Abstract
Most dairy cows suffer uterine microbial contamination postpartum. Persistent endometritis often develops, associated with reduced fertility. We used a model of differential feeding and milking regimes to produce cows in differing negative energy balance status in early lactation (mild or severe, MNEB or SNEB). Blood hematology was assessed preslaughter at 2 wk postpartum. RNA expression in endometrial samples was compared using bovine Affymetrix arrays. Data were mapped using Ingenuity Pathway Analysis. Circulating concentrations of IGF-I remained lower in the SNEB group, whereas blood nonesterified fatty acid and β-hydroxybutyrate concentrations were raised. White blood cell count and lymphocyte number were reduced in SNEB cows. Array analysis of endometrial samples identified 274 differentially expressed probes representing 197 recognized genes between the energy balance groups. The main canonical pathways affected related to immunological and inflammatory disease and connective tissue disorders. Inflammatory response genes with major upregulation in SNEB cows included matrix metalloproteinases, chemokines, cytokines, and calgranulins. Expression of several interferon-inducible genes including ISG20, IFIH1, MX1, and MX2 were also significantly increased in the SNEB cows. These results provide evidence that cows in SNEB were still undergoing an active uterine inflammatory response 2 wk postpartum, whereas MNEB cows had more fully recovered from their energy deficit, with their endometrium reaching a more advanced stage of repair. SNEB may therefore prevent cows from mounting an effective immune response to the microbial challenge experienced after calving, prolonging the time required for uterine recovery and compromising subsequent fertility.
Keywords: microarray, bovine, endometrium, innate immune system, antimicrobial peptides
high yielding dairy cows are under considerable metabolic stress in early lactation, as they cannot meet the energetic demands for milk production entirely from feed intake (1). Gestational changes cause insulin resistance in late pregnancy (2). Immediately after calving high rates of body condition score loss are associated with a severe negative energy balance (SNEB) status, indicated by alterations in blood metabolite and hormone profiles. Both nonesterified fatty acids (NEFAs) and β-hydroxybutyrate (BHB) concentrations are elevated, indicative of lipid mobilization and fatty acid oxidation (73). The liver coordinates the extensive biochemical and morphological modifications required via upregulation of genes involved in fatty acid oxidation and gluconeogenesis, and downregulation of triacylglycerol synthesis (42). However, excessive fat accumulation postpartum impairs liver function, compromising glucose production and increasing inflammatory responses. Production of insulin and IGF-I are also reduced at this time (14, 17).
Cows are expected to rebreed within 2–3 mo of calving for optimum economic return. Poor fertility is a serious economic consideration in the dairy industry and is strongly linked to the animal's health around calving. Excessive lipid mobilization is associated with metabolic and reproductive disorders (57). Cows with a low nadir in IGF-I in the first 2 wk postpartum take longer to resume estrous cyclicity and are less likely to conceive when the breeding period is reached (6, 70). Immune function is also suppressed over the periparturient period (7, 32, 43), and poor energy balance (EB) status and fatty liver can impair peripheral blood neutrophil function (24, 39, 77).
Following calving, the uterus must undergo extensive remodeling to reduce in size, remove cellular debris, and restore normal architecture (21, 39, 65). Microbial contamination of the uterus is almost universal during the first week postpartum and continues for ∼2 wk in ∼90% of animals (18, 40, 66). The most common recognized pathogens are Arcobacterium pyogenes, Escherichia coli, Fusobacterium necrophorum, Prevotella melaninogenicus, and Proteus spp. Uterine defenses rely initially on classical innate immunity and mucosal defense systems rather than adaptive immunity (33, 66). Failure in this defense system results in uterine disease. Metritis is present in 40% of cows within 2 wk of calving, and 15% have a persistent endometritis in the 3–6 wk postpartum period (40, 66). Subclinical endometritis, from 6 wk postpartum onward, is characterized by an extensive leukocytic infiltration of the endometrium and chronic inflammation (18, 66) and is associated with longer intervals to conception and a greater likelihood of culling (19, 22, 40).
The experiment described here was designed to test the hypothesis that metabolic changes in postpartum cows can delay uterine repair mechanisms and promote a state of chronic inflammation, resulting in an unfavorable uterine environment likely to contribute to reduced fertility. To this end we developed a model of differential feeding and milking regimes to produce cows in differing negative energy balance (NEB) status in early lactation (mild or severe, MNEB or SNEB) as confirmed by markedly divergent metabolic and endocrine profiles (17). Uterine tissue samples were collected at 2 wk postpartum. This time point was chosen as infection would have had sufficient time both to manifest and to be influenced by the differences in energy demand between groups (77). It was, however, before cows had ovulated, to avoid differences between cows associated with exposure to luteal progesterone, which can influence uterine resistance to infection (60).
MATERIALS AND METHODS
Animals and management.
All procedures were carried out under license in accordance with the European Community Directive, 86-609-EC. Multiparous Holstein-Friesian cows with a mean parity of 4.7 and an average previous lactation yield of 6,477 ± 354 kg were used. Cows were blocked 2 wk prior to expected calving according to parity, body condition score, and previous yield and were randomly allocated to two treatments (each n = 6 cows) designed to produce mild or severe NEB. From day 2 after calving, MNEB cows were fed ad libitum grass silage with 8 kg/day of a 21% crude protein dairy concentrate and milked once daily; SNEB cows were fed 25 kg/day silage with 4 kg/day concentrate and milked three times daily. The chemical composition of silage and concentrate offered (as previously described, Ref. 53) was the same across treatment groups. Daily measurements of milk yield, milk composition, dry matter intake, body weight, and dietary energy intake were used to calculate EB, based on the French net energy for lactation (NEL) system. Net EB was calculated as UFL/day in which 1 unité fourragère lait (UFL) is the NEL equivalent of 1 kg of standard air-dry barley as described previously (29). Samples of endometrium were collected from all cows following slaughter at 14 ± 0.4 days postpartum as described below. Array data from one MNEB cow failed the interarray quality control analysis (see below) so this animal was excluded from all analyses, leaving five cows in the MNEB group.
