Abstract
Increased energy demands to support lactation, coupled with lowered feed intake capacity results in negative energy balance (NEB) and is typically characterized by extensive mobilization of body energy reserves in the early postpartum dairy cow. The catabolism of stored lipid leads to an increase in the systemic concentrations of nonesterified fatty acids (NEFA) and β-hydroxy butyrate (BHB). Oxidation of NEFA in the liver result in the increased production of reactive oxygen species and the onset of oxidative stress and can lead to disruption of normal metabolism and physiology. The immune system is depressed in the peripartum period and early lactation and dairy cows are therefore more vulnerable to bacterial infections causing mastitis and or endometritis at this time. A bovine Affymetrix oligonucleotide array was used to determine global gene expression in the spleen of dairy cows in the early postpartum period. Spleen tissue was removed post mortem from five severe NEB (SNEB) and five medium NEB (MNEB) cows 15 days postpartum. SNEB increased systemic concentrations of NEFA and BHB, and white blood cell and lymphocyte numbers were decreased in SNEB animals. A total of 545 genes were altered by SNEB. Network analysis using Ingenuity Pathway Analysis revealed that SNEB was associated with NRF2-mediated oxidative stress, mitochondrial dysfunction, endoplasmic reticulum stress, natural killer cell signaling, p53 signaling, downregulation of IL-15, BCL-2, and IFN-γ; upregulation of BAX and CHOP and increased apoptosis with a potential negative impact on innate and adaptive immunity.
Keywords: nonesterified fatty acids, butyrate, spleen
successful genetic selection programs over the past 30–40 yr together with improved nutrition has resulted in the modern dairy cow, which is biologically efficient at producing large volumes of milk exceeding 8,500 l per 305-day lactation. This increase in milk production has been accompanied, however, by a corresponding steady decline in fertility, with conception to first service in many dairy cow herds now <40% (65). Increased milk production also causes severe metabolic demands for increased energy resulting in extensive mobilization of body fat reserves in early lactation. This period of negative energy balance (NEB) can persist for many weeks, and fatty acid catabolism can result in the accumulation of triglycerides in the liver and increased systemic concentrations of lipid metabolites in the blood resulting in a period of oxidative stress.
This early postpartum period is also associated with a dramatic increase in the rate of liver blood flow and metabolism that can compromise liver function and result in production diseases such as ketosis and fatty liver (30, 73). Fatty liver can affect up to 50% of cows in early lactation (36) and can impair liver function such as synthesis and biotransformation of metabolites and proteins and the elimination of toxic waste products (6).
The catabolism of fatty acids results in a number of metabolic changes. Nonesterified fatty acids (NEFAs) and ketone bodies including β-hydroxy butyrate (BHB) are produced by the liver as the fatty acids are metabolized and their systemic concentrations increase in proportion to the degree of fat mobilization. Short-chain volatile fatty acids (acetate, propionate, and butyrate), nutrients especially critical to ruminant mammals, are also formed during the ruminal fermentation of the dietary fiber in the gastrointestinal tract of mammalian species and are directly absorbed at the site of production (8).
Oxidation of NEFAs in the liver result in the increased production of reactive oxygen species (ROS), decreased paraoxonase activity, and the onset of oxidative stress (77). This may be linked to depression of the immune system in early lactation (38, 39, 54, 80), resulting in dairy cows becoming more vulnerable to bacterial infections at this time.
Cows with high serum NEFA prepartum had an increased incidence of mastitis and retained placenta postpartum (15) and neutrophil and lymphocyte function has been shown to decrease following calving (39, 64). In cattle, neutrophil CD18 expression is highest at calving but decreases thereafter, whereas neutrophil CD62L expression decreases gradually for 2 wk before parturition and then markedly decreases at calving (42). These changes in neutrophil CD18 and CD62L expression may contribute to increased susceptibility to mastitis and other diseases. Populations of blood leukocytes expressing MHC class II are highest before calving but decrease immediately after parturition (79), possibly contributing to periparturient disease susceptibility. Cows with fatty liver also have higher blood concentrations of the cytokine TNF-α before calving and higher concentrations of the acute phase protein serum amyloid A and haptoglobin (1) after calving.
The spleen is an important component part of the hematopoietic system and is one of the largest secondary lymphoid organs after the bone marrow and the thymus gland. In contrast to the lymph nodes, it is blood not lymph that flows through the spleen, and its chief functions are the production of mature lymphocytes, the probable formation of antibodies, and the destruction of worn-out red blood cells (RBC) (57).
The objective of this study was to 1) compare global patterns of gene expression in spleen tissue in lactating dairy cows managed to undergo moderate or severe NEB during the early postpartum period and 2) to determine the effect of NEB on stress and immune pathways.
MATERIALS AND METHODS
Animals.
All procedures were carried out under license in accordance with the European Community Directive 86-609-EC. The animal model has been described previously (20). Briefly, multiparous Holstein-Friesian cows (n = 24) were blocked 2 wk prior to expected calving according to parity, body condition score, and previous lactation yield (average lactation 6,477 ± 354 kg) and randomly allocated to mild (MNEB) (n = 12) or severe (SNEB) (n = 12) groups. MNEB cows were fed ad libitum grass silage and 8 kg/day concentrates and milked once daily; SNEB cows were fed 25 kg/day silage and 4 kg/day concentrate and milked three times daily. The chemical composition of silage and concentrate offered [as previously described (58)] was the same across treatment groups. Daily measurements of milk yield, milk composition, dry matter intake, body weight, and dietary energy intake were used to calculate energy balance (EB), based on the French net energy for lactation system. Net EB was calculated as unité fourragére lait (UFL) day 1, in which 1 UFL is the net energy for lactation equivalent of 1 kg of standard air-dry barley as described previously (33). EB data are presented as UFL/day. Measurements of body condition score and EB were used to select six cows from each group for slaughter between days 11 and 16 of lactation, which showed extremes in EB.
Tissue collection.
At slaughter the entire spleen was removed and weighed, and samples weighing ∼1 g were dissected, rinsed in RNase-free phosphate buffer, snap frozen in liquid nitrogen, stored for ∼4 h in dry ice, and subsequently stored at −80°C.
Blood sampling and metabolite assays.
Unclotted (EDTA-treated) whole blood samples were collected on the day of slaughter by jugular venipuncture for hematological analysis. RBC number, white blood cell (WBC) number, granulocyte monocyte and lymphocyte numbers, packed cell volume, hemoglobin concentration, mean corpuscular volume, mean corpuscular hemoglobin concentration and platelet numbers were determined with an automated cell counter (Celltac MEK-6108K; Nihon-Kohdon, Tokyo, Japan) within 6 h of blood sampling.
RNA extraction and quality analysis.
