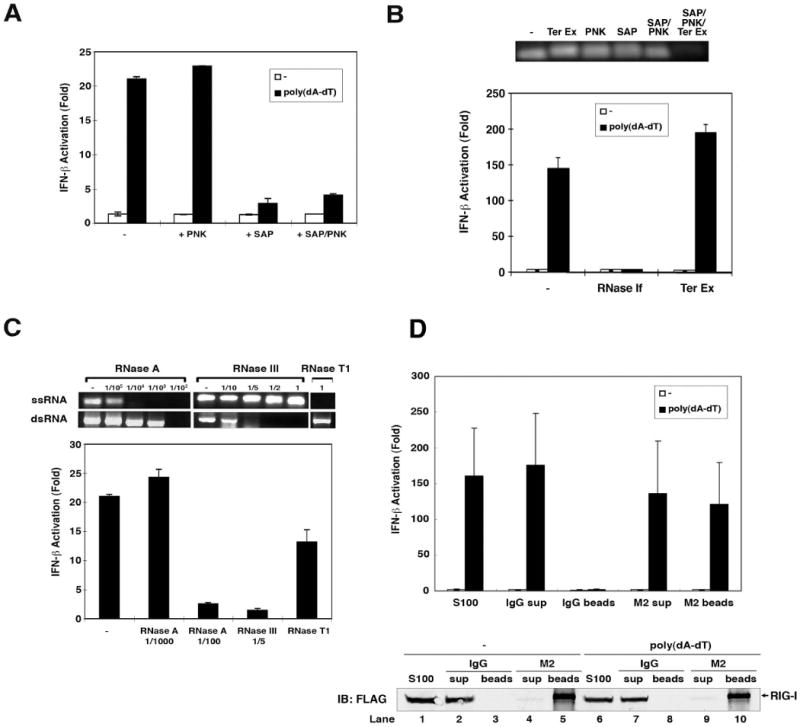
(A) RNAs extracted from poly(dA-dT) or mock transfected cells were treated with shrimp alkaline phosphatases (SAP) or polynucleotide kinase (PNK) at 37°C for 1 hour. An aliquot of SAP-treated RNA was further treated with PNK at 37°C for 1 hour to phosphorylate the DNA. RNAs were precipitated by ethanol precipitation and then transfected into HEK293-IFNβ-luciferase reporter cells. (B) Upper panel: poly(A-U) RNA transcribed by T7 polymerase was treated with terminator exonuclease (Ter Ex), PNK, SAP at 37°C for 1 hour. Two aliquots of SAP-treated RNA were phosphorylated with PNK before one of the aliquots was further treated with Ter Ex. Lower panel: RNAs extracted from poly(dA-dT)- or mock-transfected cells were treated with RNAse-If or Ter Ex and then transfected into HEK293-IFNβ-luciferase reporter cells. (C) Upper panels: single-stranded RNA (ssRNA) or double-strand RNA (dsRNA) generated by T7 polymerase was digested with various amounts of RNase A, RNase III, or RNase T1 as indicated, separated on 1% agarose gel and then visualized by ethidium bromide staining. Lower panel: RNAs extracted from poly(dA-dT) transfected cells were digested with RNAse A, RNase III, or RNase T1 and then transfected into HEK293-IFNβ-luciferase reporter cells. (D) Poly(dA-dT) was transfected into HEK293 cells stably expressing FLAG-tagged RIG-I, which was subsequently immunoprecipitated with an anti-FLAG antibody (M2) or control IgG. Nucleic acids in the cell extracts (S100), unbound supernatant (sup) and immunoprecipitates (beads) were extracted and then transfected into HEK293-IFNβ-luciferase reporter cells (upper panel). An aliquot of S100, sup and beads was separated by SDS-PAGE and immunoblotted with a FLAG antibody (lower panel).
Next, to determine the structure of the intermediary RNA, we treated the RNA with RNases known to be specific for ssRNA or dsRNA. The specificity of the RNases was tested with in vitro synthesized ssRNA or dsRNA (Fig. 2C, upper panel). RNase III specifically digests dsRNA, but not ssRNA, whereas RNase T1 has the opposite specificity. ssRNA was more sensitive to digestion by low-concentration of RNase A than dsRNA. When the RNA from poly(dA-dT) transfected cells was treated with RNase III and a high concentration of RNase A that degrades dsRNA, it lost its ability to induce IFN-β. By contrast, RNase T1 or a low concentration of RNase A, which degrades ssRNA, had no effect on the ability of the RNA to induce IFN-β (Fig. 2C, lower panel). These results suggest that the IFN-inducing RNA from poly(dA-dT) transfected cells is double-stranded RNA containing 5′-triphosphate. To determine if the RNA is a RIG-I ligand, we immunoprecipitated RIG-I from HEK293 cells stably expressing FLAG-tagged RIG-I using a FLAG-specific antibody (M2). The RNA in the immunoprecipitates was extracted and tested for IFN-β induction (Fig. 2D). The RNA extracted from the RIG-I complex immunoprecipitated from poly(dA-dT) transfected cells induced IFN-β, whereas the RIG-I complex from mock transfected cells did not contain IFN-inducing RNA. Taken together, these results indicate that poly(dA-dT) instructs or stimulates the production of dsRNA containing 5′-triphosphate, which binds to and activates RIG-I.
There are at least two potential mechanisms by which poly(dA-dT) leads to the generation of IFN-inducing RNA. Poly(dA-dT) might activate certain enzymes such as RNA kinases which modify endogenous RNAs and convert them into RIG-I ligands. Alternatively, poly(dA-dT) might serve as a template to direct the de novo synthesis of RNA by some enzymes, such as a DNA-dependent RNA polymerase. To determine if RNA polymerase II (RNA Pol-II) is involved in the synthesis of the IFN-inducing RNA, poly(dA-dT) was transfected into the HEK293-IFNβ - Luc reporter cell line which was pretreated with the RNA Pol-II inhibitor α-amanitin, and then total RNA was extracted from the cells and transfected again into the IFN-β reporter cell line. α-amanitin blocked the transcription of IFN-β induced by poly(dA-dT) (Supplementary Fig. S3A), but did not inhibit the generation of the RNA, which, when extracted from the drug-treated cells and re-transfected into HEK293-IFN-β reporter cells, was still capable of inducing IFN-β (Fig. S3B). These results indicate that RNA polymerase II is not involved in transcribing the intermediary RNA from poly(dA-dT). Actinomycin D, which intercalates GC-rich double-stranded DNA and blocks transcriptional elongation, also did not prevent the generation of IFN-inducing RNA in poly(dA-dT) transfected cells (Fig. S3B), suggesting that either actinomycin D did not inhibit transcription from poly(dA-dT) or the RNA was not generated through de novo synthesis.