Blood sampling, hormone and metabolite assays.
Blood samples were collected after morning milking (08:00 AM) by jugular venipuncture twice weekly throughout the 2 wk treatment period up to and including the day of slaughter. Samples were collected into lithium heparin-primed vials and were immediately placed on ice before centrifugation at 2,000 g for 10 min. Plasma was decanted and stored at −20°C for subsequent analysis of hormones and metabolites.
IGF-I was measured using human OCTEIA IGF-I kits (IDS, Tyne and Wear, UK). The interassay coefficient of variation (CV) was 8.7%, while the intra-assay CVs were 8.7 and 16.8% for samples with mean values of 81.5 and 4.7 ng/ml, respectively. The sensitivity of the assay was 1.9 ng/ml. Insulin was assayed by a solid-phase radioimmunoassay (RIA; Coat-a-Count, Diagnostics Products). The inter- and intra-assay CV were 14.2 and 9.8%, and 9.8 and 3.9%, for control samples with a mean insulin concentration of 6.4 and 13.1 μIU/ml, respectively. The minimum detectable concentration of the assay was 1.6 ± 0.03 μIU/ml.
Concentrations of the PGF2α metabolite PGFM were quantified using a charcoal-dextran RIA method as described previously (56). In brief, the tritiated tracer of PGFM {13,14-dihydro-15-keto-[5, 6, 8, 9, 11, 12, 14(n)-3H]-prostaglandin F2α} was from Amersham International (Amersham, Bucks, UK). The standards were supplied by Sigma. The PGFM antiserum was a kind gift from Dr. R. Kelly (University of Edinburgh, Edinburgh, UK). The limit of detection was 1 pg/tube and the intra-assay and interassay CVs were 7.6 and 14.3%. The cortisol RIA was based on a similar methodology (56) using plant cortisol (H4001; Sigma-Aldrich) as standard, sheep antiserum to cortisol (Diagnostics Scotland), and [1, 2, 6, 7-3H] cortisol (Amersham Biosciences Biosciences) as tracer. Plasma samples were first extracted with diethyl ether, and the extracts were reconstituted in assay buffer. Separation was by charcoal-dextran. The sensitivity was 0.7 nmol/l plasma, the recovery was 103%, and the intra-assay CVs for samples with a mean of 3.6 and 51.4 cortisol nmol/l of plasma were 5.6 and 6.1%, respectively. The interassay CV was 18.5%. Measurement of plasma estradiol concentrations used the Estradiol MAIA assay kit (BioStat Diagnostic Systems, Stockport, Cheshire, UK). The intra-assay CVs for samples with a mean of 1.1 and 4.6 estradiol pg/ml of plasma were 15.8 and 7.3%, respectively. The interassay CV for the same samples was 0.6 and 5.6%, respectively.
Serum amyloid A (SAA) concentrations were measured in serum using the Tridelta Phase range SAA solid phase ELISA kit according to the manufacturer's guidelines (Tridelta, Kildare, Ireland). The intra- and interassay CV for low, medium, and high concentrations within the effective range were all <12 and <18%, respectively.
Samples of blood plasma obtained on the day of slaughter were also analyzed for glucose, NEFAs, BHB, and urea using appropriate kits and an ABX Mira auto-analyzer (ABX Mira, Cedex 4, France).
Hematology.
Blood samples were taken the morning before slaughter from each cow. Blood parameters were determined in unclotted (EDTA treated) whole blood samples using an electronic particle Nihon Kohden hematology analyzer (Celltac MEK-610K; Nikon-Kohdon, Tokyo, Japan).
Uterine tissue collection and RNA isolation.
The uterus was opened, and samples of intercaruncular endometrial tissue weighing ∼1 g were dissected from the midportion of the previously gravid horn ∼1 cm anterior to the bifurcation of the uterus. These were rinsed in RNase-free phosphate buffer, snap-frozen in liquid nitrogen, and stored at −80°C. Total RNA was prepared from 200–300 mg of fragmented frozen endometrial tissue and homogenized in TRI reagent (Molecular Research Centre, Cincinnati, OH). RNA concentration and purity were determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA integrity was confirmed for all samples using automated capillary gel electrophoresis on a Bioanalyzer 2100 with RNA 6000 Nano Labchips according to manufacturers' instructions (Agilent, Waldbronn, Germany).
Microarray analysis.
Microarray hybridization and data acquisition were carried out in ARK-Genomics (Roslin Institute, Edinburgh, UK) using 24 K Affymetrix GeneChip Bovine Genome Arrays based on the established ARK-Genomics protocols (http://www.ark-genomics.org/protocols). The acquired data were analyzed using S+ Array Analyzer 2.1 built in S-Plus Enterprise Developer 7.0 software package (Insightful, Seattle, WA). The probe level expression data generated by the scanner (.CEL files) were imported into the Array Analyzer. They were filtered out if the detection was absent or if the pairs used <7 (11 pairs in total). The probe pairs were summarized into a single value per gene using robust multichip analysis with a primary Quantiles normalization. After this filtration and summarization, ∼20,000 probes/genes were available. The interarray quality control analysis using MvA and box plots showed that the sample from one cow did not meet the requirements so this animal's data were excluded from all the analyses. The summarized data were further normalized with median interquartile range. The differentially expressed genes were identified using a local pooled error test at P = 0.05 with Bonferroni false discovery rate adjustment. The significantly expressed genes were loaded into the Affymetrix website for annotation (http://www.affymetrix.com). The GEO-deposited data can be accessed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15544.
Pathway analysis.
The annotated genes were organized using Entrez Gene combined with gene symbols as identifiers and fold changes and adjusted P values as observations. They were loaded into Ingenuity Pathway Analysis (IPA) V7.5 software server (Ingenuity, Redwood City, CA) for mapping into relevant functional groups and pathway analysis. Among 274 significantly expressed probes, 197 were found to be previously reported genes with recognized gene symbols. About half of the identified genes (n = 103) had known involvement with immune and inflammatory functions. These were selected and reloaded into IPA for further analysis of immune and inflammatory pathways using gene symbols as identifiers and fold changes and adjusted P values as observations.
