Abstract
We measured gene expression of paracrine regulators involved in adipocyte differentiation within the stromovascular fraction of abdominal subcutaneous adipose tissue from obese individuals with (n = 30) and without (n = 18) type 2 diabetes mellitus (T2DM). Despite similar adiposity by design, subjects with T2DM had larger adipocytes (0.92 ± 0.28 vs. 0.75 ± 0.17 μl, p < 0.05) than controls. Gene expression of the adipogenic marker aP2 was lower (0.35 ± 0.16 vs. 0.58 ± 0.27 arbitrary units, p < 0.05) whereas the expression of matricellular peptidase, MMP2 was higher (1.65 ± 0.17 vs. 1.27 ± 0.21, p = 0.02) in T2DM vs. controls. The gene expression levels between the aP2 and MMP2 were inversely correlated (r = −0.32, p = 0.03). We conclude that early steps of adipogenesis may be impaired in T2DM independently of obesity due, in part, to an upregulation of the MMP2 transcription.
Keywords: Obesity, Type 2 diabetes, Adipocyte, Fatty acid binding protein aP2, Matrix metalloproteinase
Obesity is associated with an extensive reorganization of adipose tissue including changes in adipogenesis, angiogenesis, extracellular matrix composition, and macrophage content. More specifically, the adipocyte differentiation in subcutaneous adipose tissue decreases with obesity [1]. Based on gene expression data, we suggest that obese individuals with type 2 diabetes (T2DM) have inhibited adipocyte differentiation when compared to obese non-diabetic controls [2]. However, the molecular mechanism of the impaired preadipocyte differentiation in T2DM is not known. It is well established that pro-inflammatory cytokines, chemokines, and proteolytic factors modulate adipocyte differentiation by paracrine mechanisms. Such factors are produced primarily by distinct cellular populations that comprise the stromovascular fraction (SVF) of adipose tissue (preadipocytes, mesenchymal stem cells, endothelial cells, smooth muscle cells, and infiltrating macrophages) and, to a lesser extent, by adipocytes [3,4]. Modified sensitivity of preadipocytes to adipogenic growth factors or hormones seems to also play a role [5]. In the present study, we asked whether the genes of selected pro-inflammatory and matricellular proteins, hormones, and/or molecules from selected signaling pathways are differentially expressed in the SVF of subcutaneous abdominal adipose tissue in obese diabetic vs. obese non-diabetic individuals. The selected genes were interleukin-6 (IL-6); tumor necrosis factor α (TNFα); CD68 (a member of the scavenger receptor superfamily expressed in macrophages and mast cells); monocyte chemotactic protein-1 (MCP1); macrophage migration inhibitory factor (MIF); macrophage inflammatory protein-1-α (MIP1A); stress-activated protein kinase JNK-1; IKKβ (an IκB kinase that activates the nuclear factor NFκB, a regulator of cytokine and chemokine gene transcription); matrix metalloproteinase-2 (MMP2, a gelatinase A or a type IV collagenase); and 11 β-hydroxysteroid dehydrogenase type 1 (11β-HSD1; enzyme that converts cortisone to cortisol). Furthermore, we hypothesized that these differentially expressed genes would relate to the expression of adipogenic markers, such as human adipocyte fatty acid binding protein (aP2).
Research design and methods
Subjects
Thirty Caucasian individuals with T2DM (fasting blood glucose < 180 mg/dl) with no history of insulin or thiazolidinediones treatment and 18 individuals without T2DM were recruited in research centers in Baton Rouge, Pittsburgh, and New York. All subjects were aged 45–75, and were overweight or obese (BMI 27–40 kg/m2). Both groups were balanced for sex representation. The T2DM subjects were participants in the multi-center Look AHEAD randomized clinical trial [6]. The protocol was approved by each center’s Institutional Review Board.
Experimental protocol
Total percent body fat and the proportion of visceral to subcutaneous abdominal fat were measured by dual energy X-ray absorptiometry (DXA; Hologics QDR 4500A) and single-slice computed tomography of the abdomen, respectively. Approximately 250–350 mg of subcutaneous adipose tissue was obtained by needle biopsy as previously described [2]. A 50–100 mg sample was frozen for RNA isolation, ~50 mg was placed in osmium tetraoxide for adipocyte sizing, and the remaining adipose tissue was enzymatically digested to isolate SV cells. Insulin sensitivity was assessed by a 3-h hyperinsulinemic–euglycemic clamp with a primed-constant insulin infusion at 80 mU/m2/min. After adjustment for differences in plasma glucose and insulin concentrations at steady state, the glucose disposal rate was normalized for fat-free mass.