A cell-free system that generates IFN-inducing RNA
To dissect the mechanism by which poly(dA-dT) leads to the production of IFN-inducing RNA, we tested whether the DNA can trigger the RNA generation in a cell-free system. HeLa cytosolic extracts (S100) were incubated with poly(dA-dT), ATP, MgCl2 and RNase inhibitors at 30°C for 1 hour followed by phenol/chloroform extraction and DNase I treatment. The RNA was then tested for IFN-β induction. Remarkably, the RNA generated in this cell-free system potently activated the IFN-β promoter (Fig 3A). This activity was abolished by RNase, but not DNase treatment, indicating that the IFN-inducing activity was not due to contaminating poly(dA-dT). (Fig 3A). Similar to the cell-based assays, poly(dG-dC), poly(dI-dC) or calf thymus DNA was incapable of triggering the production of IFN-inducing RNA in the in vitro system (Supplementary Fig S4A). The cell lysates from human (HEK293 and THP1) and mouse (MEF and Raw264.7) cell lines were also capable of generating IFN-inducing RNA in the presence of poly(dA-dT) (Fig S4B). Furthermore, the RNA generated in vitro lost its ability to induce IFN-β after treatment with SAP or the enzymes that digest dsRNA (RNase III and high concentration of RNase A), but not those that digest ssRNA (RNase T1 and RNase A at low concentration) (Fig S4C and S4D). The in vitro generated IFN-inducing RNA was also resistant to terminator exonuclease, indicating that it does not contain 5′-monophosphate (Fig. S4E). These results indicate that the cell-free system that we established faithfully recapitulates the generation of IFN-inducing RNA in poly(dA-dT) transfected cells.
Figure 3. In vitro generation of IFN-inducing RNA requires ATP and UTP.
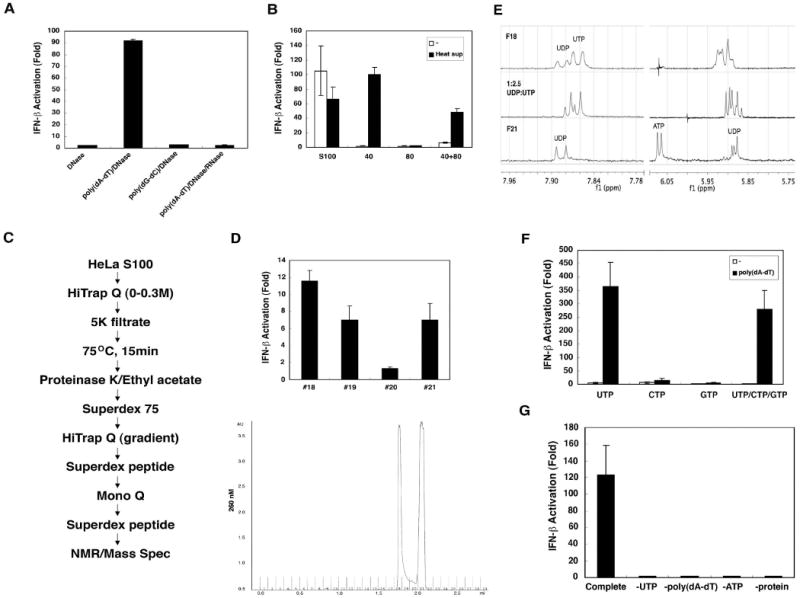
(A) HeLa S100 was incubated with poly(dA-dT) or poly(dG-dC) (20 μg/ml) and ATP (2mM) at 30°C for 1 hour. RNA was extracted with phenol/chloroform after DNase I (0.2 U/μl) and/or RNase A (0.1mg/ml) treatment, then transfected into HEK293-IFNβ-luciferase reporter cells. (B) HeLa S100 was precipitated with 40% ammonium sulfate, then the supernatant was further precipitated with 80% ammonium sulfate. The precipitates were dialyzed and incubated with poly(dA-dT) and ATP in the presence or absence of the supernatant from heat-treated HeLa S100 (“heat sup”). RNA from the in vitro reaction was extracted and transfected into HEK293-IFNβ-luciferase reporter cells. (C) Scheme of purification of heat-resistant factor required for the generation of IFN-inducing RNA. (D) Chromatogram (A260) of the small molecules on Superdex peptide column (last step; lower panel). Factions from the Superdex peptide column were incubated with a protein fraction from 40% ammonium sulfate precipitation, poly(dA-dT) and ATP, and then the RNA were extracted for IFN-β reporter assays (upper panel). (E) NMR spectra of fractions 18 (F18; top spectrum) and 21 (F21; bottom). The middle spectrum is a mixture of authentic UDP and UTP standards (1:2.5). (F) UTP, CTP, GTP or UTP/CTP/GTP (1.5 mM) was used to substitute for the “heat sup” in the in vitro reaction, and then RNA was extracted for IFN-β reporter assays. (G) Reactions containing a crude protein fraction (40% ammonium sulfate precipitate), poly(dA-dT), ATP and UTP (complete reaction) or lacking one of the components as indicated were carried out in vitro, and then RNA was extracted for IFN-β reporter assays.
ATP and UTP are required for the generation of IFN-inducing RNA in vitro
To identify the factors required for the generation of IFN-inducing RNA, we first divided HeLa cytosolic extracts (S100) into two parts by 40% and 40-80% ammonium sulfate precipitation. The precipitates were dialyzed (5kDa cut-off) and then assayed for their ability to generate IFN-inducing RNA in the presence of poly(dA-dT) and ATP. Surprisingly, the 40% and 40-80% fractions did not have the activity either alone or in combination (Fig 3B), suggesting that some factors were lost during dialysis or were not precipitated by 80% ammonium sulfate. To test whether HeLa S100 contains some small molecules or other heat-resistant factors required for the production of IFN-inducing RNA, we treated HeLa S100 at 75°C for 15 minutes to precipitate most proteins, and the supernatant (“heat sup”) was tested for its ability to support the generation of IFN-inducing RNA in the presence of the dialyzed fractions from ammonium sulfate precipitation. Adding the “heat sup” to the reaction containing 40% ammonium sulfate precipitated proteins led to a robust production of IFN-inducing RNA (Fig 3B). The activity in the “heat sup” was resistant to proteinase K, DNase I and RNase If (Fig. S5A), and could penetrate through a filter with 5 kDa molecular weight cut-off (data not shown), suggesting that some small molecules were required for generating IFN-inducing RNA. We purified the small molecules through the purification scheme indicated in Fig 3C and found two distinct peaks of activity in the last step of purification using a gel filtration column (Superdex peptide; Fig 3D). The fractions from both peaks had maximum absorption at 254 nanometer (data not shown), suggesting that they might be nucleotides or derivatives of nucleotides.
Two active fractions (18 and 21) from the Superdex peptide column were further analyzed by LC-MS (Supplementary Fig. S6) and NMR (Fig. 3E). LC-MS analysis (see Supplementary Information for details) of fraction 21 (F21) indicated the presence of UDP with an m/z of 403.0 [M–H]+, although the signal was partially masked by the presence of HEPES buffer (Fig. S6). The 1H NMR spectrum of F21 measured in D2O at 600 MHz showed the characteristic signals of a uridine nucleotide: two aromatic doublets at δ 7.89 and 5.91 ppm, an anomeric doublet at 5.87 ppm and overlapping multiplets of the ribose (Fig 3E). Comparison to an authentic standard of UDP in D2O with 10 μM HEPES and 40 μM KCl showed an excellent overlap in the 1H NMR spectrum, unequivocally establishing the active component of F21 as UDP.