Quantitative real-time PCR.
Total RNA from each sample was treated for potential genomic DNA carryover in a single reaction with DNase based on the guidelines supplied by Promega (Promega, Madison, WI). From this reaction, precisely 1 μg of DNase-treated RNA was reverse transcribed into cDNA using random hexamer primers and processed accordingly (Reverse Transcription System Kit; Promega). A master-mix of reagents was prepared for the above reaction to minimize potential variation from pipetting. Selected negative control samples were also prepared by including all reagents as above, minus the reverse transcriptase. Assays were designed for 22 genes of interest (see Supplementary Table S11 ). Three housekeeping genes were also analyzed: 18S rRNA, ribosomal protein L19 (RPL19), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences were designed online using the Primer3 web software (http://frodo.wi.mit.edu/primer3/input.htm) based on the target sequences of Affymetrix GeneChip Bovine Genome Array probes (http://www.affymetrix.com). Primer alignment specificity and compatibility were checked using the BLAST search tool (National Center for Biotechnology Information) and Amplify software (v. 3.1), respectively. Gene symbols, sequence information, accession numbers, and expected product lengths are provided in Supplementary Table S1.
Gene transcripts were quantified as described in detail previously (17). Briefly, for each assay a mastermix was prepared that contained a final concentration of 1× absolute qPCR SYBR Green Mix (ABgene, Epsom, Surrey, UK), 500 nM forward and reverse primers, and nuclease-free water. Primer annealing and amplicon-specific melting temperatures were determined using the gradient function of the DNA Engine Opticon 2 thermal cycler (MJ Research, Waltham, MA). Equivalent amounts of sample cDNA were added to each reaction in duplicate. To minimize variation, all samples included in each analysis were derived from the same RT batch prepared under the same conditions and were analyzed on a single plate. Thermal cycling conditions applied to each assay consisted of an initial Taq activation step at 95°C for 15 min followed by 38 cycles of denaturation (95°C), annealing (range 50.0–64.2°C), extension (72°C), and an amplicon-specific fluorescence acquisition reading (range 74–84°C). A melting curve analysis was performed for each amplicon between 50 and 95°, and as such any smaller nonspecific products such as dimers were melted (if present) prior to fluorescence acquisition. All qPCR results were recorded with the Opticon Monitor Analysis Software (V2.02; MJ Research). Values for r2 and amplification efficiency (E) were derived from the linear regression analysis of Log (input cDNA) vs. cycle number at threshold (Ct) plot. The slope created for each set of standards was used to determine E according to: E = 10−slope − 1, where an E value of 1.0 would correspond to 100% cDNA replication at each cycle. Variation within each assay was determined from the average standard deviation across the whole quantification range and converted to percentage [ng input cDNA] using the equation ± % [ng input cDNA] = [(E+1)SD − 1] × 100% (62). For comparison of expression data, absolute values were derived from standard curves generated from purified cDNAs identical to amplified products and expressed as fg/μg reverse-transcribed RNA.
Data analysis.
Gene expression data from quantitative real-time PCR (qPCR) and values for EB, endocrine, metabolic, and hematological parameters were compared between the two EB groups using independent samples t-test in SPSS for Windows, v. 16.0. Data were log transformed if necessary to achieve homogeneity of variance. Differences were deemed significant where P < 0.05. Array data were analyzed as described above.
RESULTS
Metabolic, hormonal, and EB status of cows in mild and severe NEB.
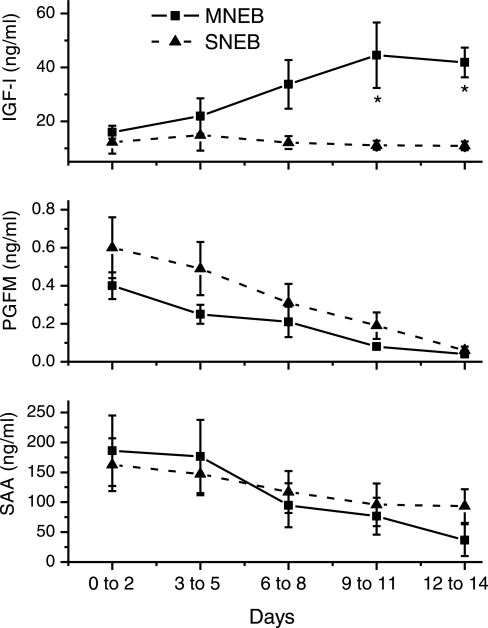
All cows achieved a state of NEB between calving and the time of tissue collection 2 wk later (17). Cows exposed to a restricted dietary intake and increased milking frequency (SNEB) were in a lower EB status compared with MNEB cows at tissue collection (P < 0.02; Table 1). The divergence in EB status at tissue collection was confirmed by changes in measured metabolites: systemic NEFA and BHB were elevated (P < 0.001), while glucose was reduced (P < 0.001) in the SNEB compared with the MNEB group (Table 1). Concentrations of IGF-I were low following calving in both groups, then started to increase again after 1 wk in the MNEB cows, but remained basal in the SNEB group, resulting in a significant difference at tissue collection (P < 0.05, Fig. 1). Although insulin and estradiol levels were also numerically lower in SNEB cows, this did not achieve statistical significance. Concentrations of both SAA and PGFM were elevated at calving in all cows and then fell by the time of tissue collection, but values did not differ significantly between groups (Fig. 1). No cows had ovulated again postpartum at the time of tissue collection, so progesterone levels remained low in all animals.
Table 1.