Total RNA was prepared from 100–200 mg of fragmented frozen spleen tissue using the TRIzol reagent (Sigma-Aldrich Chemical, Dorset, UK). Tissue samples were homogenized in 3 ml of TRIzol reagent and chloroform and subsequently precipitated using isopropanol (Sigma-Aldrich). RNA samples were stored at −80°C. Twenty micrograms of total RNA from each sample was treated for genomic DNA contamination with the RNase-free DNase set (QIAGEN, Crawley, West Sussex, UK) and purified using the RNeasy mini kit in accordance with guidelines supplied (QIAGEN). RNA quality and quantity were assessed by automated capillary gel electrophoresis on a Bioanalyzer 2100 with RNA 6000 Nano Labchips according to manufacturers instructions (Agilent, Waldbronn, Germany).
Microarray hybridization.
Gene expression was determined using a 24,027 probe set bovine oligonucleotide array (Affymetrix, High Wycombe, UK), representing ∼23,000 bovine transcripts based on the original mapping using Unigene build 57 (March 24, 2004). Hybridization of samples to arrays and scanning was carried out by the German Resource Centre for Genomics Research, Germany, according to the manufacturer's instructions.
Microarray analysis.
All microarray analyses including, preprocessing, normalization and statistical analysis was carried out using R (61) version 2.6 and Bioconductor (24) version 2.1. Data were quality assessed before and after normalization using a number of in-built quality control methods implemented in the Bioconductor affycoretools and associated packages to identify problems if they existed with array hybridization, RNA degradation, and data normalization. Microarray data were preprocessed using the mmgMOS normalization method, and differential gene expression was analyzed using the pumaDE method, both implemented in the Bioconductor package puma (49, 59). Puma uses a Bayesian hierarchical model to calculate the probability of positive likelihood ratio of differential gene expression which was converted into “P-like values” using the recommended formula prior to subsequent analysis. The number of differentially expressed genes (DEG) detected by the puma method was also compared with that detected by the eBayes (Limma) (74) and Rank Product (5) methods. In a recent review of methods that did not include the puma method the eBayes implemented in the limma method proved the most robust overall, while the Rank Product method was ranked best when the number of samples was low or when the data are noisy (34). In a more recent comparison of methods, however, the puma method was ranked best of 35 methods tested (59).
As many of the original annotations for the Affymetrix bovine chip have been found to be erroneous (13, 23), Affyprobeminer (47) redefines the chip definition files (CDFs) for Affymetrix chips, taking into account the most recent genomic sequence information. The remapped annotations used in this study were determined using the bovineccdscdf annotation file downloaded from the Affyprobeminer website, which returned Entrez Gene gene name identifiers (17, 53). This remapped annotation included mappings to all RefSeq (mature RNA protein coding transcripts and validated complete coding sequences) in GenBank. Annotations were also supplemented by interrogating the Ensembl bos-taurus database version 46 using the biomaRt package in Bioconductor and also by manual annotation with recent entries in Entrez Gene. The Entrez Gene IDs of differentially expressed genes were then submitted to DAVID (14, 32) to determine gene function and to cluster genes into functionally significant clusters and also to determine significant biological pathways by interrogating KEGG (37). The official gene names corresponding to the Entrez Gene of the remapped annotations was used as the “population” background gene list in DAVID as opposed to the default DAVID bos-taurus gene list. This was to ensure that the calculated EASE scores were not overly conservative resulting in a failure to detect significant differences.
Pathway analysis.
To examine the molecular functions and genetic networks, the microarray data were explored using Ingenuity Pathways Analysis (IPA ver. 5.5; Ingenuity Systems, Mountain View, CA; http://www.ingenuity.com). A data set containing gene identifiers and corresponding expression and P-like values was uploaded into the application. Each identifier was mapped to its corresponding gene object in the Ingenuity knowledge base. A P-like value of P < 0.05 from the puma analysis was set to identify genes whose expression was significantly differentially up- or downregulated. These genes, called “focus” genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity. Network analysis returns a score that ranks networks according to their degree of relevance to the network eligible molecules in the dataset. The network score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher's exact test. The score is the negative log of this P value. A score of 2 thus indicates a P value of 10−2 or 0.01 and is considered statistically significant. Only those molecules that demonstrate relationships to other genes, proteins or endogenous chemicals were integrated into the analysis.
Quantitative real-time RT-PCR.
Using the same RNA samples that were analyzed in microarray studies, first-strand cDNA was synthesized using the Reverse Transcription system according to manufacturer's instructions (Promega UK Southampton, UK). We reverse transcribed 1 μg of purified total RNA into cDNA using random hexamers. The converted cDNA was quantified by absorbance at 260 nm, diluted to 50 ng/μl working stocks and stored at −20°C for subsequent analyses.
Analysis of putative reference genes for real-time RT-PCR studies was carried out using the GeNorm version 3.4 Microsoft Excel add-in (78). Genes analyzed included those that were shown to be stable in microarray analysis and were thus shown not to be differentially expressed among MNEB and SNEB groups. Selected genes included cyclin B1, integrin-β2, mitochondrial ribosomal protein L19, ubiquitin-conjugating enzyme E2K, ubiquitin-conjugating enzyme E2J2, and myosin light chain 3 (Supplementary Table S11 ). The ubiquitin-conjugating enzyme E2K gene exhibited the greatest stability during real-time RT-PCR analysis of spleen mRNA samples analyzed with M values ranging from 0.10 to 0.23. Based on a recommended cut-off V value of 0.15; ubiquitin-conjugating enzyme E2K was selected as a single standard reference gene for these experiments. Using more than one of these reference genes did not contribute to a more accurate normalization factor.
Primers were designed to measure gene expression of the 22 selected genes using the Primer3 software program (66). Details of primer sets used in the current study including all reference genes are listed in Supplementary Table S1. Primers for real-time RT-PCR were commercially synthesized (Sigma-Aldrich Ireland, Dublin, Ireland) and were first tested using end-point PCR. All amplified PCR products generated in this study were also sequenced to verify their identity. In the case of all genes examined in this study, DNA sequences were 100% identical to published sequences.
Real-time PCR reactions were carried out in a total volume of 20 μl with 1 μl cDNA (10–50 ng/μl), 10 μl Power SYBR master mix (Applied Biosystems, Warrington, UK), 1 μl forward and reverse primers (10 ng of each), and 8 μl nuclease-free H2O. Dissociation curves were examined for the presence of a single PCR product. Conditions were optimized to ensure that cDNA concentration, primer concentration, and efficiency of reactions were optimal. Real-time RT-PCR was performed using a ABI 7500 FAST quantitative PCR system (Applied Biosystems, Warrington, UK) with the following cycling parameters: 95°C for 10 min and 40 cycles of 95°C for 15 s, 60°C for 60 s, followed by amplicon dissociation (95°C for 1 min, 50°C for 45 s, increasing 0.5°/cycle until 95°C was reached). Gene expression results were calculated using the 2−ΔΔCT method (50).
Statistical analysis.
Differences in quantitative PCR and blood hematological data between the two energy balance groups was analyzed by analysis of variance using the MIXED procedure of SAS (69). Blood cell counts were log10 transformed and proportions (%) were transformed using the arsine of the square root of the proportion prior to analysis. Correlations between microarray and quantitative RT-PCR data was determined using the CORR procedure of SAS. A P value <0.05 was considered significant.