Average adipocytes size
The adipocyte diameter was measured using a Multisizer-3 (Beckman Coulter, Fullerton, CA, USA) as previously described [7], and the average adipocyte volume (μL) of cells with diameters >22 μm was calculated.
Isolation of the stromal vascular fraction
The tissue was minced and digested with collagenase. Following centrifugation, the adipocyte-containing supernatant was removed and the stromovascular cells localized to the pellet were frozen for RNA isolation.
Semi-quantitative real time PCR [(q)RT-PCR]
Total RNA was extracted using Trizol as per the manufacturer’s instructions (Invitrogen, Carlsbad, CA), purified using an RNeasy Mini kit (Qiagen Inc., Valencia, CA) and measured spectrophotometrically at 260/280 nm. Semi-quantitative real time (q)RT-PCR reactions were performed as previously described [2] on an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA). A list of measured transcripts is provided in Table 1 and of the primers and probes used in Supplementary Table 2. To compensate for RNA input variation and the efficiency of reverse transcription, expression data were normalized by dividing the amount of target genes by the amount of cyclophilin B mRNA and expressed as arbitrary units (A.U.). Each sample was run in duplicate and mean values were used for the expression of each gene.
Table 1.
Selected characteristics of the subjects and expression of selected genes
| Obese T2DM (15 men, 15 women) | Obese controls (9 men, 9 women) | |
|---|---|---|
| Anthropometric and metabolic characteristics | ||
| Age (y) | 59 ± 9 | 55 ± 9 |
| Weight (kg) | 96 ± 10 | 97 ± 14 |
| BMI (kg/m2) | 33.5 ± 2.5 | 32.1 ± 1.6 |
| Body fat (%) | 36 ± 7 | 37 ± 8 |
| Ratio VAT/SAT | 0.96 ± 0.58 | 0.83 ± 0.55 |
| Adipocyte size (μL) | 0.92 ± 0.28a | 0.75 ± 0.17b |
| Fasting glucose (mg/dL) | 146.2 ± 25.9a | 99.9 ± 7.7c |
| Fasting insulin (μU/ml)d | 12.5 ± 0.2 (5.5,25.1)a | 9.4 ± 0.2 (3.8,40.5)b |
| HbA1c (%) | 6.8 ± 1.0a | 5.3 ± 0.8c |
| Glucose disposal (mg/min/kg FFM) | 5.5 ± 2.0a | 10.0 ± 3.6c |
| Gene expression in stromovascular cells (A.U.) | ||
| aP2 | 0.35 ± 0.16a | 0.58 ± 0.27b |
| JNK-1 | 1.20 ± 0.68 | 1.09 ± 0.73 |
| MIFd | 1.5 ± 0.2 (0.7,3.7) | 1.3 ± 0.2 (0.7,3.7) |
| MMP2d | 1.7 ± 0.2 (0.7,4.3)a | 1.3 ± 0.2 (0.4,3.2)b |
| IL-6d | 0.2 ± 0.4 (0.1,1.1) | 0.2 ± 0.5 (0.1,2.1) |
| CD68d | 2.3 ± 0.2 (0.8,6.0) | 2.6 ± 0.2 (1.1,9.2) |
| MCP1d | 1.3 ± 0.3 (0.3,4.2) | 1.4 ± 0.3 (0.6,8.9) |
| 11β-HSD1 | 1.71 ± 0.51 | 1.79 ± 0.83 |
| IKKβd | 1.2 ± 0.2 (0.5,2.7) | 1.0 ± 0.2 (0.4,3.2) |
| MIP1A | 2.07 ± 1.74 | 1.94 ± 1.41 |
| TNFαd | 4.8 ± 0.8 (0.002,35.5) | 5.2 ± 0.7 (0.1,48.1) |
| Cyclophilin B | 2.59 ± 1.08 | 2.36 ± 1.45 |
| Gene expression in adipose tissue (A.U.) | ||
| Pref-1 | 1.35 ± 1.23 | 1.37 ± 0.88 |
Data are means ± SD or geometric means ± SD (range) as appropriate;
p < 0.05;
p < 0.0001;
log transformed; A.U., arbitrary units.
Statistical analyses
Analyses were performed using SAS (version 9.1.3, SAS Institute). Data were checked for normal distribution and, when necessary, logarithmically transformed and presented as either mean (SD) or geometric mean (SD and range). Differences between diabetic and non-diabetic subjects were analyzed using a two-way ANOVA with fixed effects of the groups, sex, and their interaction. A p < 0.05 was considered statistically significant. Relationships between metabolic variables and gene expression were assessed by simple linear regression analysis.