Mean detectable concentrations of circulating hormones and metabolites at the time of tissue collection in cows maintained under mild or severe NEB
| MNEB | SNEB | P | ||
|---|---|---|---|---|
| No. cows | 5 | 6 | ||
| Days postpartum | 14±0.73 | 14±0.68 | ||
| Energy | Net EB, UFL/day | −1.7±1.17 | −6.1±1.03 | 0.018 |
| Endocrine | IGF-I, ng/ml | 51.4±10.33 | 10.6±1.09 | 0.002 |
| Insulin, μIU/ml | 0.2±0.07 | 0.1±0.02 | 0.12 | |
| PGFM, pg/ml | 34.0±8.72 | 55.0±17.65 | 0.34 | |
| Cortisol, ng/ml | 13.2±5.80 | 5.5±1.61 | 0.24 | |
| Estradiol, pg/ml | 2.2±0.33 | 1.6±0.24 | 0.14 | |
| Metabolism | Glucose, mmol/l | 4.1±0.15 | 2.7±0.15 | <0.001 |
| NEFA, mmol/l | 0.3±0.05 | 1.4±0.14 | <0.001 | |
| BHB, mmol/l | 0.5±0.09 | 3.7±0.20 | <0.001 | |
| SAA, mg/ml | 62±37.1 | 133±29.2 | 0.16 |
Values are means ± SE. EB, energy balance; MNEB, mild negative energy balance; SNEB, severe negative energy balance; UFL, unité fourragère lait; IGF-I, insulin like-growth factor-I; PGFM, 13,14-dihydro-15-keto-prostaglandin F2α; NEFA, nonesterified fatty acid; BHB, β-hydroxybutyrate; SAA, circulating serum amyloid A.
Fig. 1.
Changes in circulating concentrations of: IGF-I (top), PGFM (middle), and serum amyloid A (SAA, bottom) following calving. IGF-I concentrations increased to significantly higher values in mild negative energy balance (MNEB) cows P < 0.05, but PGFM and SAA values did not differ between groups. SNEB, severe negative energy balance.
Hematology.
Blood samples taken on the day before slaughter showed several significant differences in hematological parameters between groups (Table 2). White blood cell (WBC) count and lymphocyte number were both reduced in SNEB cows. The red blood cell distribution width was higher and the platelet crit and mean platelet volume were lower in SNEB cows. Other parameters were not significantly altered.
Table 2.
Comparison of hematological parameters between groups
| MNEB | SNEB | P | |
|---|---|---|---|
| n | 5 | 6 | |
| White blood cell count | 9.4±0.62 | 6.0±0.69 | 0.006 |
| Granulocytes, n | 0.6±0.30 | 0.1±0.04 | 0.12 |
| Granulocytes, % | 6.9±3.92 | 2.2±0.55 | 0.23 |
| Lymphocyte, n | 8.9±0.84 | 5.8±0.70 | 0.021 |
| Lymphocytes, % | 92±4.2 | 95±1.5 | 0.59 |
| Monocytes, n. | 0.06±0.02 | 0.15±0.06 | 0.26 |
| Monocytes, % | 0.7±0.34 | 3.0±1.27 | 0.14 |
| Red blood cell count | 6.3±0.43 | 7.4±0.17 | 0.23 |
| Hemoglobin concentration | 9.8±0.36 | 10.4±0.19 | 0.12 |
| Hematocrit | 26±1.5 | 29±0.6 | 0.11 |
| Mean corpuscular volume | 42±1.1 | 43±0.5 | 0.73 |
| Mean corpuscular hemoglobin | 15.8±0.64 | 15.4±0.33 | 0.53 |
| Red blood cell distribution width | 16.0±0.58 | 17.9±0.52 | 0.035 |
| Platelet crit | 0.23±0.02 | 0.15±0.02 | 0.028 |
| Mean platelet volume | 5.3±0.21 | 4.2±0.36 | 0.033 |
Values are means ± SE. Significant differences are shown in boldface.
Arrays and pathways.
Array analysis of endometrial samples identified 274 differentially expressed probes between the EB groups representing 240 genes of which 197 had recognized gene symbols. Of this total, 176 genes (73%) were significantly upregulated and 64 (27%) were downregulated. Initial analysis of the top 20 differentially expressed genes increased in the SNEB cows revealed upregulation of many key genes known to be involved in inflammatory responses including MMP1, MMP3, and MMP13, CXCL5, HLA-DQB (MHC class II antigen), S100A8, S100A9, and S100A12 (calgranulin A, B, and C), AHSG, IL1R, IL8, and IL8RB (Table 3). These were increased between 28.8- and 6.4-fold in the SNEB group. The most highly downregulated genes (real fold change down −5.9 to −2.6) represented a much more diverse group with no clear theme (Table 4). Seven of the 20 most highly downregulated genes were unidentified. Of particular interest were PLA2G10 (a phospholipase A2 involved in lipid accumulation and modification of lipoproteins), NTRK2 and CCNB1 (involved in cell differentiation and mitosis, respectively), PTHLH (important in epithelial-mesencymal interactions), SLC2A5 (also known as GLUT5, a facilitated glucose/fructose transporter), NOV (influential in cell adhesion), and MYB (involved in cell proliferation and a target for estrogen receptor signaling). IL2 (a cytokine produced by CD4+ T-helper Th1 cells) was also downregulated, contrasting with IL1B, which was increased.
Table 3.