The second active fraction, F18, was determined to be a 2:1 ratio of UTP to UDP. LC-MS clearly showed the presence of UDP but under the initial conditions we were unable to detect UTP. 1H NMR analysis showed two doublets at δ 7.85 and 7.89 ppm in a 2:1 ratio and a complex mixture of signals from δ 5.85 to 5.90 ppm (Fig 3E). The close similarity of the two sets of signals between F18 and F21 indicated the presence of a metabolite related to UDP, most likely UTP. 1H NMR of an authentic standard of 1:2.5 UDP:UTP with 10 μM HEPES and 40 μM KCl was nearly identical to F18. The small difference in chemical shift can be attributed to a difference in pH and ionic strength between F21 and the authentic standard. We originally missed the presence of UTP in F18 by LC-MS using our original LC conditions (condition 1 in experimental procedures), but changing the buffer to 0.1 M NH4OAc as a buffer (condition 2) allowed us to clearly verify the presence of UTP with an m/z of 483.0 [M–H]+ (Fig S6).
To verify the requirement of UTP and UDP in the production of IFN-inducing RNA, we replaced the “heat sup” with UTP, CTP, GTP or a mixture of these three nucleotides in the in vitro assay containing the dialyzed protein (40% ammonium sulfate precipitate), poly(dA-dT) and ATP. Only UTP could support the generation of IFN-inducing RNA (Fig 3F). UDP, UMP and uridine, but not dNTP, also supported IFN induction in the in vitro assays (Fig S5B & C), suggesting that the protein fraction might have nucleotide kinase activity to convert UDP, UMP or uridine to UTP. As the reaction mixtures always contained ATP, we removed ATP from the reaction and found that this removal abolished the production of IFN-inducing RNA (Fig 3G). Therefore, both ATP and UTP are required for the generation of IFN-inducing RNA in the in vitro system.
DNA-dependent RNA polymerase III catalyzes the generation of IFN-inducing RNA
Treatment of HeLa S100 with proteinase K completely destroyed the generation of IFN-inducing RNA in the in vitro system, indicating that one or more proteins in S100 are required for the RNA generation (Fig. 4A). We purified the active proteins from HeLa S100 through eight steps of conventional chromatography (Fig. 4B, left). Fractions from the last Superdex-200 step were analyzed for their ability to stimulate the generation of IFN-inducing RNA in the presence of poly(dA-dT), ATP and UTP (Fig. 4B, upper right). Aliquots of the fractions were also analyzed by silver staining (Fig. 4B, lower right). At least 8 visible bands co-purified with the activity. Three of these bands, p45, p40 and p23, were identified by nano-electrospray tandem mass spectrometry. The p45 bands contained subunit D of DNA-directed RNA polymerase III (POLR3D), p40 contained POLR3F, POLR3C, POLR3D and POLR3H, and p23 contained POLR3H, POLR3G and POLR3D. Immunoblotting showed that POLR3F and POLR3G co-purified with the activity that generates IFN-inducing RNA (Supplementary Fig. S7). As these proteins are subunits of RNA polymerase III (Pol-III), we attempted to identify all the subunits of the Pol-III complex. We established a HEK293 cell line stably expressing FLAG-POLR3F and immunoprecipitated the Pol-III complex with the FLAG antibody. The Pol-III complex was eluted with a FLAG peptide and then immunoprecipitated again with an antibody against POLR3G or a control IgG (Fig. 4C). Silver staining of the immunoprecipitated proteins revealed at least 12 distinct bands that were present in the POLR3G complex, but not in the control IgG sample. Nano-electrospray mass spectrometry of these unique bands identified eleven known Pol-III subunits (POLR3A, POLR3B, POLR3C, POLR3D, POLR3E, POLR3F, POLR3G, POLR3GL, POLR3H, POLR1C and CGRP-RCP) (Hu et al., 2002) and two other proteins (TRM1-like and RPS4) whose roles in Pol-III-mediated transcription have not been reported (Supplementary Table 1). Importantly, the purified Pol-III complex immunoprecipitated with anti-POLR3G, but not the control IgG immunoprecipitate, was sufficient to catalyze the synthesis of IFN-inducing RNA in the presence of poly(dA-dT), ATP and UTP (Fig. 4D).
Figure 4. Purification and identification of Pol-III.
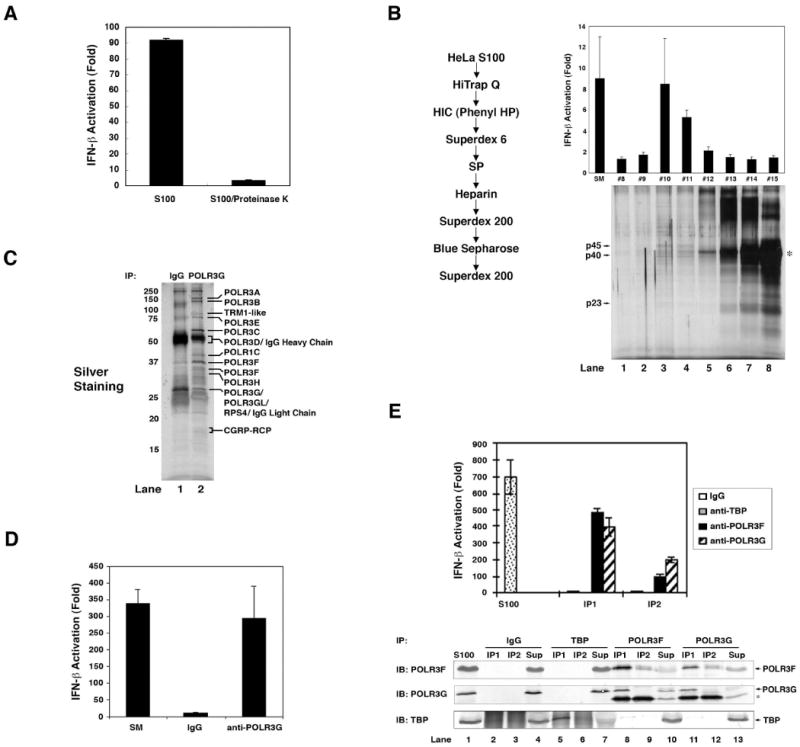
(A) HeLa S100 was incubated with or without proteinase K (2 mg/ml) at 37 °C for 1 hour and then incubated with poly(dA-dT) and ATP before RNA was extracted for IFN-β reporter assays. (B) Protein purification was carried out according to the scheme shown on the left and frations from the last Superdex 200 column were analyzed for their activity to produce IFN-inducing RNA (upper panel) and by silver staining (12% SDS-PAGE; lower panel). Arrows indicate the proteins co-purifying with the activity and analyzed by mass spectrometry. The asterisk indicates chicken albumin which was added as a carrier protein during purification. (C) Cytosolic extracts from HEK293 cells stably expressing FLAG-POLR3F were used for immunopurification of the Pol-III complex using anti-FLAG agarose (M2). Following elution with FLAG peptide, aliquots of the FLAG-Pol-III complex were subjected to immunoprecipitation with an antibody against POLR3G or control IgG, and the precipitated proteins were analyzed by silver staining. Unique bands from anti-POLR3G were identified by mass spectrometry. (D) The immunopurified Pol-III complex from (C) was incubated with poly(dA-dT), ATP and UTP before RNA was extracted for IFN-β reporter assays. (E) HeLa cell lysate (S100) was immunoprecipitated twice (IP1 and IP2) with an antibody specific for TBP, POLR3F, POLR3G or control IgG. The precipitated endogenous proteins were tested for their activity to produce IFN-inducing RNA (upper panel). Aliquots of the precipitated proteins and supernatants (“Sup”) were analyzed by immunoblotting with the indicated antibodies.
