Abstract
A novel autosomal recessive nonsyndromic hearing impairment (ARNSHI) locus, DFNB71 was localized to 8p22-21.3. In order to map the locus, a whole genome scan was carried out using DNA samples from a consanguineous seven-generational Pakistani family with profound prelingual ARNSHI. A maximum multipoint LOD score of 4.2 occurred at marker D8S261. The DFNB71 region of homozygosity and 3-unit support interval is flanked by markers D8S1130 and D8S1786. This region has a genetic distance of 19.1 cM and contains 10.6 Mb of sequence.
Keywords: 8p22-21.3, autosomal recessive non-syndromic hearing impairment, DFNB71, Pakistan
Introduction
The incidence of profound hearing impairment (HI) is estimated at 1–2 per 1,000 births and ~60% of the congenital cases can be attributed to genetic factors (1, 2). Of the genetic cases, ~30% are syndromic with the remaining cases display non-syndromic HI (NSHI) where the HI phenotype is usually sensorineural. A large number of genes attributable to NSHI have been mapped and identified. For autosomal recessive (AR) NSHI > 60 genes have been localized and 28 genes have been identified (3). Although this magnitude of progress is remarkable and significant advances have been made, many more NSHI loci remain to be discovered. In this article, a new locus, DFNB71, is shown to segregate in a seven-generational consanguineous Pakistani family with ARNSHI. The hearing impaired family members in this pedigree present with profound prelingual HI. The DFNB71 locus maps to a 10.6 Mb region on chromosome 8p22-21.3.
Materials and Methods
Family History
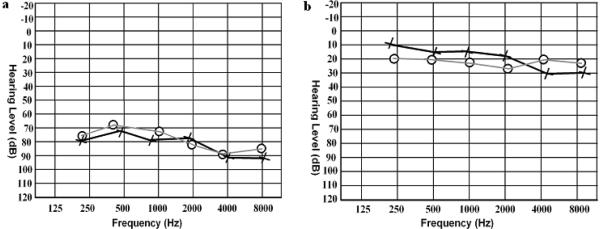
With the prior approval from the Institutional Review Board of Quaid-I-Azam University, Islamabad, Pakistan and Baylor College of Medicine and Affiliated Hospitals the study was initiated and informed consent was obtained from all family members who participated in the study. Pedigree 4216 was constructed on the basis of information collected from the family members (Figure 1). Hearing impaired and non-hearing impaired family members underwent audiometric testing. Hearing impaired family members have prelingual profound HI which affects all frequencies. Figure 2 displays the audiogram for a hearing impaired family member (individual 34) that displays bilateral, profound HI that affects all frequencies and the audiogram for a family member without hearing impairment (individual 38) whose hearing is within the normal range (Figure 2). The hearing impaired members of pedigree 4216 use sign language as their primary means of communication. The family members with hearing impairment also underwent a physical examination for defects in ear morphology, vision, mental retardation and other clinical features that could indicate that the HI is syndromic. There was no evidence in this kindred that the HI belongs to a syndrome or that there is gross vestibular involvement.
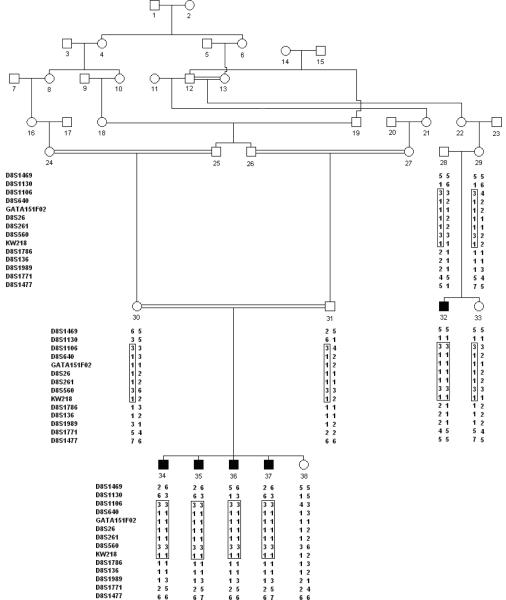
Figure 1.
Drawing of pedigree 4216. Black symbols represent individuals with hearing impairment and clear symbols represent unaffected individuals. Haplotypes are shown beneath each genotyped individual. The DFNB71 haplotype is enclosed in a rectangle.