RESULTS
Animal performance.
One of the MNEB animals was excluded because of an abnormally lower feed intake relative to all other animals in the group. One animal was removed from the SNEB group following microarray quality assessment, which indicated that hybridization did not proceed optimally (data not shown). Therefore results are presented based on five cows per group.
The effect of treatment on feed intake, milk yield, and metabolic profile of the animals has been reported previously (20). In brief, three-times-a-day versus once-a-day milking was effective in decreasing (P < 0.020) EB from day 2 postcalving to slaughter (SNEB −8.7 ± 0.9 UFL/day compared with MNEB −4.9 ± 0.9 UFL/day). In addition, on the day of slaughter, animals in the SNEB group had higher systemic concentrations of NEFAs (1.41 ± 0.136 vs. 0.55 ± 0.216 mM, P < 0.01) and BHBs (3.71 ± 0.201 vs. 0.59 ± 0.097 mM, P < 0.001) than the MNEB animals.
Blood cell counts.
WBC counts were lower (P = 0.021) in SNEB compared with MNEB animals; the back-transformed means and 95% confidence limits were 5.8 (4.43–7.59) vs. 9.3 (7.11–12.19), respectively. Lymphocyte numbers were also lower (P = 0.047) in SNEB vs. MNEB animals 5.6 (4.16–7.62) vs. 8.7 (6.42–11.75), respectively. There was no difference (P > 0.05) between SNEB and MNEB groups in any of the other hematological variables measured.
Differential gene expression.
A total of 5,788 genes were expressed in the spleen. A cut-off P-like value of P < 0.05 resulted in a total of 545 DEG using the puma method. Of these 300 were upregulated, while 245 were downregulated. A comparison between different preprocessing/normalization/statistical methods showed that the puma method captured >50% of the DEG detected by the eBayes (Limma) method and >60% of the DEG detected by the Rank Product method (data not shown). The puma method, however, was more sensitive in detecting a greater number of DEG than either of the other methods.
Biological theme analysis.
Analysis of DEG using the online tool DAVID indicated the themes that were statistically overrepresented in SNEB versus MNEB spleen (Table 1). The overriding theme associated with the DEG set was protein synthesis, and this was supported by the finding that the ribosome pathway was significantly overrepresented in the set of DEG.
Table 1.
DAVID biological themes for DEG genes
| GO Category | Gene Category | Count | % | P Value (BH) |
|---|---|---|---|---|
| Molecular function | structural constituent of ribosome | 28 | 5.4 | 5.80E-06 |
| Cellular component | ribosome | 28 | 5.4 | 1.70E-06 |
| Molecular function | structural molecule activity | 34 | 6.5 | 7.90E-06 |
| Biological process | protein biosynthesis | 35 | 6.7 | 2.60E-05 |
| Biological process | macromolecule biosynthesis | 36 | 6.9 | 2.60E-05 |
| Cellular component | ribonucleoprotein complex | 31 | 5.9 | 4.60E-06 |
| Cellular component | nonmembrane-bound organelle | 39 | 7.5 | 1.50E-05 |
| Cellular component | intracellular nonmembrane-bound organelle | 39 | 7.5 | 1.50E-05 |
| Sp pir keywords | ribosomal protein | 20 | 3.8 | 1.10E-03 |
| Biological process | cellular biosynthesis | 44 | 8.4 | 9.00E-04 |
| Sp pir keywords | ribonucleoprotein | 17 | 3.3 | 1.90E-03 |
| Biological process | biosynthesis | 46 | 8.8 | 2.50E-03 |
| Cellular component | cytoplasm | 63 | 12 | 4.70E-04 |
| Cellular component | protein complex | 48 | 9.2 | 2.30E-03 |
| Cellular component | intracellular organelle | 77 | 14.7 | 2.80E-03 |
| Cellular component | organelle | 77 | 14.7 | 2.70E-03 |
| KEGG pathway | ribosome | 11 | 2.1 | 3.70E-02 |
| Cellular component | intracellular | 89 | 17 | 2.20E-02 |
The count and % represent the number and % of differentially expressed genes (DEG) in the specific gene ontology (GO) category. The P value is the Benjamini and Hochberg (BH) corrected P value.
Pathway analysis.
A total of 3,623 genes on the array could be mapped to the IPA database, including 248 DEG that were upregulated (Supplementary Table S2) and 190 DEG that were downregulated (Supplementary Table S3). Biological categories with the greatest number of DEG were cellular growth and proliferation, cell death, cellular movement, cellular development, and cell-to-cell signaling and interaction (Supplementary Fig. S1). Free radical scavenging had the greatest ratio of up- to downregulated genes in all categories.
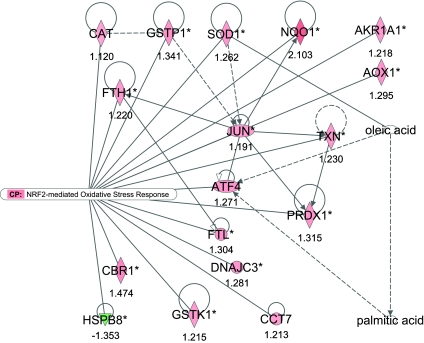
Canonical signaling pathway analysis revealed that NF-E2-related factor-2 (NRF2)-mediated oxidative stress response, mitochondrial dysfunction, amyotrophic lateral sclerosis signaling, endoplasmic reticulum (ER) stress pathway, aryl hydrocarbon receptor signaling, p53 signaling, protein ubiquitination pathway, and antigen presentation pathway were associated with the greatest number of upregulated genes, while natural killer (NK) cell signaling was associated with the greatest number of downregulated genes. Overall a greater number of upregulated genes were associated with metabolic and/or signaling pathways (Table 2). Palmitic and oleic acid, the major NEFAs produced as a result of NEB, have known links with the NRF2 pathway (Fig. 1).
Table 2.