To determine if endogenous Pol-III could catalyze the synthesis of IFN-inducing RNA from poly(dA-dT), we purified the Pol-III complex from HeLa S100 by immunoprecipitation. Proteins immunoprecipitated with the antibodies against POLR3F or POLR3G, but not control IgG, had the ability to generate IFN-inducing RNA (Fig. 4E). We also tested the involvement of TATA-binding protein (TBP), which is known to recognize the dT-dA sequence present in most Pol-II and some Pol-III promoters. Proteins precipitated with an antibody against TBP did not contain POLR3F or POLR3G, nor did they catalyze the synthesis of IFN-inducing RNA. Taken together, these results show that the core RNA Pol-III complex catalyzes the generation of IFN-inducing RNA from poly(dA-dT) in vitro.
Pol-III catalyzes the synthesis of poly(A-U) RNA using poly(dA-dT) as a template
Poly(dA-dT), but not poly(dG-dC), leads to the generation of IFN-inducing RNA in transfected cells (Fig. 1E) and crude cell lysates (Fig. S4A). Similarly, the Pol-III complex purified from HEK293 stable cells expressing FLAG-tagged POLR3D or POLR3F catalyzed the synthesis of IFN-inducing RNA from poly(dA-dT), but not poly(dG-dC) (Fig. 5A). Ethidium bromide staining showed that poly(dA-dT), but not poly(dG-dC), served as the template for RNA synthesis in vitro (Fig. 5B). In vitro transcription experiments showed that Pol-III catalyzed the incorporation of α-32P-UTP into RNA when poly(dA-dT), but not calf thymus DNA, was used as the template (Fig. 5C). Northern blot analysis using a radiolabeled poly(A-U) RNA probe confirmed that the RNA synthesized by Pol-III in the presence of poly(dA-dT) was indeed poly(A-U) (Fig. 5D, lane 2).
Figure 5. Pol-III catalyzes the synthesis of poly(A-U) RNA using poly(dA-dT) as the template.
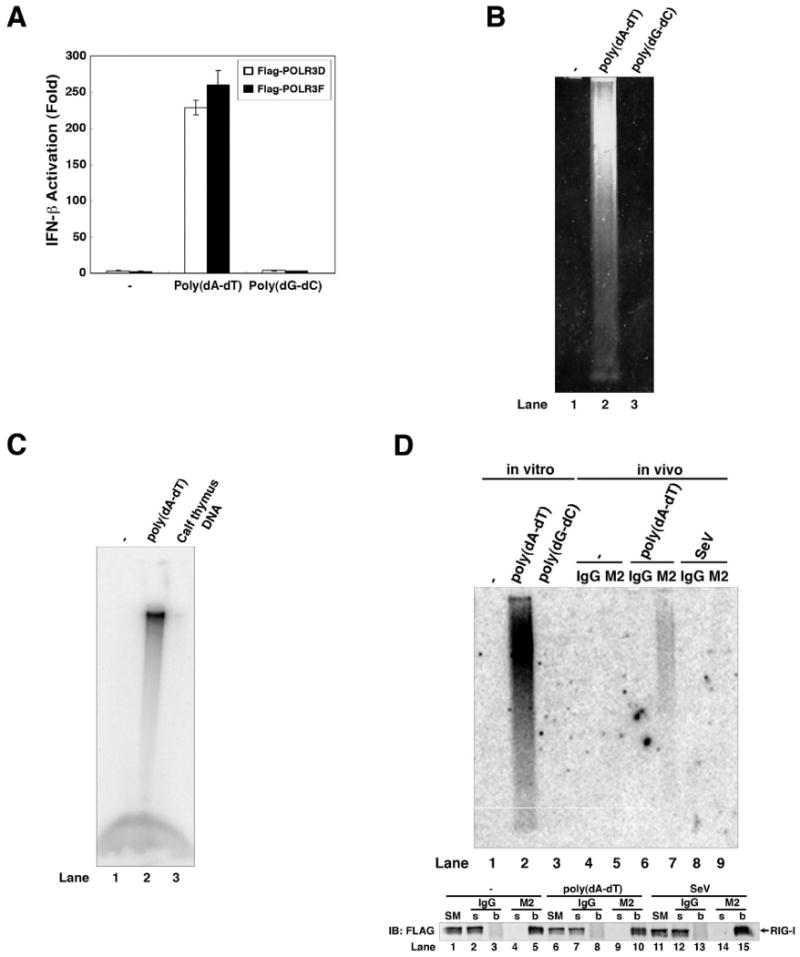
(A) Pol-III complex was immunopurified from HEK293 cells stably expressing FLAG-tagged POLR3D or POLR3F, and then incubated with poly(dA-dT) or poly(dG-dC) in the presence of NTP. RNA was extracted from the reactions for IFN-β reporter assays. (B) RNAs synthesized by in vitro transcription of poly(dA-dT) or poly(dG-dC) were analyzed by agarose gel eletrophoresis followed by ethidium bromide staining. (C) RNAs synthesized by in vitro transcription of poly(dA-dT) or calf thymus DNA in the presence of α-32P-UTP and NTP were analyzed by agarose gel electrophoresis followed by autoradiography. (D) HEK293 cells stably expressing FLAG-tagged RIG-I were transfected with poly(dA-dT) (lanes 6 & 7), infected with Sendai virus (lanes 8 & 9) or mock treated (lanes 4 & 5). RIG-I was subsequently immunoprecipitated with the FLAG antibody (M2) or control IgG, and RNA in the precipitates was extracted and analyzed by Northern blotting using 32P-labeled poly(A-U) RNA as a probe. In lanes 1-3, RNAs synthesized by in vitro transcription of poly(dA-dT) or poly(dG-dC) or in the absence of DNA were analyzed by Northern blotting. Bottom panel: Immunoprecipitated proteins were analyzed by immunoblotting with a FLAG antibody. SM: starting material (cell lysates); s: supernatant; b: bound proteins on the beads.
To test whether poly(dA-dT) directs the synthesis of poly(A-U) RNA in cells, we transfected poly(dA-dT) into HEK293 cells stably expressing FLAG-tagged RIG-I, immunoprecipitated RIG-I with the FLAG antibody, then analyzed the RIG-I bound RNA by Northern blotting using the radiolabeled poly(A-U) RNA probe (Fig. 5D; lanes 4-9). Poly(A-U) RNA was present in the RIG-I complex isolated from poly(dA-dT) transfected cells, but not mock-transfected or Sendai virus –infected cells. These results strongly suggest that poly(dA-dT) serves as a template to direct the synthesis of poly(A-U) RNA by Pol-III. Due to its sequence complementarity, poly(A-U) likely forms a double-stranded RNA, and it apparently retains the 5′-triphosphate, rendering it a strong RIG-I ligand.
Pol-III is necessary for the generation of IFN-inducing RNA
To determine if Pol-III is the major enzyme responsible for poly(dA-dT)-dependent RNA synthesis in HeLa S100, we depleted Pol-III from the extract by repeated immunoprecipitation using an antibody against POLR3F. The Pol-III depleted S100 was largely devoid of the activity to synthesize IFN-inducing RNA, whereas mock depletion with a control IgG did not remove the activity (Fig. 6A). Consistent with the notion that the cytoplasmic Pol-III generates IFN-inducing RNA in the cytosol, RNA isolated from the cytosol of poly(dA-dT)-transfected cells strongly induced IFN-β, whereas nuclear RNA isolated from the same cells did not (Fig.6B).
Figure 6. Pol-III is required for the production of IFN-inducing RNA.