Top 20 genes ranked by real fold change up in endometrium of cows in SNEB compared with MNEB (all P < 0.0001)
| Fold Change Up | Entrez Gene ID | Unigene ID | Gene Symbol | Entrez Gene Name |
|---|---|---|---|---|
| 28.8 | 281308 | Bt.3417 | MMP1 | matrix metallopeptidase 1 |
| 14.8 | 281735 | Bt.7165 | CXCL5 | chemokine (C-X-C motif) ligand 5 |
| 14.0/12.9* | 281309 | Bt.18504 | MMP3 | matrix metallopeptidase 3 (stromelysin 1) |
| 11.3 | 539241 | Bt.350 | HLA-DQB1 | MHC class II, DQ β1 |
| 9.4 | 616818 | Bt.9360 | S100A8 | S100 calcium binding protein A8 |
| 9.4 | 281914 | Bt.39 | MMP13 | matrix metallopeptidase 13 (collagenase 3) |
| 8.3 | 532569 | Bt.87249 | S100A9 | S100 calcium binding protein A9 |
| 8.3 | 515640 | Bt.9175 | IL1R | interleukin 1 receptor |
| 8.2 | 282467 | Bt.357 | S100A12 | S100 calcium binding protein A12 |
| 8.2 | 505080 | Bt.13628 | TGM3 | transglutaminase 3 |
| 8.1 | 280828 | Bt.49740 | IL8 | interleukin 8 |
| 8.0/7.7/6.9* | 280988 | Bt.23250 | AHSG | α-2-HS-glycoprotein |
| 8.0 | 522269 | Bt.13633 | ACTN4 | actinin, α4 |
| 7.6 | 505518 | Bt.6410 | C15H11ORF34 | chromosome 11 open reading frame 34 ortholog |
| 7.4 | 505317 | Bt.19959 | TRPA1 | transient receptor potential cation channel, subfamily A, member 1 |
| 7.4 | 504598 | Bt.5878 | MATN4 | matrilin 4 |
| 7.4/6.1* | 514346 | Bt.56517 | SDS | serine dehydratase |
| 7.3 | 515200 | Bt.23199 | CTSL1 | cathepsin L1 |
| 7.2 | † | Bt.65714 | † | † |
| 6.4 | 281863 | Bt.4208 | IL8RB | interleukin 8 receptor-β |
Represented by >1 probe on the array;
unidentified gene.
Table 4.
Top 20 genes ranked by real fold change down in endometrium of cows in SNEB compared with MNEB (all P < 0.0001)
| Fold Change Down | Entrez Gene ID | Unigene ID | Gene Symbol | Entrez Gene Name |
|---|---|---|---|---|
| −5.9 | † | Bt.1296 | † | † |
| −4.6 | † | Bt.24179 | † | † |
| −4.5 | 613966 | Bt.22381 | PLA2G10 | phospholipase A2, group X |
| −4.0 | 505824 | Bt.64757 | NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 |
| −3.9 | 617336 | Bt.22389 | SH1SA2 | shisa homolog 2 (Xenopus laevis) |
| −3.8 | † | Bt.69297 | † | † |
| −3.8 | † | Bt.26467 | † | † |
| −3.7 | 327679 | Bt.15980 | CCNB1 | cyclin B1 |
| −3.6 | † | Bt.92107 | † | † |
| −3.4 | 286767 | Bt.12848 | PTHLH | parathyroid hormone-like hormone |
| −3.4 | 528174 | Bt.37396 | GPR133 | G protein-coupled receptor 133 |
| −3.3 | 507243 | Bt.22741 | CLIC6 | chloride intracellular channel 6 |
| −3.1 | † | Bt.31265 | † | † |
| −3.1 | 511596 | Bt.41310 | IL2 | interleukin 2 |
| −3.1 | † | Bt.17034 | † | † |
| −2.9/−2.6/−2.6* | 533044 | Bt.13588 | PSAT1 | phosphoserine aminotransferase 1 |
| −2.9 | 282868 | Bt.19805 | SLC2A5 | solute carrier family 2 (facilitated glucose/fructose transporter) member 5 |
| −2.8 | 505727 | Bt.27716 | NOV | nephroblastoma overexpressed gene |
| −2.8 | 317776 | Bt.12781 | MYB | v-myb myeloblastosis viral oncogene homolog (avian) |
| −2.7 | † | Bt.84625 | † | |
| −2.6 | 504879 | Bt.15901 | MGC127236 | amyloid P component-like |
Represented by >1 probe on the array;
unidentified gene.
IPA was used to place all the differentially expressed genes into different function and disease categories. This confirmed that the main canonical pathways and biofunctions affected related to immunological and inflammatory disease, connective tissue disorders and cellular growth, proliferation and interaction (Table 5).
Table 5.
Main functions identified using IPA (all with P value <0.0001)
| Top Canonical Pathways | Ratio |
|---|---|
| Hepatic fibrosis/hepatic stellate cell activation | 10/135 (0.074) |
| Complement system | 6/36 (0.167) |
| Acute phase response signaling | 11/178 (0.062) |
| HIF1α signaling | 8/105 (0.076) |
| Leukocyte extravasation signaling | 10/195 (0.051) |
| Top Biological Functions | Molecules |
|---|---|
| Diseases and disorders | |
| Cancer | 93 |
| Inflammatory response | 58 |
| Reproductive system disease | 48 |
| Renal and urological disease | 26 |
| Hematological disease | 49 |
| Molecular and cellular functions | |
| Cellular movement | 52 |
| Antigen presentation | 53 |
| Cell-to-cell signaling and interaction | 51 |
| Cellular growth and proliferation | 75 |
| Cell death | 70 |
| Physiological system development and function | |
| Tissue morphology | 57 |
| Hematological system development and function | 64 |
| Immune cell trafficking | 40 |
| Cell-mediated immune response | 70 |
| Humoral immune response | 55 |
IPA, Ingenuity Pathway Analysis.
All genes identified as belonging to immune or inflammatory pathways (n = 103, listed in Supplementary Table S2) were further classified into immune subpathways (Table 6). Pathways with the greatest number of molecules represented included acute phase response, complement system, pattern recognition receptors, and leukocyte extravasation signaling. These are all consistent with the processes known to be required to remodel the postpartum uterus and to clear any microbial infections likely to be present (21, 40, 65). In addition the subpathway “hepatic fibrosis” was highly significant. This pathway includes several matrix metalloproteinases (MMPs), as the condition is associated with increased deposition and reduced degradation of collagen in diseased liver.
Table 6.