Figure 2.
Audiograms for a hearing impaired (panel a) and unimpaired family member (panel b) from pedigree 4216. Circles and crosses represent air conduction for the right and left ear, respectively. Panel (a) displays the audiogram for individual 34 at 24 years of age and panel (b) displays the audiogram from individual 38 at 17 years of age.
Genotyping
Venous blood samples were obtained from 11 family members including 5 individuals with profound prelingual HI. Genomic DNA was extracted from whole blood following a standard protocol (4) A genome scan was carried out on all DNA samples at the Center for Inherited Disease Research (CIDR). A total of 396 fluorescently labeled short tandem repeat (STR) markers were genotyped. These markers are spaced ~10 cM apart and are located on the 22 autosomes and the X and Y chromosomes. For fine mapping, microsatellite markers were PCR amplified according to standard procedure in a total volume of 25 μl with 40 ng genomic DNA, 240 nM dNTP, and 1 unit of Taq DNA Polymerase (Fermentas Life Sciences, Burlington, ON, Canada) in GeneAmp® PCR System 9700 (Applied Biosystems, Applera Corp., Foster City, CA, USA). PCR products were resolved on 8% non-denaturing polyacrylamide gel and genotypes were assigned by visual inspection.
Linkage Analysis
PEDCHECK(5) was used to evaluate the genotype data for Mendelian inconsistency and MERLIN (6) was used to assess the data for occurrence of unlikely genotypes, which are most likely due to genotyping error. Two-point linkage analysis was carried out using the MLINK program of the FASTLINK computer package (7). LOD scores were calculated using a fully penetrant autosomal recessive model with a disease allele frequency of 0.001. Multipoint linkage analysis was performed using SUPERLINK (8, 9) and SIMWALK2 (10, 11) utilizing map distances from the Rutgers combined linkage-physical map of the human genome (12). The allele frequencies for the genome scan markers were estimated from the data by means of both observed and reconstructed genotypes of founders from this pedigree and 35 additional families from Pakistan that underwent a genome scan at CIDR at the same time. In order to fine map the DFNB71 loci 12 additional markers were genotyped in the 8p region of which 8 were informative (D8S640, GATA151F02, D8S26, D8S261, KW218, D8S1786, D8S136 and D8S1989). Two-point linkage analysis was carried for the fine mapping markers and multipoint linkage analysis was performed again using both the genome scan and fine mapping markers. Equal allele frequencies were used for the fine mapping markers, since they were only genotyped in family 4216. A sensitivity analysis was carried out by performing multipoint linkage analysis and varying the allele frequency for the alleles segregating with the HI phenotype from 0.2 to 0.6 for the fine mapping markers, because analyses carried out with potentially incorrect allele frequencies (e.g. equal allele frequencies) can increase type I error (13). Haplotypes in the 8p22-21.3 region were reconstructed using SIMWALK2.
Candidate Gene Sequencing
Primers were designed for the exons of the hairless (HR) gene (MIM 602302) isoform a (NM_005144) and isoform b (NM_018411), the platelet-derived growth factor receptor-like (PDGFL) gene (MIM 604584) and the tumor suppressor candidate 3 (TUSC3) gene (MIM 601385) isoform a (NM_006765) and isoform b (NM_178234) using Primer3 software (14). DNA from one unaffected and two HI pedigree members were diluted to 5 μg/nl, amplified by PCR under standard conditions and purified with ExoSAP-IT ® (USB Corp., Cleveland, OH, USA). Sequencing was performed with the BigDye ® Terminator v3.1 Cycle Sequencing Kit together with an Applied Biosystems 3700 DNA Analyzer (Applera Corp., Foster City). Sequence variants were identified via Sequencher™ Version 4.1.4 software (Gene Codes Corp., Ann Arbor, MI, USA).