Gene classification according to canonical signaling pathways using IPA
| Pathway | P Value | %DEG | Genes |
|---|---|---|---|
| NRF2-mediated oxidative stress response | 0.0004 | 9.4 | TXN, AKR1A1, ATF4, SOD1, FTH1, DNAJC3, PRDX1, GSTP1, CBR1, AOX1, GSTK1, FTL, JUN, CAT, NQO1, CCT7, HSPB8 |
| Mitochondrial dysfunction | 0.0028 | 9.7 | NDUFS7, NDUFB7, PRDX5, NDUFA6, UQCRC2, PARK7, SDHB, NDUFS4, CAT, NDUFV1, COX6A1, NDUFA2, APH1A, [bi]NDUFB5, UCP2, [bi]PRDX3 |
| Amyotrophic lateral sclerosis signaling | 0.0224 | 6.8 | RAB5B, SSR4, BAX, SOD1, CAT, CCS, BIRC2 |
| Endoplasmic reticulum stress pathway | 0.0251 | 16.7 | ATF4, DNAJC3, XBP1 |
| Natural killer cell signaling | 0.0263 | 6.4 | TYROBP, FCGR3A, SH2D1A, LCP2, FYN, KIR3DL1, NCR3 |
| Aryl hydrocarbon receptor signaling | 0.0263 | 6.6 | ALDH9A1, GSTK1, BAX, JUN, CCND2, NQO1, GSTP1, ESR1, RELA, FASN |
| p53 signaling | 0.0282 | 6.9 | BAX, JUN, CCND2, FASN, CTNNB1, SERPINE2 |
| Protein ubiquitination pathway | 0.0437 | 4.9 | PSMB6, UBE2S, B2M, UCHL3, PSMD3, PSMB1, PSMD4, BIRC2, IFNG, PSMD1 |
| Antigen presentation pathway | 0.0468 | 7.7 | PSMB6, CALR, B2M |
The %DEG is the proportion of DEG relative to the total number of genes in the specific canonical pathway. Downregulated genes are highlighted in boldface; upregulated genes are in lightface.
Fig. 1.
Ingenuity pathway analysis shows 17 genes from the 180 genes associated with the NF-E2-related factor-2 (NRF-2)-mediated oxidative stress response pathway. A network of genes are associated with the transcription factors JUN and activating transcription factor 4 (ATF4). The network has been overlaid with the links between palmitic and oleic acid [the major nonesterified fatty acids (NEFAs) produced as a result of negative energy balance (NEB)] and superoxidase dismutase (SOD1) and ATF4. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in severe negative energy balance (SNEB) vs. mild negative energy balance (MNEB) spleen. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction (Supplementary Fig. S2).
Canonical metabolic pathways analysis revealed that the pentose phosphate pathway, oxidative phosphorylation, ubiquinone biosynthesis, and methane metabolism were associated with the greatest number of upregulated genes, while phospholipid degradation was associated with the greatest number of downregulated genes (Supplementary Table S4).
Network analysis.
A total of 33 networks were identified by IPA, 21 of these had a score [−log(P value)] of 2 or greater, 19 of which had 10–34 focus genes among the DEG (Supplementary Table S5). Only the first three networks will be considered here in the context of SNEB and immune function.
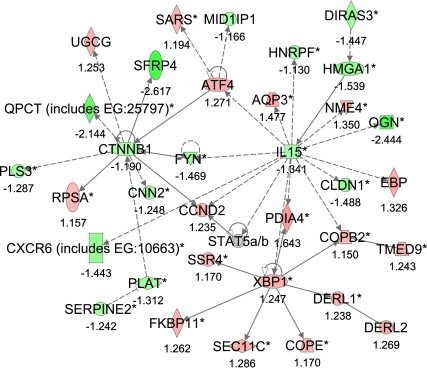
The first network (#1) with a score of 59 and 34 focus genes indicated links between the cytokine interleukin 15 (IL-15) and the transcription factors catenin β-1 (CNNB1), activating transcription factor 4 (ATF4), and X-box binding protein-1 (XBP1). The main functions are in cell growth and proliferation, hematological system development and function, and immune response (Fig. 2, Supplementary Table S5).
Fig. 2.
Gene Network 1. Ingenuity pathway analysis shows a network of 34 “focus” genes with a score of 59. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB spleen. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product, and the lines indicate the type of interaction (Supplementary Fig. S2).
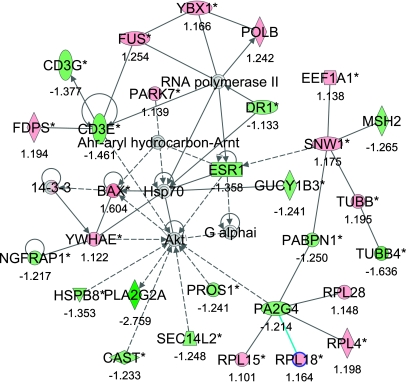
The second network (#2) with a score of 46 and 29 focus genes indicated gene clusters centered around the transcription factors downregulation of transcription (DR1), eukaryotic translation elongation factor 1α (EEF1A1), fusion (FUS), proliferation-associated 2G4 (PA2G4), and SNW domain containing 1 (SNW1). This network has main functions in immune response, cell death, and immunological disease (Fig. 3, Supplementary Table S5).
Fig. 3.
Gene Network 2. Ingenuity pathway analysis shows a network of 29 focus genes with a significant score of 46. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB spleen. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product, and the lines indicate the type of interaction (Supplementary Fig. S2).
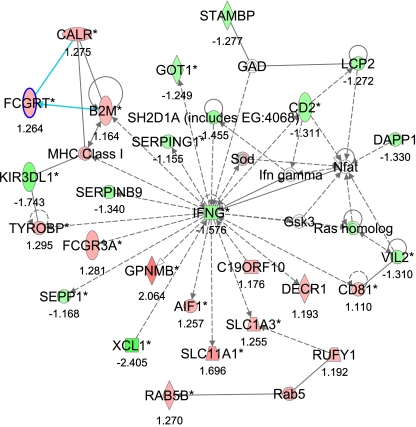
The third network (#3) with a score of 41 and 27 focus genes indicated gene clusters centered around interferon-γ (INFG), with main functions in immune response cell death and immunological disease (Fig. 4, Supplementary Table S5).
Fig. 4.
Gene Network 3. Ingenuity pathway analysis shows a network of 27 focus genes with a significant score of 41. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB spleen. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product, and the lines indicate the type of interaction (Supplementary Fig. S2).
Quantitative PCR analysis.
A total of 22 genes including interleukin-2 (IL-2) that was not annotated by Affyprobeminer were analyzed by quantitative PCR (Table 3). Genes were chosen to be representative of those in the top pathway and three IPA networks and the more biologically interesting of the most up- and downregulated genes also. Among genes, the direction and magnitude of expression between methods were highly consistent. The magnitude of differential expression tended to be higher by quantitative PCR, and the differences between qPCR and microarray tended to be greater for those genes most differentially expressed. Within genes there was good correlation between microarray and qPCR expression. The expression of IL-2, determined by quantitative PCR only was not significantly different between MNEB and SNEB groups (P > 0.5).
Table 3.