Top 20 canonical subpathways from IPA analysis (all P < 0.001) associated with immune response differentially influenced by energy balance status in bovine endometrium*
| Subpathway | Gene Symbol |
|---|---|
| Hepatic fibrosis | IL8, VCAM1, CCR5, MMP13, IL1B, IL1R, CCR7, MMP1, MMP9,IGF1 |
| Acute phase response | C3, AHSG, IL1R, MAPK13, HP, SOD2, IL1RN, CFB, IL1B,PIK3R1, CRABP1 |
| Complement system | C3, CFB, C1QA, C1QB, C1QC, CFH |
| Pattern recognition receptors | IFIH1, C3, IL1B, C1QA, C1QB, CIQC,PIK3R1 |
| IL-17 signaling | IL8, MMP3, MAPK13, CXCL5, PTGS2PIK3R1 |
| Airway pathology in chronic obstructive pulmonary disease | IL8, MMP1, MMP9 |
| Leukocyte extravasation signaling | VCAM1, MMP3, MMP13, MAPK13, ACTN4, MMP1, MMP9,PIK3R |
| Primary immunodeficiency signaling | ZAP70, IGHM, CD79A, IGLL1,IGHG1, |
| HIF1α signaling | MMP3, MMP13, MAPK13, MMP9, MMP1,PIK3R |
| IL-10 signaling | CCR5, IL1RN, IL1B, MAPK13, IL1R |
| LXR/RXR activation | IL1RN, CD36, IL1B, IL1R, MMP9 |
| Oncostatin M signaling | MMP3, MMP13, CH13L1, MMP1 |
| Glucocorticoid receptor signaling | VCAM1, IL8, IL1RN, IL1B, MAPK13, MMP1,FKBP4, PIK3R1 |
| Bladder cancer signaling | IL8, MMP3, MMP13, MMP9, MMP1 |
| IL-6 signaling | IL8, IL1RN, IL1B, MAPK13, IL1R |
| NF-κB signaling | IL1RN, ZAP70, IL1B, IL1R, MAP3K8,PIK3R1 |
| HMGB1 signaling | IL8, VCAM1, IL1R, MAPK13,PIK3R1 |
| P38 MAPK signaling | IL1RN, IL1B, MAPK13, IL1R,PLA2G10 |
| Dendritic cell maturation | IL1RN, IL1B, MAPK13, CCR7,PIK3R1, IGHG1 |
| IL-8 signaling | IL8, VCAM1, IL8RA, PTGS2, MMP9,PIK3R1 |
Based on significant differential expression of 103 immune-related genes using Bovine Affymetrix arrays. Genes in boldface decreased in SNEB; remainder increased.
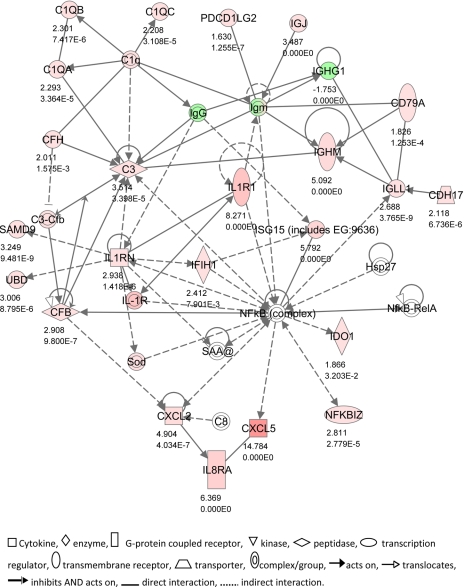
The four top networks identified by IPA are illustrated in Figs. 2–5. These were: 1) antigen presentation, cell-mediated immune response, humoral immune response (score 49, 24 focus molecules); 2) cellular movement, hematological system development and function, immune cell trafficking (score 40, 20 focus molecules); 3) hematological system development and function, humoral immune response, tissue morphology (score 29, 17 focus molecules; and 4) posttranslational modification, cell death, connective tissue development and function (score 23, 13 focus molecules).
Network 1 (Fig. 2) featured a number of genes associated with activation of the complement system (C1QA, C1QB, C1QC, C3, C8, CFB, CFH), which were all upregulated in SNEB cows. CD79A is a B lymphocyte antigen receptor whose activation precedes rearrangement of heavy chain immunoglobulins during B cell ontogeny. This network also included IL1R and the antagonist IL1RN. IL-1 receptor signaling can activate the NF-κB signaling pathway. This is important in macrophages during the respiratory burst and is thought to be a key link between this and other inflammatory responses (28). PDCD1LG2 is a programmed cell death ligand involved in antigen receptor signaling. CXCL5 was one of the most highly upregulated genes in SNEB cows. This is produced with IL-8 in response to stimulation by IL-1 or TNF-α and is a potent chemokine. Expression of another chemokine CXCL2 was also increased, which is important for neutrophil recruitment.
Fig. 2.
IPA Network 1. Differentially regulated genes in endometrium involved in antigen presentation, cell-mediated immune responses, and humoral immune response, with 24 focus molecules and a score of 49. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB endometrium. The fold value and P values are indicated under each node. The shapes of nodes indicate the functional class of the gene product as shown in the key.
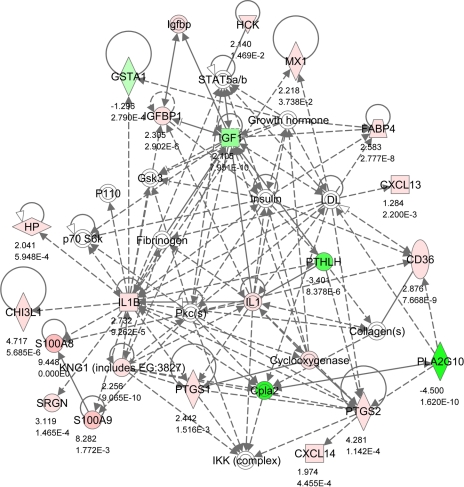
Network 2 (Fig. 3) showed strong associations with the EB model used as the basis of the experiment, in which plasma concentrations of IGF-I were reduced, while NEFA was greatly elevated. Local endometrial expression of IGF-I was also reduced, whereas IGFBP1 was increased in SNEB cows. Associations between these genes are shown with plasma GH and insulin and STAT5, a transcription factor that is phosphorylated in response to cytokine signaling. Network 2 also featured signaling of the proinflammatory cytokine IL-1 and linked this to prostaglandin production, as PTGS1 and PTGS2 were both higher in SNEB cows. CD36 encodes a protein found on platelets that acts as a receptor for thrombospondins, which are involved in cell adhesion processes. CD36 also binds to collagen, anionic phospholipids and oxidized LDL and may play a role in fatty acid transport. The fatty acid binding protein FABP4 was also upregulated whereas GSTA1 (glutathione S-transferase) was decreased. GSTA1 can detoxify products of oxidative stress, providing protection from reactive oxygen species and the products of peroxidation. These changes may be related to the raised NEFA concentrations. CXCL13 and CXCL14 are attractants for B cells and monocytes, respectively. This network also included the antimicrobial calgranulin genes S100A8 and S100A9, which were both highly increased in SNEB.