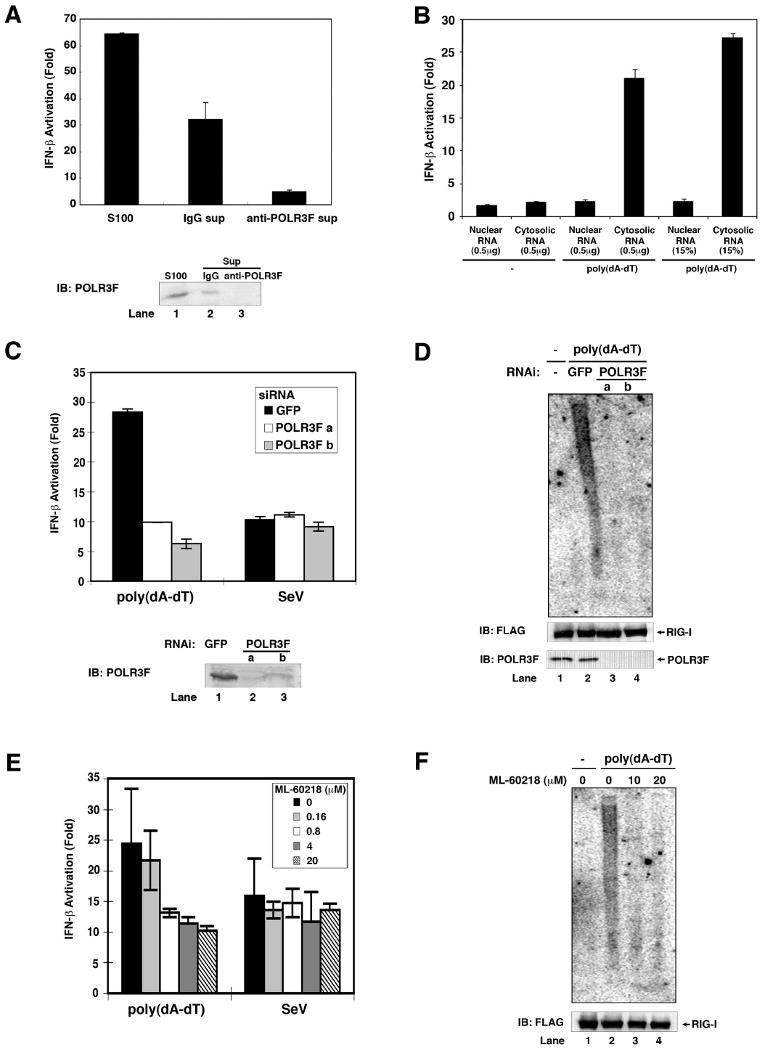
(A) HeLa cell lysate (S100) was immunodepleted five times with a POLR3F specific antibody or control IgG. S100 and the immunodepleted supernatants were incubated with poly(dA-dT), UTP and ATP before RNA was extracted for IFN-β reporter assays (upper panel). The efficiency of immunodepletion was analyzed by immunoblotting with the POLR3F antibody (lower panel). (B) HEK293 cells were transfected with or without poly(dA-dT) for 4 hours. Nuclear and cytosolic lysates were prepared for RNA extraction. 0.5 μg or 15% of nuclear or cytosolic RNAs were transfected into HEK293-IFNβ-luciferase reporter cells to measure IFN-β induction. (C) Two distinct siRNA oligos against POLR3F (a and b) or a control siRNA against GFP were transfected into HEK293 cells. 48 hours after transfection, the cells were transfected with poly(dA-dT) or infected with Sendai virus for 2 hours before RNAs were extracted for IFN-β reporter assays (upper). The efficiency of RNAi was analyzed by immunoblotting with an antibody against POLR3F (lower). (D) HEK293/RIG-I-FLAG stable cells were transfected with siRNA as described in (C), then transfected with poly(dA-dT) for 3 hours. The RIG-I complex was immunoprecipitated using a FLAG antibody, then the RNAs associated with RIG-I were extracted and analyzed by Northern blotting using 32P-labeled poly(A-U) RNA as a probe. The RIG-I in the immunoprecipitates and POLR3F in cell lysates were analyzed by immunoblotting. (E) HEK293 cells were treated with the indicated concentrations of the Pol-III inhibitor ML-60218 for 10 hours before transfection with poly(dA-dT) or infection with Sendai virus for 2 hours. RNAs were extracted for IFN-β reporter assays. (F) HEK293/RIG-I-FLAG stable cells were treated with ML-60218 and then transfected with poly(dA-dT). RNAs associated with the RIG-I complex were analyzed by Northern blotting as described in (D).
To test whether Pol-III is required for the production of IFN-inducing RNA in poly(dA-dT) transfected cells, we took two independent approaches. In the first approach, we used two different pairs of siRNA oligos against POLR3F to silence the expression of this essential Pol-III subunit in HEK293 cells, then transfected the cells with poly(dA-dT) or infected the cells with Sendai virus. RNA from these cells were extracted and then transfected into the HEK293-IFNβ-luciferase reporter cell line. Both siRNA oligos, but not the control GFP siRNA, strongly inhibited the generation of IFN-inducing RNA in poly(dA-dT) transfected cells (Fig. 6C). In contrast, the production of Sendai virus RNA, which is largely carried out by the viral RNA-dependent RNA polymerase, was not affected by the knockdown of POLR3F. Northern blot analysis showed that RNAi of POLR3F blocked the production of RIG-I-associated poly(A-U) RNA, demonstrating that Pol-III is required for the transcription of poly(dA-dT) in cells (Fig. 6D). In the second approach, we treated HEK293 cells with a specific chemical inhibitor of RNA Pol-III, ML-60218 (Wu et al., 2003), before transfection with poly(dA-dT) or infection with Sendai virus. This inhibitor selectively inhibited the production of IFN-inducing RNA in cells transfected with poly(dA-dT), but not in cells infected with Sendai virus (Fig. 6E & F). The partial blockade of IFN-β induction might be due to incomplete knockdown and partial chemical inhibition of Pol-III in cells. Nevertheless, it is clear from these results that Pol-III is required for the transcription of poly(dA-dT) into poly(A-U) RNA, which induces IFN-β.
Pol-III is required for IFN-β induction by Legionella pneumophila
A previous study showed that RNAi of MAVS inhibits IFN-β induction in a human lung epithelial cell line by Legionella pneumophila (Opitz et al., 2006), a gram-negative intracellular bacterium that infects macrophages and causes Legionnaires' disease. However, the role of MAVS in immune defense against Legionella and the underlying mechanism are largely unknown. We infected BMDM isolated from wild-type and Mavs-/- mice with L. pneumophila and then measured IFN-β expression by quantitative PCR (qPCR; Fig. 7A). The induction of IFN-β by the bacteria was markedly reduced in Mavs-/- macrophages. Strikingly, the Pol-III inhibitor ML-60218 strongly inhibited IFN-β induction by L. pneumophila in a dose-dependent manner (Fig. 7B). When the RNA was extracted from the drug-treated and bacteria-infected cells and then transfected into Raw264.7 macrophages again, IFN-β induction by RNA extracted from Pol-III inhibited cells was reduced to a level similar to that observed with RNA from mock-infected cells (Fig. 7C), indicating that Pol-III is required for the generation of IFN-inducing RNA by Legionella. The RNA isolated from the cytoplasm of Legionella-infected cells contained IFN-inducing activity, whereas the nuclear RNA from the same cells did not induce IFN (Fig. 7D). Importantly, ML-60218 treatment significantly increased the bacterial load in the macrophages (Fig. 7E), indicating that inhibition of the host Pol-III promotes rather than impedes bacterial growth. IFN-β treatment restored inhibition of Legionella replication in cells treated with the Pol-III inhibitor (Fig. 7F), demonstrating that Pol-III - induced IFN-β production is important for suppressing the bacterial replication. Taken together, these results strongly suggest that Pol-III and MAVS mediate interferon induction and immune defense against Legionella.