Results for qPCR assays and correlation with microarray data
| Gene Name | qPCR Fold Change | P Value | Array Fold Change | Correlation | n |
|---|---|---|---|---|---|
| AFT 4 | 1.82 | 0.0041 | 1.27 | 0.88 | 8 |
| BAX | 1.80 | 0.0345 | 1.60 | 0.57 | 9 |
| BCL2A1 | −1.86 | 0.0147 | −1.34 | 0.84 | 8 |
| CAT | 2.08 | 0.0051 | 1.12 | 0.8 | 7 |
| CTNNB1 | −2.26 | 0.0164 | −1.19 | 0.68 | 8 |
| ESR1 | −1.85 | 0.0126 | −1.36 | 0.66 | 8 |
| FUS | 1.41 | 0.0235 | 1.25 | 0.52 | 8 |
| GPNMB | 2.30 | 0.0431 | 2.06 | 0.87 | 9 |
| HBB | −14.94 | 0.0252 | −7.32 | 0.68 | 8 |
| IFNG | −3.68 | 0.0080 | −1.58 | 0.65 | 9 |
| IL2 | 1.91 | 0.5055 | nd | nd | |
| IL15 | −2.37 | 0.0312 | −1.34 | 0.62 | 8 |
| MAL | −4.19 | 0.0044 | −4.06 | 0.39 | 8 |
| NQO1 | 1.98 | 0.0125 | 2.10 | 0.82 | 7 |
| PA2G4 | −3.31 | 0.0041 | −1.21 | 0.95 | 8 |
| PLAUR | 11.44 | 0.0108 | 8.41 | 0.75 | 9 |
| PRDX1 | 2.18 | 0.0307 | 1.32 | 0.74 | 8 |
| SERPINB9 | −1.96 | 0.0106 | −1.34 | 0.57 | 8 |
| SOD1 | 1.91 | 0.0005 | 1.26 | 0.66 | 9 |
| UCP2 | −3.63 | 0.0366 | −1.58 | 0.98 | 6 |
| XBP1 | 1.92 | 0.0070 | 1.25 | 0.63 | 7 |
| XCL1 | −2.71 | 0.0018 | −2.41 | 0.83 | 8 |
nd, Not determined.
DISCUSSION
Effect of milking frequency and nutrition on EB.
The main aim of this study was to investigate the effects of NEB on genes affecting immune function in the postpartum dairy cow. In this study, estimated daily EB, blood metabolite, and hematological data indicate that three-times-a-day versus once-a-day milking combined with differential nutrition was effective in creating a significant difference in EB between the MNEB and SNEB groups. Blood concentrations of NEFA and BHB were higher in SNEB compared with MNEB animals and consistent with that reported previously for animals managed under a similar regime (58). Blood hematological measurements indicated that SNEB was associated with a decrease in WBC and in lymphocyte numbers indicating a depression in immune function as has been reported following calving in dairy cows (38, 39, 64).
Effect of NEB on gene expression in the spleen.
This is the first study to our knowledge to explore the effects of SNEB on gene expression in the spleen of the postpartum dairy cow. The results indicate that the spleen responds to SNEB with an overall increase in gene expression. The predominant gene ontology (GO, 76) categories affected by SNEB were those associated with protein synthesis and the KEGG ribosome pathway. This is not surprising in that changes in protein synthesis are an indispensable functional consequence of changes in gene expression.
Pathway analysis.
Analysis of DEG using DAVID revealed the significant biological processes affected by NEB; however, network analysis using IPA revealed how the individual DEG cooperate in a variety of metabolic and signaling pathways, potentially revealing those genes that are below the limit of detection of conventional microarrays (72). In addition, network analysis enables the identification of biological mechanisms, pathways, and functions most relevant to the genes of interest (51) and enables the subtle but pleiotropic effects of metabolic diseases (44) such as SNEB to be determined.
The two most significant pathways were those associated with the NRF2-mediated stress response and mitochondrial dysfunction. NRF2 is a member of the cap'n'collar family of bZIP transcription factors expressed in a wide variety of tissues. Antioxidant defense genes such as superoxide dismutase (SOD1), catalase (CAT), peroxiredoxin-1 (PRDX1), and NAD(P)H:quinone oxidoreductase (NQO1) were all found to be upregulated in SNEB by microarray analysis and confirmed by quantitative PCR. Their expression is dependent on NRF2 activity (43) acting as a heterodimer with other bZIP transcription factors such as ATF4 (29) or c-JUN (81) to defend against oxidative stress (12). SOD1 detoxifies superoxide (O2−) resulting in the creation of H2O2, which in turn is further metabolized to H2O by CAT or PRDX1. The increased expression of antioxidant genes and genes involved in free radical scavenging in SNEB animals in this study is likely a response to mitigate or reduce the damaging effects of increased concentrations of ROS.
A significant number and proportion of genes involved in ER stress pathway were also upregulated in SNEB. The ER is required for the folding, processing, and export of newly synthesized proteins. Disruption of ER function, termed ER stress, results in the accumulation of misfolded proteins in the ER (71) and activates the unfolded protein response signaling pathway, which includes upregulation of two key transcription factors, ATF4 and XBP1, and the molecular chaperone DNAJB3, which is likely to be involved in protein folding.
In addition to oxidative stress due to lipid peroxidation by the liver, a consequence of ER stress is the additional accumulation of ROS. Activation of the NRF2 and ATF4 transcription factors, initiates the convergence of ER stress and oxidative stress signaling (70) with widespread alteration of multiple cellular processes that, although distinct, also overlap (68). The two major products of fatty acid breakdown in SNEB are palmitate and oleate (67). Of these only palmitate triggered an ER stress response in MIN6 cells (a murine pancreas β-cell line) (41). Prolonged or severe ER stress is widely accepted to trigger apoptosis (26). The balance between apoptosis occurring or not depends on the anti- (e.g., B-cell lymphoma/leukemia-2; BCL-2) and proapoptotic BCL-2 family of proteins including BCL associate protein (BAX). ER stress upsets the balance between pro- and antiapoptotic pathways, and the outcome depends on the duration of exposure to stress and whether a response to the initial stress is adequate (70). ATF4 and XBP1 are known to indirectly repress BCL-2 transcription (56) by inducing the expression of CCAAT/enhancer binding protein (C/EBP) homologous protein (CHOP) also called FUS (see Fig. 3). In this study, BAX, ATF4, XBP1, and FUS were upregulated in SNEB animals, while BCL2A1 was downregulated. SNEB therefore was of a sufficient duration in this study to create a cellular environment favoring apoptosis of immune-related cell types in the spleen and thus rendering animals in SNEB more susceptible to infection.
In this study systemic concentrations of BHB increased in SNEB relative to MNEB animals. Among the short chain fatty acids butyrate is a potent inducer of apoptosis and inhibitor of cell proliferation, differentiation, and motility (11, 16, 22, 44). The mechanisms by which butyrate induces cellular differentiation and suppresses growth have not been elucidated; however, the level of BCL-2 expression can regulate the apoptotic effect of butyrate in vitro (55).
Similar to ER stress, mitochondrial function also depends on the BCL-2 family of proteins, and proapoptotic stimuli can interfere with mitochondrial function (45). Mitochondria are essential for the production of energy required for metabolism in the form of ATP, which is generated by oxidative phosphorylation. The mitochondria are a significant source of ROS (27). Genes involved in oxidative phosphorylation and ubiquination were, in the main, upregulated in this study, indicating that SNEB results in increased splenic metabolic activity probably in an effort to redress the negative effects of oxidative stress and in the proteasome degradation of misfolded proteins as a result of ER stress. In contrast, Loor et al. (51) found that ubiquitination and oxidative phosphorylation were decreased in the liver of cows with nutritionally induced ketosis and were associated with an overall decrease in liver function.