Results
Subjects
Subject characteristics are provided in Table 1. The subjects were clamped at plasma glucose concentrations of ~~100 mg/dL and plasma insulin concentrations of ~170 μU/ml. As expected, glucose disposal rates during the clamp were lower and hemoglobin A1c (HbA1c) was higher in diabetic than in non-diabetic individuals. There was no difference in the adiposity level between the groups, but the average adipocyte size was significantly larger in diabetics (0.92 ± 0.28 vs. 0.75 ± 0.17 μl; p < 0.05).
Gene expression (Table 1)
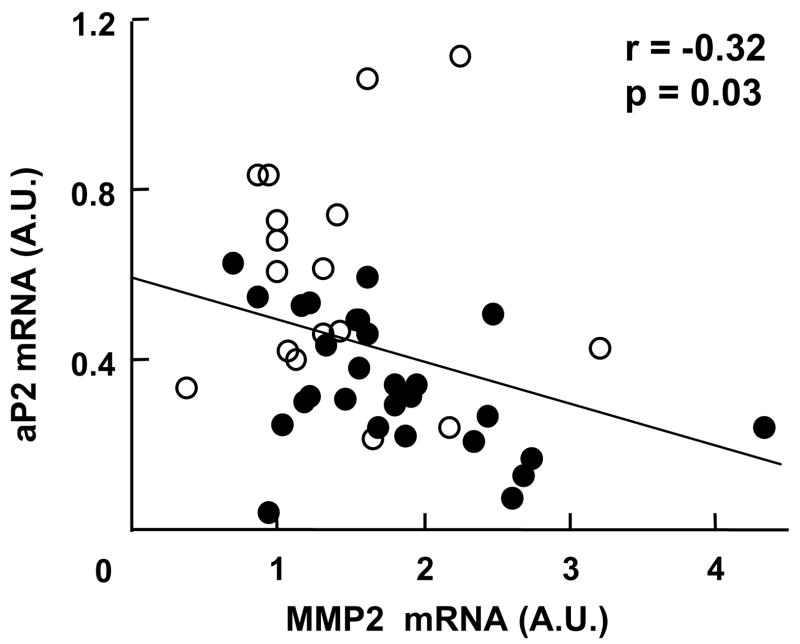
Both MMP2 mRNA and MIF expression levels were higher in the SVF of adipose tissue in diabetic than in non-diabetic individuals, (p < 0.02 and p = 0.08, respectively). In contrast, the gene expression of aP2 was lower in subjects with T2DM (p < 0.05). The gene expression of aP2 correlated negatively with HbA1c (r = −0.31, p < 0.05), whereas that of MMP2 correlated positively (r = 0.35, p = 0.02). We observed a negative association between aP2 and MMP2 mRNA levels (Fig. 1). Of note, additional measurement of the levels of the gene encoding the preadipocyte-specific marker preadipocyte factor-1 (Pref-1) in adipose tissue was not different between the groups. Finally, the expression of the remaining genes in the SVF was not different between groups.
Fig. 1.
Relationship between aP2 and MMP2 mRNA gene expression levels in the stromovascular cells from subcutaneous abdominal adipose tissue in obese individuals with T2DM (solid circles) and obese controls (open circles).
Discussion
Although evidence supports an impairment of adipocyte differentiation in obese individuals with T2DM independently of adiposity [2], no mechanism has been proposed. The two major hypotheses to explain such impairment include: (1) reduced sensitivity of preadipocytes to adipogenic hormones (i.e. glucocorticoids) and (2) increased secretion of cytokines, chemokines, and ECM components by the SV cells of adipose tissue. In an attempt to identify factors that play a role in the impairment of adipogenesis in T2DM, we studied mRNA levels of selected proteins in the SVF of subcutaneous abdominal adipose tissue in obese diabetic and obese non-diabetic individuals.
We found lower levels of aP2 mRNA in subjects with T2DM as compared to obese controls. Using fluorescent staining of freshly isolated SV cells from adipose tissue for aP2, lipids, and the macrophage marker CD68, we have previously identified that aP2 is expressed in preadipocytes, immature adipocytes containing small amount of multi-locular lipid droplets, and in a small proportion of macrophages [8]. However, in the present study, the quantification of these cellular types was not performed and thus definitive conclusion cannot be drawn. Since the gene expression of inflammatory markers TNFα and IL-6 as well as of molecules pertaining to their regulatory signaling pathways (JNK and NFκB) was similar between the groups, one cannot imply an increased amount of macrophages in the subcutaneous adipose tissue of obese T2DM vs. obese controls. Additional measurement of the expression of the preadipocyte-specific marker Pref-1 in adipose tissue showed similar levels in both groups suggesting a comparable number of preadipocytes. Together, the above observations foster the interpretation that the reduced levels of aP2 mRNA reflect either lower number of small adipocytes or decreased expression of aP2 per cell in the small adipocytes present in the SVF suggesting impaired preadipocyte differentiation. It is possible that the adipocyte hypertrophy found in T2DM may be linked to the impaired preadipocyte differentiation.