Figure 7. Pol-III is required for IFN-β induction by intracellular bacteria and DNA viruses.
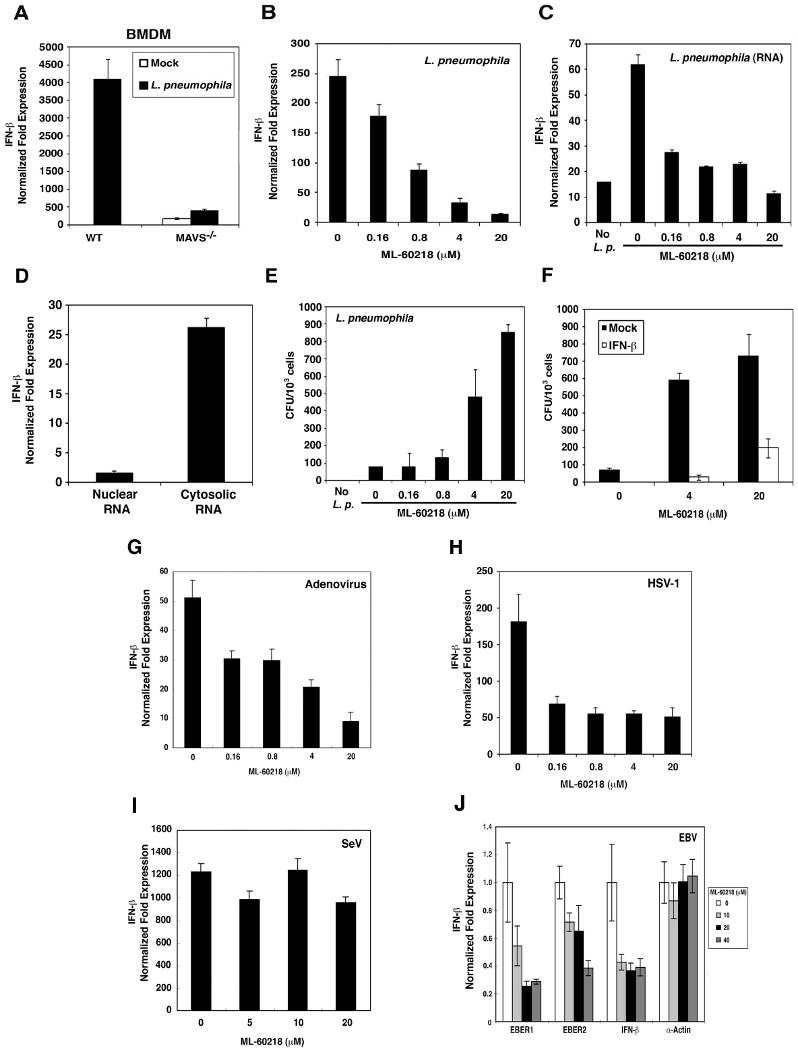
(A) Wild-type or MAVS-deficient BMDM cells were infected with Legionella pneumophila for 8 hours. Total RNAs were extracted for qPCR. The expression level of IFN-β gene was normalized with that of β-actin gene. (B) Raw264.7 cells were treated with the indicated concentrations of ML-60218 for 10 hours and then infected with Legionella pneumophila. Total RNAs were extracted after 8 hours of infection for qPCR of IFN-β and β-actin. (C) RNAs extracted from ML-60218-treated and Legionella pneumophila-infected Raw264.7 cells as described in (B) were transfected into Raw264.7 cells for 8 hours. The expression of IFN-β and β-actin was measured by qPCR. (D) Raw264.7 cells were infected with Legionella pneumophila for 8 hours. Nuclear and cytosolic lysates were prepared to extract RNAs, which were transfected into Raw264.7 cells for 8 hours. The expression of IFN-β was measured by qPCR and normalized to the level of β-actin. (E) Raw264.7 cells were treated with ML-60218 for 10 hours and infected with Legionella pneumophila for 24 hours. Cells were lysed in 0.1 % saponin and the lysates were spread on BCYE agar plates. Bacterial colonies were counted 2 days after incubation and shown as CFU per 1000 cells. (F) Raw264.7 cells were treated with ML-60218 in the presence or absence of mouse IFN-β (1000 U/ml) for 10 hours and subsequently infected with Legionella pneumophila for 24 hours. Bacterial colonies were determined as described in (E). (G-I) Raw264.7 cells were treated with ML-60218 for 10 hours and then infected with adenovirus (AdV; G), Herpes Simplex Virus-1 (HSV-1; H) or Sendai virus (SeV; I). Total RNAs were extracted for qPCR of IFN-β and β-actin. (J) The Epstein-Bar virus (EBV)-producing B95-8 cells were treated with ML-60218 for 24 hours and then total RNAs were prepared for qPCR. The expression levels of EBER1, EBER2, IFN-β and α-actin genes were normalized with that of the β-actin RNA. Both α- and β-actin are Pol-II transcribed genes.
Pol-III mediates IFN-β induction by DNA viruses
Some DNA viruses, including adenovirus, herpes simplex virus 1 (HSV-1) and Epstein-Barr virus (EBV), induce IFN-β production in a RIG-I dependent matter (Cheng et al., 2007; Rasmussen et al., 2007; Samanta et al., 2006). To determine whether Pol-III plays a role in IFN induction by DNA viruses, we treated Raw264.7 cells with ML-60218 and then infected the cells with adenovirus, HSV-1 or Sendai virus (RNA virus as a control). IFN-β induction by adenovirus (Fig. 7G) and HSV-1 (Fig. 7H) was markedly reduced by ML-60218 treatment. By contrast, ML-60218 treatment had no effect on IFN-β induction by Sendai virus (Fig. 7I), suggesting that the effect of ML-60218 was not due to general inhibition of IFN-β synthesis. ML-60218 also inhibited the production of IFN-inducing RNA in macrophages infected with HSV-1, but not Sendai virus (Supplementary Fig. S8A&B).
IFN induction by EBV has been linked to human autoimmune diseases, including systemic lupus erythematosus (SLE) (Ronnblom and Pascual, 2008). Interestingly, EBV-encoded small RNA 1 and 2 (EBER1 and EBER2) are non-coding, non-polyadenylated RNAs that form dsRNA-like stem-loop structures (Lerner et al., 1981). These abundant small RNAs, which contain 5′-triphosphate, are present in RNP particles that can be precipitated with anti-La antibody from the SLE patients. We tested the effects of the Pol-III inhibitor on IFN-β production in an EBV-producing human B cell line, B95-8. These cells were treated with different amounts of ML-60218 before total RNA was extracted for qPCR. The levels of EBER1, EBER2 and IFN-β RNA were reduced by the treatment with ML-60218 (Fig 7J). By contrast, the expression of a Pol-II dependent gene, α-actin, was not affected by the Pol-III inhibitor. ML-60218 also inhibited the production of IFN-inducing RNA in EBV-infected cells (Supplementary Fig. S8C). Collectively, these results suggest that Pol-III mediates IFN-β induction in EBV-infected B cells, possibly through transcription of EBV-encoded small RNAs.