Ubiquinone biosynthesis is required for the oxidation of NADH (83), which is essential for the adequate supply of redox species required to mount a response to oxidative stress. In contrast the gene encoding uncoupling protein-2 (UCP2), an important regulator of mitochondrial ROS (2), was downregulated in SNEB animals, facilitating increased ROS production. UCP2 is reported to be highly expressed in spleen (21) and immune tissue (40), suggesting a role for UCP2 in immunity or inflammatory responsiveness (21).
Overall the main signaling pathways affected by SNEB suggest that the spleen is subjected to increased oxidative stress and responds by mobilizing genes and proteins to mitigate the effects of oxidative stress and possibly also butyrate, to limit their effects on apoptosis. Failure to redress the effects of oxidative stress results in activation of apoptotic pathways and cell death.
Network analysis.
Network analysis using IPA revealed a number of interacting gene networks; the one with the highest score (see Fig. 2) shows links between the cytokine IL-15 and the transcription factors CNNB1, ATF4, and XBP1 with main functions in cell growth and proliferation, hematological system development and function, and immune response. From an immune perspective, which was the primary focus of this study, the finding that IL-15 was downregulated in SNEB is significant, and the remaining discussion on network analysis will be centered on this.
IL-15 was first identified because of its IL-2-like activity in inducing T-lymphocyte proliferation (25). Unlike IL-2, which is mainly produced by activated T-lymphocytes, IL-15 is expressed constitutively in most tissues and in cattle is highly expressed in spleen and skeletal tissue (9). IL-15 is produced in activated monocytes, macrophages, dendritic cells, bone marrow stromal cells, and thymic epithelium, whereas T-lymphocytes do not typically produce IL-15 (19, 25, 82).
IL-15 and IL-2 share very little primary protein and cDNA sequence homology; however, molecular modeling suggests that they belong to the same 4 α-helix bundle cytokine family (25). Functional studies have shown that IL-15 also uses the IL-2Rβ-receptor subunit and the common gamma chain (γc) subunit, but it is the distribution of distinct IL-R15α and IL-2Rα that determine their specific actions (7, 19).
IL-15 is a pleiotropic proinflammatory cytokine and, like IL-2, is a potent stimulator of T-lymphocyte proliferation, inflammatory (CD4+), helper (CD4+T), and cytotoxic T-cells (e.g., CD8+T), and NK cells (52). IL-15 is capable of priming NK cytotoxicity (31) and maintaining NK cell survival in vitro (10) and can also selectively stimulate the proliferation of CD8+ memory T-cells in contrast to IL-2, which inhibits CD8+ T-cell proliferation (84). IL-15 is thought to act independently of IL-2 but can act synergistically in stimulating the cellular immune response. In a recent report IL-15 is also thought to act together with IL-12 and IL-18 to in turn regulate interferon-γ (IFNG)(Fig. 4), which ensures maturation of B-cells in the spleen prior to their homing to the lymph nodes or inflammatory sites (28).
IL-15 signaling in lymphocytes activates Janus kinase (JAK) and signal transducer and activator of transcription (STAT) pathways (35). JAK/STAT signaling is critical in the generation of inflammatory responses to infection and in T-cell proliferation, and Li et al. (46) suggests that IL-15 is the critical growth factor for all T-cell proliferation.
In this study a sample of splenic tissue was harvested that included capsular, red pulp and most likely white pulp tissue. Although the sample was taken from the same location across animals no account was taken for the possibility of changes in the different splenocyte populations between animal groups. However, of all the blood cell populations measured only lymphocyte numbers were significantly decreased (by ∼33%) in SNEB, and only activated monocytes and macrophages are known to express IL-15 mRNA. T-lymphocytes, a major source of IL-2, express little if any IL-15 mRNA (25). IL-2 expression determined by RT-PCR was found to be numerically higher in SNEB but this difference was not significant (P > 0.5). Hemoglobin beta (HBB), a gene known to be expressed in mouse macrophages (48), and T-lymphocyte maturation-associated protein ER of T-cells (62), were two of the most downregulated genes in SNEB animals. Plasminogen activator urokinase receptor (PLAUR), which is highly expressed in human macrophages (75), was the most upregulated gene in SNEB animals which might suggest an increase in macrophage or chemotactic activity (63). PLAUR is also expressed, however, in other immune cell types (85) including neutrophils (60), and its expression in macrophages can be stimulated by exposure to increased concentrations of free fatty acids (3). Increased systemic concentrations of NEFAs in this study may have contributed to the observed increase in PLAUR activity seen in SNEB animals; however, this requires further study.
The effects of decreased lymphocyte numbers are evident in Fig. 4 where IFNG, which is mainly produced by T-lymphocytes and NK cells, is downregulated in SNEB animals. IFNG has wide-ranging effects on gene expression (4), is specifically produced by CD4+ T and CD8+T cells (18), and has potent antiviral and immunomodulatory functions including the induction of immunoglobulin secretion by B-cells and enhancing NK-cell and macrophage activity. In this study both IL-15 gene expression and lymphocyte numbers were decreased in SNEB animals, indicating for the first time, to our knowledge, a functional link between SNEB, splenic IL-15 expression, and suppression of immune function in dairy cows.
In conclusion, SNEB in the postpartum dairy cow resulted in an increase in products of lipid catabolism including NEFAs and BHB. This in turn was shown to be associated with a reduction in circulating blood lymphocyte numbers and in pleiotropic effects in splenic gene expression associated with increased oxidative stress and apoptosis, negatively impacting immune function.
GRANTS
This work was funded by the Wellcome Trust and the Irish National Development Plan.
Supplementary Material
Acknowledgments
The authors thank the Teagasc Moorepark farm staff and the skilled technical assistance of Jonathan Kenneally.
Address for reprint requests and other correspondence: D. G. Morris, Teagasc, Mellows Campus, Athenry, Co. Galway, Ireland (e-mail: dermot.morris@teagasc.ie).