Results
Analysis of the genome scan markers revealed a maximum multipoint LOD score of 3.1 which occurred at marker D8S1106. The region of homozygosity for the genome scan markers is flanked by markers D8S1130 and D8S1771 which spans a 23.4 cM region according to the Rutgers combined linkage-physical map of the human genome (12) and contains 13.6 Mb of sequence (15). Analysis of the fine mapping markers produced a maximum two-point LOD score of 3.04 (θ=0.00) at marker D8S640 (Table 1) and the maximum multipoint LOD score (4.2) occurred at marker D8S261. The 3-unit support interval spanned from markers D8S1130 to D8S1786. Haplotypes were constructed to determine the critical recombination events. The 8p22-21.3 haplotype for pedigree 4216 is presented in Figure 1. All HI family members are homozygous for a common haplotype, which is also flanked by markers D8S1106 and D8S1786, and agrees with the 3-unit support interval (Table 1). This region is 19.1 cM according to the Rutgers combined linkage-physical map of the human genome and contains 10.6 Mb of sequence. Sensitivity analysis revealed that varying the allele frequencies caused the LOD score to fluctuate. But, a LOD score of greater than 3.3 was maintained within the region of homozygosity for all allele frequencies tested. Sequencing the exons and promoter regions of three candidate genes, TUSC3 (MIM: 601385), PDGFRL (MIM: 604584) and HR (MIM: 602302) using DNA from two HI pedigree members did not reveal any potentially functional sequence variants.
Table 1.
Two-point LOD score results between DFNB71 and chromosome 8 markers.
|
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genetic map | Physical map | Two Point LOD score at θ = | |||||||
| Marker | PositionI | PositionII | 0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 |
| D8S1469 | 19.38 | 9,127,124 | − ∞ | −2.76 | −0.87 | −0.22 | 0.15 | 0.13 | 0.01 |
| D8S1130 | 22.31 | 11,871,501 | − ∞ | −2.53 | −0.65 | −0.01 | 0.36 | 0.31 | 0.13 |
| D8S1106 | 24.76 | 12,880,323 | 1.58 | 1.54 | 1.36 | 1.15 | 0.77 | 0.43 | 0.16 |
| D8S640 | 24.76 | 12,979,668 | 3.04 | 2.96 | 2.65 | 2.27 | 1.54 | 0.87 | 0.32 |
| GATA151F02 | 26.35 | 13,296,758 | 1.16 | 1.12 | 0.99 | 0.83 | 0.55 | 0.30 | 0.10 |
| D8S26 | 27.04 | 13,750,569 | 1.69 | 1.65 | 1.50 | 1.30 | 0.92 | 0.56 | 0.23 |
| D8S261 | 32.78 | 17,870,469 | 1.69 | 1.65 | 1.50 | 1.30 | 0.92 | 0.56 | 0.23 |
| D8S560 | 39.68 | 21,646,126 | 1.76 | 1.72 | 1.56 | 1.36 | 0.97 | 0.60 | 0.26 |
| KW218 | 40.71 | 22,024,717 | 2.38 | 2.32 | 2.08 | 1.78 | 1.21 | 0.68 | 0.21 |
| D8S1786 | 41.41 | 22,489,213 | 1.62 | 1.59 | 1.46 | 1.29 | 0.94 | 0.59 | 0.25 |
| D8S136 | 41.41 | 22,489,339 | 1.39 | 1.37 | 1.25 | 1.11 | 0.81 | 0.51 | 0.21 |
| D8S1989 | 45.23 | 24,697,056 | − ∞ | −0.51 | 0.63 | 0.91 | 0.85 | 0.53 | 0.16 |
| D8S1771 | 45.71 | 25,497,030 | −2.46 | −0.29 | 0.31 | 0.48 | 0.47 | 0.31 | 0.10 |
| D8S1477 | 55.27 | 32,186,709 | − ∞ | −4.14 | −2.11 | −1.28 | −0.56 | −0.23 | −0.06 |
Markers in italic flank the region of homozygosity and the 3-unit support interval. Genome scan markers are displayed in bold.
The March 2006 human reference sequence (NCBI Build 36.1, Kent et al. 2002)
The Rutgers Combined Linkage-Physical Map of The Human Genome Build 36 (Matise et al. 2007)
Discussion
There are 54 known genes within the 10.6 Mb region of the DFNB71 locus. Expression information on three of these genes indicated that they may be important in hearing impairment physiology. TUSC3 was originally associated with prostate cancer (16) but is of interest as a candidate gene for HI due to its expression in the inner ear (17–19). The platelet-derived growth factor receptor polypeptides alpha, beta, c and d are expressed in the inner ear (17–19), although none of these genes reside in the DFNB71 interval, a related gene the platelet-derived growth factor receptor like (PDGFRL) gene is within the DFNB71 region. HR is a homolog of the mouse HR which displays expression in the mouse organ of corti (20). Sequencing of the exons and promoter regions of TUSC3, PDGRFL and HR and their isoforms did not reveal any potential functional variants and therefore their involvement in the etiology of HI due to DFNB71 is not supported. Eventual identification the gene responsible for DFNB71 will enhance our knowledge of the molecular mechanisms behind hearing and HI.