Discussion
In this report, we delineate a mechanism by which AT-rich cytosolic DNA induces IFN-β production in human and mouse cell lines. We found that poly(dA-dT) serves as a template for the de novo synthesis of poly(A-U) RNA by DNA-dependent RNA polymerase III. The IFN-inducing activity of the RNA is resistant to terminal exonuclease and RNase T1 but sensitive to alkaline phosphatase and RNase III, indicating that this is a double-stranded RNA containing more than one phosphate at the 5′-ends. Although at present we cannot definitively determine whether the RNA contains 5′-triphosphate or 5′-diphosphate, Pol-III is known to synthesize RNA containing 5′-triphosphate. Our results that Pol-III is responsible for the synthesis of the RNA from poly(dA-dT) (Fig. 4 & 6), which binds directly to RIG-I (Fig. 2D & 5D), strongly suggest that the RNA contains 5′-triphosphate.
The RNA Pol-III complex is both necessary and sufficient to catalyze the synthesis of poly(A-U) from poly(dA-dT) in vitro. By employing RNAi and a specific inhibitor of Pol-III, we show that Pol-III is required for the production of IFN-inducing RNA by poly(dA-dT) in cells. Furthermore, Pol-III inhibition significantly reduced the induction of IFN-β by intracellular bacteria (Legionella pneumophila) and DNA viruses (adenovirus, HSV-1 and EBV), but not RNA viruses (Sendai virus). Pol-III inhibition also led to a significant increase of bacterial replication (Legionella pneumophila). These results strongly suggest that Pol-III plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses.
Cytosolic RNA Pol-III is a DNA sensor that triggers innate immune response
Pol-III was purified from cytosolic extracts (Fig. 4), consistent with its role as a cytosolic DNA sensor. Cytosolic RNA isolated from cells transfected with poly(dA-dT) or infected with Legionella strongly induced IFN-β, whereas RNA isolated from the nucleus of the same cells did not (Fig. 6B & 7D), further supporting our model that cytosolic Pol-III is responsible for production of IFN-inducing RNA which is then recognized by RIG-I in the cytosol. Previous studies utilizing in vitro transcription assays showed that two-thirds of the Pol-III activity is in the cytoplasm, whereas only 16 and 11 percent of the Pol-I and -II activities are in the cytoplasm, respectively (Jaehning and Roeder, 1977). These pioneering studies divided Pol-III into two chromatographic forms (IIIA and IIIB). Only Pol-IIIA is present in the nucleus while approximately equal amounts of Pol-IIIA and IIIB are found in the cytoplasm. It has been a long-standing mystery why the majority of Pol-III activity is present in the cytosol where DNA should be avoided. Our finding that cytosolic Pol-III is a DNA sensor that mediates innate immune response provides a potential answer to this conundrum. The cytosol is a strategic location for Pol-III to function in immune defense, because RNA synthesized in the cytosol contains 5′-triphosphate that can be detected by RIG-I, whereas RNA synthesized in the nucleus is likely to be modified by enzymes such as the capping enzymes (e.g, mRNA) or processed to generate 5′-monophosphate (e.g, tRNA), which escapes detection by RIG-I. Therefore, we propose that nuclear and cytoplasmic RNA polymerases serve opposing functions: while the nuclear polymerases produce the host RNA that should be modified to avoid triggering autoimmune response through the RIG-I pathway, the cytosolic Pol-III is specialized to detect foreign and aberrant DNA to mount an immune response to defend the integrity of the host cytoplasm.
Recognition of AT-rich DNA sequences by RNA Pol-III
At least three different complexes have been identified as transcription factors for polymerase III (TFIIIA, TFIIIB and TFIIIC) (Schramm and Hernandez, 2002). TBP is one of the subunits in TFIIIB and binds to the TATA box. The nature of poly(dA-dT) sequence might provide the TATA motifs for TBP binding. However, TBP is not present in the highly purified Pol-III complex that catalyzes in vitro transcription of poly(dA-dT), and TBP immunoprecipitated from cytosolic extracts does not have the transcription activity (Fig. 4C & E). Therefore, TBP may not be responsible for the preferential recognition of dA-dT-rich sequences by Pol-III, although our results cannot rule out the possibility that TBP might play a role in DNA sequence recognition by cytosolic Pol-III in vivo. The Pol-III complex we purified contains at least 13 subunits; some of these subunits may determine the optimal sequences that permit transcription by Pol-III.
A recent study suggests that homopolymeric ribonucleotide composition, such as A-U rich sequence, is required for RNA to bind and activate RIG-I (Saito et al., 2008). Thus, it is possible that a GC-rich DNA could still be transcribed into an RNA, but the RNA lacks the ability to activate RIG-I. However, we found that Pol-III could not transcribe poly(dG-dC) or poly(dG-dC-dA-dT) DNA (Fig. 5). Moreover, if we used T7 RNA polymerase to transcribe an RNA containing GCAU sequence or a random sequence with 50% GC composition, this RNA could efficiently induce IFN-β (Supplementary Fig. S9), indicating that the failure of GC-rich DNA to induce IFN-β was due to the lack of the production of the RNA rather than the inability of the GC-rich RNA to induce IFN-β.
Detection of viral and bacterial DNA by cytosolic RNA Pol-III
Although many DNA viruses encode DNA-dependent RNA polymerases and some use the cellular RNA polymerases for viral gene transcription, the majority of the viral RNA is modified by cellular and virus-encoded modification enzymes such as the capping enzymes. However, some viral RNAs, such as the VA-I and VA-II RNAs from adenovirus and EBER1 and EBER2 RNAs from EBV, are non-coding small RNAs containing 5′-triphosphate (Lerner et al., 1981; Rosa et al., 1981). Our results and previous studies suggest that these viral RNAs are transcribed by cellular Pol-III. EBER genes contain internal control sequence motifs (Box A and B) that are recognized by TFIIIB and TFIIIC, which direct Pol-III transcription (Schramm and Hernandez, 2002). In addition, the upstream promoters of EBER genes contain a TATA-like sequence which specifies Pol-III, but not Pol-II, transcription (Howe and Shu, 1993). Thus, the transcription of cytosolic DNA, some of which is derived from bacteria and viruses, is controlled not only by recognition of dA-dT rich sequence by Pol-III, but also by other transcription factors associated with Pol-III, such as TFIIIA, B and C.
While the VA and EBER small RNAs are abundant Pol-III transcripts that contain 5′-triphosphate, they are normally in complex with cellular proteins to form RNP particles that predominantly reside in the nucleus (Lerner et al., 1981). Therefore, these viral RNAs are segregated from RIG-I in the cytosol, providing an explanation for why cells containing these abundant viral RNAs do not produce copious amounts of IFN. This may be a mechanism by which the virus evades the host immune system, and it may also be a mechanism by which the host avoids excessive autoimmune responses. However, the VA and EBER RNP particles react with antibodies from SLE patients (Lerner et al., 1981; Rosa et al., 1981). Both the sera from SLE patients and cells chronically infected with EBV have elevated levels of type-I interferons, and epidemiology studies have suggested a link between EBV infection and SLE (Ronnblom and Pascual, 2008). It is possible that under certain pathological conditions, some of the viral RNA gains access to RIG-I, triggering IFN production that contributes to autoimmune diseases.