Fig. 3.
IPA Network 2. Differentially regulated genes in endometrium involved in cellular movement, hematological system development and function and immune cell trafficking with 20 focus molecules and a score of 40. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB endometrium. The fold value and P values are indicated under each node. The shapes of nodes indicate the functional class of the gene product as shown in the key given in Fig. 2. Solid lines indicate a direct interaction, and dotted lines an indirect interaction.
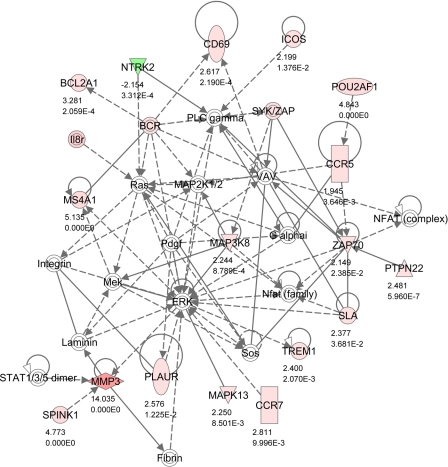
Network 3 (Fig. 4) was also identified as having a role in hematological system development and function. It featured a number of genes involved in T cell development and activation (CD69, CCR5, CCR7, PTPN22, ZAP70). T cells play a key role in inflammation and tissue remodeling through activation of MMPs that break down extracellular matrix. MMP3 encodes an enzyme that degrades fibronectin, laminin, and several types of collagen and is thought to be of particular importance in wound repair. MMP3 is linked to PLAUR, the receptor for urokinase plasminogen activator. PLAU converts plasminogen to plasmin, the active form of a proteolytic enzyme that plays an important initial step in MMP activation. This pathway is activated through a MAP kinase signaling cascade and MAK3K8 and MAPK13 were both upregulated in SNEB.
Fig. 4.
IPA Network 3. Differentially regulated genes in endometrium involved in hematological system development and function, humoral immune response, and tissue morphology with 17 focus molecules and a score of 29. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB endometrium. The fold value and P values are indicated under each node. The shapes of nodes indicate the functional class of the gene product as shown in the key given in Fig. 2. Solid lines indicate a direct interaction, and dotted lines an indirect interaction.
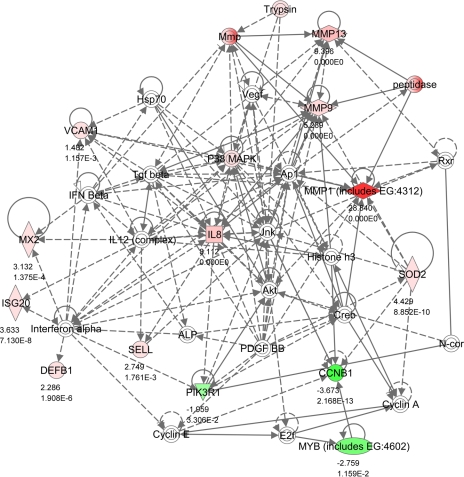
Network 4 (Fig. 5) also relates to connective tissue remodeling. MMP1, MMP9, and MMP13 were all highly upregulated in SNEB endometrium. The uterus must undergo extensive remodeling during the postpartum period. IL-8 is a major mediator of inflammatory responses and is also a potent angiogenic factor. Other genes associated with inflammation were VCAM1 and SELL, which are both involved with leukocyte-endothelial cell adhesion. MYB encodes a transcription factor that regulates progression through the cell cycle, and both this and the cyclin CCNB1 were downregulated in SNEB. Network 4 also featured the interferon-responsive genes MX2 and ISG20, in addition to DEFB1, which can also respond to viral infections. Differential expression of IFN genes themselves between groups was not detected, but three other IFN-inducible genes were differentially increased in endometrium of SNEB cows (MX1, IFIH1, and Loc512486; see Supplementary Table S2).
Fig. 5.
IPA Network 4. Differentially regulated genes in endometrium involved in posttranslational modification, cell death, and connective tissue development and function with 13 focus molecules and a score of 23. The network is displayed graphically as nodes (gene/gene products) and edges (the biological relationship between nodes). The node color intensity indicates the expression of genes: red upregulated, green downregulated in SNEB vs. MNEB endometrium. The fold value and P values are indicated under each node. The shapes of nodes indicate the functional class of the gene product as shown in the key given in Fig. 2. Solid lines indicate a direct interaction, and dotted lines an indirect interaction.
Antimicrobial genes.
A separate search of the entire bovine Affymetrix array was made to identify genes representing antimicrobial peptides. From this 16 genes were identified (5 different β-defensins, 6 different cathelicidins, LAP, LEAP-2, TAP, CAMP, and SLP1). Of these two were differentially expressed using the array analysis: DEFB1 and TAP were increased 2.29-fold and 3.56-fold, respectively, in endometrium of SNEB cows (see Supplementary Table S2). There were trends for several other of these genes to be more highly expressed in SNEB cows [LAP, DEFB5, DEFB7, BBD(C7), SLP1], but these did not achieve statistical significance. On the other hand, none of the cathelicidins (CATHL1, CATHL2, CATHl4, CATHL5, CATHL6, or CATHL7) showed any change in expression between the two groups (data not shown).