Acknowledgements
We wish to thank the family members for their invaluable participation and cooperation. The work was funded by Higher Education Commission (HEC), Government of Pakistan and the NIH National Institute of Deafness and other Communication Disorders grant DC03594. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403. Electronic-database information
The URLs for data presented herein are as follows:
The Hearing Research Group at Brigham and Women's Hospital (2006a) Human Cochlear cDNA library and EST database: http://www.brighamandwomens.org/bwh_hearing/human-cochlear-ests.aspx.
The Hearing Research Group at Brigham and Women's Hospital (2006b) Mouse Organ of Corti cDNA Array: http://www.brighamandwomens.org/bwh_hearing/InnerEarcDNAArrays.aspx.
Van Camp G, Smith RJH (2008) Hereditary Hearing Loss Homepage: http://webh01.ua.ac.be/hhh/, August 2008
UCSC Human Genome Project Working Draft, March 2006 assembly, http://genome.cse.ucsc.edu/ (the physical positions of STR loci). Rutgers Combined Linkage-Physical Map of the Human Genome http://compgen.rutgers.edu/maps/index.shtml
References
- 1.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 2.Gorlin RJ, Toriello HV, Cohen MM. Hereditary hearing loss and its syndromes. Oxford University Press; New York: 1994. [Google Scholar]
- 3.Van Camp G, Smith RJH. Hereditary Hearing Loss. 2008. Homepage: http://webh01.ua.ac.be/hhh/ [Google Scholar]
- 4.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17(20):8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 7.Cottingham RW, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53(1):252–63. [PMC free article] [PubMed] [Google Scholar]
- 8.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics (Oxford, England) 2002;18(Suppl 1):S189–98. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 9.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. American journal of human genetics. 2006;78(6):922–35. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weeks DE, Sobel E, O'Connell JR, Lange K. Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet. 1995;56(6):1506–7. [PMC free article] [PubMed] [Google Scholar]
- 11.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58(6):1323–37. [PMC free article] [PubMed] [Google Scholar]
- 12.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17(12):1783–6. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freimer NB, Sandkuijl LA, Blower SM. Incorrect specification of marker allele frequencies: effects on linkage analysis. Am J Hum Genet. 1993;52(6):1102–10. [PMC free article] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. Humana Press; Totowa, N.J.: 2000. pp. xi–500. [DOI] [PubMed] [Google Scholar]
- 15.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bova GS, MacGrogan D, Levy A, Pin SS, Bookstein R, Isaacs WB. Physical Mapping of Chromosome 8p22 Markers and Their Homozygous Deletion in a Metastatic Prostate Cancer. Genomics. 1996;35(1):46–54. doi: 10.1006/geno.1996.0321. [DOI] [PubMed] [Google Scholar]
- 17.Masmoudi S, Tlili A, Majava M, Ghorbel AM, Chardenoux S, Lemainque A, et al. Mapping of a new autosomal recessive nonsyndromic hearing loss locus (DFNB32) to chromosome 1p13.3–22.1. Eur J Hum Genet. 2003;11(2):185–8. doi: 10.1038/sj.ejhg.5200934. [DOI] [PubMed] [Google Scholar]
- 18.Robertson NG, Khetarpal U, Gutierrez-Espeleta GA, Bieber FR, Morton CC. Isolation of novel and known genes from a human fetal cochlear cDNA library using subtractive hybridization and differential screening. Genomics. 1994;23(1):42–50. doi: 10.1006/geno.1994.1457. [DOI] [PubMed] [Google Scholar]
- 19.Hearing Research Group at Brigham and Women's Hospital Human Cochlear cDNA library and EST database. 2006. http://www.brighamandwomens.org/bwh_hearing/human-cochlear-ests.aspx.
- 20.Hearing Research Group at Brigham and Women's Hospital Mouse Organ of Corti cDNA Array. 2006. http://www.brighamandwomens.org/bwh_hearing/InnerEarcDNAArrays.aspx.