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Ametaj BN, Bradford BJ, Bobe G, Beitz DC. Acute phase response indicates inflammatory conditions may play a role in the pathogenesis of fatty liver in dairy cows. J Dairy Sci 85: 189, 2002. [Google Scholar]
- 2.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, Miroux B, Couplan E, Alves-Guerra MC, Goubern M, Surwit R, Bouillaud F, Richard D, Collins S, Ricquier D. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet 26: 435–439, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Assmann A, Möhlig M, Osterhoff M, Pfeiffer AFH, Spranger J. Fatty acids differentially modify the expression of urokinase type plasminogen activator receptor in monocytes. Biochem Biophys Res Commun 376: 196–199, 2008. [DOI] [PubMed] [Google Scholar]
- 4.Boehm T, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Ann Rev Immunol 15: 749–795, 1977. [DOI] [PubMed] [Google Scholar]
- 5.Breitling R, Herzyk P. Rank-based methods as a non-parametric alternative of the T-statistic for the analysis of biological microarray data. J Bioinform Comput Biol 3: 1171–1189, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Breukink HJ, Wensing T. Pathophysiology of the liver in high yielding dairy cows and its consequences for health and production. Isr J Vet Med 52: 66–72, 1977. [Google Scholar]
- 7.Budagian V, Bulanova E, Paus R, Bulfone-Paus S. IL-15/IL-15 receptor biology: a guided tour through an expanding universe. Cytokine Growth Factor Rev 17: 259–280, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Bugaut M. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol 86B: 439–472, 1987. [DOI] [PubMed] [Google Scholar]
- 9.Canals A, Gasperre LC, Boyd PC, Almeria S, Zarlengan DS. Cloning and expression of bovine interleukin-15: analysis and modulation of transcription by exogenous stimulation. J Interf Cytokine Res 17: 473–480, 1997. [DOI] [PubMed] [Google Scholar]
- 10.Carson WE, Fehniger TA, Haldar S, Eckhert K, Lindemann MJ, Lai CF, Croce CM, Baumann H, Caligiuri MA. A potential role for interleukin-15 in the regulation of human natural killer cell survival. J Clin Invest 99: 937–943, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JS, Faller DV, Spanjaard RA. Short-chain fatty acid inhibitors of histone deacetylases: promising anticancer therapeutics? Curr Cancer Drug Targets 3: 219–236, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Chen XL, Dodd G, Thomas S, Zhang X, Wasserman MA, Rovin BH, Kunsch C. Activation of Nrf2/ARE pathway protects endothelial cells from oxidant injury and inhibits inflammatory gene expression. Am J Physiol Heart Circ Physiol 290: H1862–H1870, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acid Res 33: e175, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003. [PubMed] [Google Scholar]
- 15.Dyk PB. The association of prepartum non-esterified fatty acids and body condition with peripartum health problems on 95 Michigan dairy farms (M.S. thesis). East Lansing, MI: Michigan State Univ., 1995.
- 16.Emenaker NJ, Calaf GM, Cox D, Basson MD, Qureshi N. Short-chain fatty acids inhibit invasive human colon cancer by modulating uPA, TIMP-1, TIMP-2, mutant p53, Bcl-2, Bax, p21 and PCNA protein expression in an in vitro cell culture model. J Nutr 131: 3041S–3046S, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO gene nomenclature database, 2006 updates. Nucleic Acids Res 34: D319–D321, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Ann Rev Immunol 11: 571–611, 1993. [DOI] [PubMed] [Google Scholar]
- 19.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood 97: 14–31, 2001. [DOI] [PubMed] [Google Scholar]
- 20.Fenwick MA, Fitzpatrick R, Kenny DA, Diskin MG, Patton J, Murphy JJ, Wathes DC. Interrelationships between negative energy balance (NEB) and IGF regulation in liver of lactating dairy cows. Domest Anim Endocrinol 34: 31–44, 2008. [DOI] [PubMed] [Google Scholar]
- 21.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin MF, Surwit RS, Ricquier D, Craig H, Warden CH. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nature Genet 15: 269–272, 1997. [DOI] [PubMed] [Google Scholar]
- 22.Gassull MA, Cabre E. Nutrition in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care 4: 561–569, 2001. [DOI] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Deetling M, Duidot S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabstein KH, Eisenham J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, Johnson L, Alderson MR, Watson JD, Anderson DM, Giri JG. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science 264: 965–968, 1994. [DOI] [PubMed] [Google Scholar]
- 26.Green DR. Apoptotic pathways: ten minutes to dead. Cell 121: 671–674, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Green DR, Reed JC. Mitochondria and apoptosis. Science 281: 1309–1312, 1998. [DOI] [PubMed] [Google Scholar]
- 28.Hart G, Avin-Wittenberg T, Shachar I. IL-15 regulates immature B-cell homing in an Ly49D-, IL-12-, and IL-18-dependent manner. Blood 111: 50–59, 2008. [DOI] [PubMed] [Google Scholar]
- 29.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 276: 20858–20865, 2001. [DOI] [PubMed] [Google Scholar]
- 30.Holtenius P, Holtenius K. New aspects of ketone bodies in energy metabolism of dairy cows: a review. J Vet Med A 43: 479–587, 1996. [DOI] [PubMed] [Google Scholar]
- 31.Horng T, Bezbradica JS, Medzhitov R. NKG2D signalling is coupled to the interleukin 15 receptor signalling pathway. Nat Immunol 8: 1345–1352, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrige J. INRAtion V2.7. In: Microsoft Computer Program of Ration Formulation For Ruminant Livestock, edited by Agabriel J, Champciaux P, Espinasse C. Dijon, France: CNERTA, 1989.
- 34.Jeffery IB, Higgins DG, Culhane AC. Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics 7: 359, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston JA, Bacon CM, Finbloom DS, Rees RC, Kaplan D, Shibuya K, Ortaldo JR, Gupta Chen YQ S, Giri JD, O'Shea JJ. Tyrosine phosphorylation and activation of STAT5, STAT3 and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci USA 92: 8705–8709, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorritsma R, Jorritsma H, Schukken YH, Bartlett PC, Wensing T, Wentink GH. Prevalence and indicators of post partum fatty infiltration of the liver in nine commercial dairy herds in The Netherlands. Livest Prod Sci 68: 53–60, 2001. [Google Scholar]
- 37.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keane CJ, Hanlon AJ, Roche JF, Burton JL, Mee JF, O'Doherty JV, Sweeney T. Short Communication: A potential phenotype in neutrophils of cows milked once daily in early lactation. J Dairy Sci 89: 1024–1027, 2006. [DOI] [PubMed] [Google Scholar]
- 39.Kehrli ME, Nonnecke BJ, Roth JA. Alterations in bovine neutrophil function during the periparturient period. Am J Vet Res 50: 207–214, 1989. [PubMed] [Google Scholar]
- 40.Kizaki T, Suzuki K, Hitomi Y, Taniguchi N, Saitoh D, Watanabe K, Onoe K, Day NK, Good RA, Ohno H. Uncoupling protein 2 plays an important role in nitric oxide production of lipopolysaccharide-stimulated macrophages. Proc Natl Acad Sci USA 99: 9392–9397, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laybutt DR, Preston AM, Akerfeldt MC, Kench JG, Busch AK, Biankin AV, Biden TJ. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabeologica 50: 752–763, 2007. [DOI] [PubMed] [Google Scholar]