A second finding of this study is the higher MMP2 gene expression in T2DM. Recently, Derosa et al. [9] reported higher plasma levels of MMP2 in T2DM compared to healthy controls reflecting generalized abnormality of the extracellular matrix metabolism. An induction of the MMP2 transcription in the adipose tissue of genetically obese mice has also been reported [10] but our finding of higher expression in T2DM, compared to obese non-diabetic subjects, is novel. MMP2 is an endopeptidase that degrades the basement membrane surrounding adipocytes and thus may facilitate hypertrophic development of adipocytes and formation of adipocyte clusters [11]. Recent studies demonstrate a direct but inconsistent effect of MMP2 on early phases of murine preadipocyte differentiation [4,12]. The negative correlation between MMP2 and aP2 gene expression in SV cells from subcutaneous adipose tissue leads us to speculate that elevated MMP2 transcript may play an inhibitory role early in the differentiation program. Alternative hypothesis is that impaired adipocyte differentiation may lead to an upregulation of MMP2 transcription. As usual, it is however impossible to imply causality from correlation analyses. Additional testing using MMP2 and its blockers in primary human preadipocyte cultures will be required.
In conclusion, the present study suggests: (1) impaired adipogenesis in T2DM and (2) a potential role of increased MMP2 transcript level in the impaired adipogenesis in T2DM.
Supplementary Material
Acknowledgments
We thank the rest of members of the Look AHEAD adipose research groups for their contribution to this project. The study was funded by DK60412 (E.R.) but also supported by the University of Pittsburgh Obesity & Nutrition Research Center (P3-DK46204), the University of Pittsburgh General Clinical Research Center (MO1-RR000056), the Pennington Biomedical Research Center Clinical Nutrition Research Unit (P30 DK072476), and the Columbia University Diabetes and Endocrinology Research Center (P30 DK63608).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2007.12.180.
References
- 1.van Harmelen V, Skurk T, Rohrig K, Lee YM, Halbleib M, Aprath-Husmann I, Hauner H. Int J Obes Relat Metab Disord. 2003;27:889–895. doi: 10.1038/sj.ijo.0802314. [DOI] [PubMed] [Google Scholar]
- 2.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E. Obesity (Silver Spring) 2006;14:1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fain JN, Madan AK, Hiler ML, Cheema P, Bahouth SW. Endocrinology. 2004;145:2273–2282. doi: 10.1210/en.2003-1336. [DOI] [PubMed] [Google Scholar]
- 4.Maquoi E, Munaut C, Colige A, Collen D, Lijnen HR. Diabetes. 2002;51:1093–1101. doi: 10.2337/diabetes.51.4.1093. [DOI] [PubMed] [Google Scholar]
- 5.Masuzaki H, Flier JS. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3:255–262. doi: 10.2174/1568008033340135. [DOI] [PubMed] [Google Scholar]
- 6.Ryan DH, Espeland MA, Foster GD, Haffner SM, Hubbard VS, Johnson KC, Kahn SE, Knowler WC, Yanovski SZ. Control Clin Trials. 2003;24:610–628. doi: 10.1016/s0197-2456(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 7.Harris RB, Martin RJ. Am J Physiol. 1986;250:R276–R286. doi: 10.1152/ajpregu.1986.250.2.R276. [DOI] [PubMed] [Google Scholar]
- 8.Tchoukalova YD, Sarr MG, Jensen MD. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1132–R1140. doi: 10.1152/ajpregu.00337.2004. [DOI] [PubMed] [Google Scholar]
- 9.Derosa G, D’Angelo A, Tinelli C, Devangelio E, Consoli A, Miccoli R, Penno G, Del Prato S, Paniga S, Cicero AF. Diabetes Metab. 2007;33:129–134. doi: 10.1016/j.diabet.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. J Biol Chem. 2003;278:11888–11896. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 11.Brown LM, Fox HL, Hazen SA, LaNoue KF, Rannels SR, Lynch CJ. Am J Physiol. 1997;272:C937–C949. doi: 10.1152/ajpcell.1997.272.3.C937. [DOI] [PubMed] [Google Scholar]
- 12.Alexander CM, Selvarajan S, Mudgett J, Werb Z. J Cell Biol. 2001;152:693–703. doi: 10.1083/jcb.152.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.