Like viruses, intracellular bacteria are potent inducers of type-I interferons. However, the mechanisms by which bacteria induce interferons are largely unknown. Interestingly, Mavs-deficient macrophages are severely defective in producing IFN-β in response to infection by Legionella pneumophila. Our finding that Pol-III is required for IFN-β induction by L. pneumophila provides a mechanism for the Mavs-dependent innate immune response against this bacterium. Our results suggest that Pol-III detects some segments of the bacterial DNA and transcribes them into RNA ligands that induce IFN-β through the RIG-I- MAVS pathway. The utilization of Pol-III as a DNA sensor for bacteria and DNA viruses may allow the host to take advantage of the RIG-I-MAVS pathway to defend against a much larger spectrum of microbial pathogens.
MAVS-dependent and -independent pathways triggered by cytosolic DNA
Our results that deletion of MAVS or inhibition of Pol-III blocked IFN-β production in response to Legionella infection suggest that the Pol-III – MAVS pathway is the major pathway responsible for interferon induction by this bacterium (Fig. 7A & 7B). Future research should reveal whether the MAVS-dependent DNA sensing pathway is the major pathway for immune defense against other pathogens, including bacteria, DNA viruses, or even parasites such as malaria. However, the Pol-III – MAVS pathway is unlikely to be the only cytosolic DNA sensing pathway that induces type-I IFNs. While the human cell lines HEK293 and HeLa solely rely on the Pol-III – MAVS pathway to induce IFN in response to poly(dA-dT), primary MEF cells possess both MAVS – dependent and –independent pathways to detect cytosolic DNA. Interestingly, we noticed that spontaneous immortalization of MEF cells by repeated passages led to a loss of the MAVS-independent pathway (Chiu, Y and Chen, Z. J., unpublished). It is not clear why immortalized or transformed cells lose the MAVS-independent pathway, but it is interesting to note that the loss of IFN induction by DNA transfection in some transformed cells such as HEK293 and HeLa may allow the widespread use of these cells in molecular cell biology. The MAVS-independent pathway appears to recognize DNA structure rather than its sequence. Although the DNA-binding protein DAI (ZBP1) has been proposed as a DNA sensor that induces type-1 IFNs, DAI-deficient mice and cells from these mice do not have apparent defects in producing type-I IFNs in response to DNA stimulation (Ishii et al., 2008; Takaoka et al., 2007). The recently discovered DNA-binding protein AIM2 is clearly a cytosolic DNA sensor that activates the inflammasome and caspase-1, but it does not induce type-I IFNs (Schroder et al., 2009). It is possible that multiple sensors exist to detect the invasion of cytosol by foreign or aberrant self DNA. The identification of these DNA sensors, including Pol-III, should provide critical insights into the mechanism of innate immune responses against a large variety of microbial pathogens including bacteria and viruses. Furthermore, this line of research will lead to a deeper understanding of the mechanism underlying debilitating autoimmune diseases such as lupus.
Experimental Procedures
Nucleic Acid Extraction
Total RNAs were extracted by TRIzol (Invitrogen) from cells according to manufacturer's instruction. To prepare nuclear and cytoplasmic RNA, cells were lysed in Buffer A (10 mM Hepes [pH7.5], 10 mM KCl, and 1.5 mM MgCl2) by douncing and centrifuged at 100 × g for 5 minutes to remove cell debris. The supernatant was centrifuged at 500 × g for 5 minutes to collect the nuclear pellet. The supernatant was further centrifuged at 20,000 × g for 10 minutes to collect the cytosolic supernatant. Nucleic acids were extracted from the nuclear and cytosolic lysates with phenol and chloroform, followed by isopropanol precipitation.
In vitro Reconstitution Assay
Cell extracts (S100) were prepared as described previously (Deng et al., 2000). S100 (2 mg/ml) was incubated with poly(dA-dT) or poly(dG-dC) (20 μg/ml) together with ATP (2 mM), MgCl2 (5 mM) and RNase inhibitor (0.4U/μl, Invitrogen) in a 50 μl reaction mixture at 30 °C for 1 hour. After incubation, 50 μl of Buffer A was added, then nucleic acids were extracted by phenol/chloroform, followed by isopropanol precipitation and DNase I digestion. The extracted RNA was tested for IFNβ induction. For reconstitution experiments, dialyzed proteins from ammonium sulfate precipitation or column fractions and heat supernatant (heat sup) were used in the reactions in lieu of S100. Heat sup was obtained by incubating HeLa S100 at 75 °C for 15 min followed by centrifugation at 20,000×g to collect the supernatant. For purification of small molecules from the heat sup, the reaction mixtures contain 40% dialyzed ammonium sulfate precipitates from S100, column fractions from heat sup, poly(dA-dT), ATP, MgCl2 and RNase inhibitor. For purification of Pol-III, the reaction mixtures contain column fractions from S100, UTP (0.2mM), poly(dA-dT), ATP, MgCl2 and RNase inhibitor.
Viral and bacterial infection
Raw264.7 cells were pre-treated with the Pol III inhibitor, ML-60218 (Calbiochem), for 10 hours and then infected with an engineered adenovirus type 5 (Ad-CMV-GFP from Vector Biolabs; this virus lacks the E1 and E3 genes) at 100 MOI, HSV-1 at 100 MOI, or Legionella pneumophila (serogroup 1 strain AA100, from ATCC) at 50 MOI for 8 hours. Cells were rinsed with PBS and lysed in Trizol for total RNA extraction.
Bacterial Replication Assay
Raw264.7 cells were pre-treated with ML-60218 in the presence or absence of mouse IFN-β (Sigma) for 10 hours and then infected with Legionella pneumophila at MOI = 50 for 24 hours. Cells were washed with PBS and then lysed in 0.1 % saponin for 5 min. Cell lysates were diluted with PBS by 40,000 Fold and then spread on buffered charcoal yeast extract (BCYE) agar plates (Opitz et al., 2006). After incubation at 37°C for 2 days, colonies were counted to determine colony-forming units (CFU).
Supplementary Material
Acknowledgments
We thank Drs. Xiang Chen and Xiao-Dong Li for assistance in preparing BMDM, Dr. Hon-Ren Huang (Harvard University) for technical advice, and Brian Skaug for critically reading the manuscript. This work was supported by grants from the NIH and the Welch Foundation (Z.J.C) and by the Chilton Foundation (J.M). Z.J.C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Cheng G, Zhong J, Chung J, Chisari FV. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci U S A. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Howe JG, Shu MD. Upstream basal promoter element important for exclusive RNA polymerase III transcription of the EBER 2 gene. Mol Cell Biol. 1993;13:2655–2665. doi: 10.1128/mcb.13.5.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Jaehning JA, Roeder RG. Transcription of specific adenovirus genes in isolated nuclei by exogenous RNA polymerases. J Biol Chem. 1977;252:8753–8761. [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, Preissner R, Slevogt H, N'Guessan PD, Eitel J, et al. Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem. 2006;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Rasmussen SB, Sorensen LN, Malmgaard L, Ank N, Baines JD, Chen ZJ, Paludan SR. Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J Virol. 2007;81:13315–13324. doi: 10.1128/JVI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. Embo J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Schroder K, Muruve DA, Tschopp J. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