- 42.Lee EK, Kehrli ME. Expression of adhesion molecules on neutrophils of periparturient cows and neonatal calves. Am J Vet Res 59: 37–43, 1998. [PubMed] [Google Scholar]
- 43.Lee JM, Calkins MJ, Chan K, Kan WK, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem 278: 12029–12038, 2003. [DOI] [PubMed] [Google Scholar]
- 44.Li CJ, Li RW, Wang YH, Elsasser TH. Pathway analysis identifies perturbation of genetic networks induced by butyrate in a bovine kidney epithelial cell line. Funct Integr Genomics 7: 193–205, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis. J Biol Chem 11:7260–7270, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Li XC, Demirci G, Ferrari-Lacraz S, Groves C, Coyle A, Malek TR, Strom TB. IL-15 and IL-2: a matter of life and death for T cells in vivo. Nat Med 7: 114–118, 2001. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC, Kane DW, Weinstein JN. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics 23: 2385–2390, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Zeng M, Stamler JS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci USA 96: 6643–6647, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Milo M, Lawrence ND, Rattray M. Probe-level measurement error improves accuracy in detecting differential gene expression. Bioinformatics 22: 2107–2113, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(-delta delta CT) method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 51.Loor JJ, Everts RE, Bionaz M, Dann HM, Morin DE, Oliveira R, Rodriguez-Zas SL, Drackley JK, Lewin HA. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics 32: 105–116, 2007. [DOI] [PubMed] [Google Scholar]
- 52.Ma A, Boone DL, Lodolce JP. The pleiotropic functions of interleukin 15: Not so interleukin 2-like after all. J Exp Med 191: 753–755, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 33: D54–D58, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallard BA, Dekkers JC, Ireland MJ, Leslie KE, Sharif S, Vankampen CL, Wagter L, Wilkie BN. Alteration in immune responsiveness during the peripartum period and its ramification on dairy cow and calf health. J Dairy Sci 81: 585–595, 1998. [DOI] [PubMed] [Google Scholar]
- 55.Mandal M, Kumar R. Bcl-2 expression regulates sodium butyrate-induced apoptosis in human MCF-7 breast cancer cells. Cell Growth Diff 7: 311–318, 1996. [PubMed] [Google Scholar]
- 56.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bc12 and perturbing the cellular redox state. Mol Cell Biol 21: 1249–1259, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol 5: 606–616, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Patton J, Kenny DA, Mee JF, O'Mara FP, Wathes DC, Cook M, Murphy JJ. Effect of milking frequency and diet on milk production, energy balance, and reproduction in dairy cows. J Dairy Sci 89: 1478–1487, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearson RD, Liu X, Sanguinetti G, Milo M, Lawrence ND, Rattray M. A comprehensive re-analysis of the Golden Spike data: Towards a benchmark for differential expression methods. BMC Bioinformatics 9: 164, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Politis I, Zavizjon B, Cheli F, Baldi A. Expression of urokinase plasminogen activator receptor in resting and activated bovine neutrophils. J Dairy Res 69: 195–204, 2002. [DOI] [PubMed] [Google Scholar]
- 61.R Development Core Team. R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; URL http://www.R-project.org, 2007.
- 62.Rancano C, Rubio T, Correas I, Alonso MA. Genomic structure and subcellular localization of MAL, a human T-cell-specific proteolipid protein. J Biol Chem 269: 8159–8164, 1994. [PubMed] [Google Scholar]
- 63.Renaud SJ, Macdonald-Goodfellow SK, Graham CH. Coordinated regulation of trophoblast invasiveness by macrophages and interleukin 10. Biol Reprod 76: 448–454, 2007. [DOI] [PubMed] [Google Scholar]
- 64.Ropstad E, Vik-Mo L, Refsdal AO. Levels of milk urea, plasma constituents and rumen liquid ammonia in relation to feeding of dairy cows during early lactation. Acta Vet Scand 30: 199–208, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Royal MD, Darwash AO, Flint APF, Webb R, Wolliams JA, Lamming GE. Declining fertility in dairy cattle: changes in traditional and endocrine parameters of fertility. Anim Sci 70: 487–502, 2000. [Google Scholar]
- 66.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386, 2000. [DOI] [PubMed] [Google Scholar]
- 67.Rukkwamsuk T, Geelen MJ, Kruip TA, Wensing T. Interrelation of fatty acid composition in adipose tissue, serum, and liver of dairy cows during development of fatty liver postpartum. J Dairy Sci 83: 52–59, 2000. [DOI] [PubMed] [Google Scholar]
- 68.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci 32: 469–674, 2007. [DOI] [PubMed] [Google Scholar]
- 69.SAS. The SAS System for Windows (release 9.1). Cary, NC: SAS Institute, 2003.
- 70.Schroder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci 65: 862–894, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789, 2005. [DOI] [PubMed] [Google Scholar]
- 72.Sharma MR, Polavarapu R, Roseman D, Patel V, Eaton E, Kishor PB, Nanji AA. Transcriptional networks in a rat model for nonalcoholic fatty liver disease: a microarray analysis. Exp Mol Pathol 81: 202–210, 2006. [DOI] [PubMed] [Google Scholar]
- 73.She P, Hippen AR, Young JW, Lindberg GL, Beitz DC, Richardson LF, Tucker RW. Metabolic responses of lactating dairy cows to 14-day intravenous infusions of glucagons. J Dairy Sci 82: 1118–1127, 1999. [DOI] [PubMed] [Google Scholar]
- 74.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3, 2004. [DOI] [PubMed] [Google Scholar]
- 75.Svensson PA, Hägg DA, Jernås M, Englund MCO, Hulten LM, Ohlsson BG, Hulthe J, Wiklund O, Carlsson B, Fagerberg B, Carlsson LMS. Identification of genes predominantly expressed in human macrophages. Atherosclerosis 177: 287–290, 2004. [DOI] [PubMed] [Google Scholar]
- 76.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turk R, Diuretic D, Gereš D, Svetina Turk A, N, Flegar-Meštrić Z. Influence of oxidative stress and metabolic adaptation on PON1 activity and MDA level in transition dairy cows. Anim Reprod Sci 108: 98–106, 2007. [DOI] [PubMed] [Google Scholar]
- 78.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034, 2002. [DOI] [PMC free article] [PubMed]
- 79.Van Kampen C, Mallard BA. Effects of peripartum stress and health on circulating bovine lymphocyte subsets. Vet Immunol Immunopathol 59: 79–91, 1997. [DOI] [PubMed] [Google Scholar]
- 80.Van Knegsel ATM, deVries Reilingh G, Meulenberg S, van den Brand H, Dijkstra J, Kemp B, Parmentier HK. Natural antibodies related to energy balance in early lactation dairy cows. J Dairy Sci 90: 5490–5498, 2007. [DOI] [PubMed] [Google Scholar]
- 81.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17: 3145–3156, 1998. [DOI] [PubMed] [Google Scholar]
- 82.Waldman TA. Interleukin-15. Encyclopedia of Hormones, edited by Henry HL, Norman AW. Academic, 2003, p. 478–484.
- 83.Yakovlev G, Reda T, Hirst J. Reevaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase. Proc Natl Acad Sci USA 104: 12720–12725, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ cells in vivo by IL-15. Immunity 8: 591–599, 1998. [DOI] [PubMed] [Google Scholar]
- 85.Zola H, Swart B, Nicholson I, Voss E. CD 87. In: Leukocytes and Stromal Cell Molecules: The CD Markers. Hoboken, NJ: Wiley, 2007, p. 184–185.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.