Other differentially expressed genes.
Those 137 genes identified from the array analysis as being differentially expressed between groups but which were not placed into the immune and inflammatory categories are listed in Supplementary Table S3. Of these genes 86 were upregulated and 51 were downregulated. They are not discussed any further here to maintain the focus of this paper.
qPCR analysis.
To validate the array data, uterine samples were analyzed by qPCR for 22 genes. The selected list included genes which were identified as being either up- or downregulated or unchanged between groups in the array analysis. Expression data for these genes were compared between the two EB groups (Table 7), and concentrations obtained by qPCR were also compared directly to the array data. Correlation between the two techniques was consistently high, with most genes having r2 values >0.8. Thirteen genes tested showed significantly different values between EB groups by both techniques, and four were nonsignificant by both. Four genes identified as significant using the array data also had fold changes of 1.6- to 2-fold by qPCR, but these were not significant largely due to some within-group variation. One gene (LAP) was shown to be increased 1.9-fold by qPCR (P < 0.04) but was not significant on the array. Expression of the housekeeping genes RPL19, GAPDH, and 18S rRNA was not altered by EB status.
Table 7.
Comparison of some genes analyzed by both qPCR and Affymetix arrays
| Gene | MNEB n = 5 cows | SNEB n = 6 cows | Fold Change in SNEB by qPCR | P for qPCR Data* | Fold Change in SNEB by Array | P for Array Data |
|---|---|---|---|---|---|---|
| MMP1 | 0.2±0.10 | 11.1±6.09 | 55.5 up | 0.003† | 28.8 up | <10−10 |
| MMP3 | 0.1±0.04 | 1.9±0.97 | 19.0 up | 0.009† | 14.0/12.9 up+ | <10−10 |
| IL8 | 2.5±1.3 | 44.3±27.8 | 17.7 up | 0.10† | 8.1 up | <10−10 |
| IL1R | 15±5.9 | 217±115 | 14.5 up | 0.016† | 8.3 up | <10−10 |
| AHSG | 0.1±0.02 | 1.2±0.52 | 12.0 up | 0.009† | 6.9 up | <10−10 |
| IL1B | 0.6±0.13 | 6.5±4.34 | 10.8 up | 0.026† | 2.7 up | <10−5 |
| S100A8 | 0.05±0.02 | 0.36±0.13 | 7.2 up | 0.004 | 9.4 up | <10−10 |
| MMP9 | 0.3±0.11 | 2.0±0.55 | 6.7 up | 0.02 | 5.3 up | <10−10 |
| IL8RB | 1,226±681 | 7486±1847 | 6.1 up | 0.017 | 6.4 up | <10−10 |
| MMP13 | 0.1±0.03 | 0.5±0.25 | 5.0 up | 0.032† | 9.4/5.3 up+ | <10−10 |
| IGFBP1 | 0.5±0.16 | 2.1±0.76 | 4.2 up | 0.062† | 2.3 up | <10−5 |
| CXCL5 | 1.1±0.41 | 3.7±1.34 | 3.4 up | 0.068† | 14.8 up | <10−10 |
| AKR1C4 | 0.8±0.13 | 2.1±0.55 | 2.6 up | 0.037† | 3.2 up | <10−6 |
| PTGS2 | 363±145 | 804±257 | 2.2 up | 0.19 (NS) | 4.3 up | <10−10 |
| IGF1 | 16±4.3 | 8±3.2 | 2.0 down | 0.19 (NS) | 2.1 down | <10−9 |
| DEFB1 | 2.8±0.70 | 4.4±0.79 | 1.6 up | 0.19 (NS) | 2.3 up | <10−5 |
| PDK4 | 29±10.0 | 53±8.6 | 1.8 up | 0.10 (NS) | 3.2 up | 0.04 |
| LAP | 24±5.5 | 45±6.55 | 1.9 up | 0.04 | NS | NS |
| TBXAS1 | 2.2±0.33 | 2.8±0.46 | 1.3 up | 0.30 (NS) | NS | NS |
| DEFB7 | 51±17.1 | 76±13.6 | 1.5 up | 0.28 (NS) | NS | NS |
| DEFB5 | 52±19.1 | 128±47.9 | 2.5 up | 0.20 (NS) | NS | NS |
| IGFBP6 | 5.8±1.12 | 3.0±0.89 | 1.9 down | 0.08 (NS) | NS | NS |
| Housekeeping genes | ||||||
| RPL19 | 28±2.3 | 24±4.5 | 0.81 | |||
| GAPDH | 19±2.6 | 27±7.1 | 0.36 | |||
| 18S rRNA | 72,079±9463 | 72187±9939 | 0.99 |
Comparison was by t-test, those indicated by
used log-transformed data to normalise variances. +, 2 probes on array; NS, not significantly different.
DISCUSSION
All cows deal with postpartum uterine infection initially using the innate immune system, but with time the adaptive response gains importance. The results of this study provided evidence for an ongoing inflammatory response in the uteri of cows in SNEB at 2 wk postpartum. Accompanying this, cows in SNEB had higher expression levels of several antimicrobial genes, notably S100A8, S100A9, and S100A12. In comparison, cows in the MNEB group had nearly recovered from their energy deficit by this stage as evidenced by increased plasma IGF-I and glucose concentrations together with lower NEFA, BHB, and WBC. Several of the genes that were highly downregulated in SNEB are involved in cell proliferation and in interactions between cells (e.g., NTRK2, CCNB1, MYB, NOV). PTHLH is of particular interest as parathyroid hormone-like hormone is known to have a role in epithelial-mesenchymal interactions, for example during development of the mammary gland (16). The postpartum uterus must replace the epithelium that has been lost following placental separation, and this step will be important in re-establishing the innate defense system. Histological analysis confirmed a greater degree of tissue repair in the uterus of MNEB cows (S. Kirton, M. A. Fenwick, D. C. Wathes, unpublished observations). A poor EB status may therefore inhibit the ability of the cow to mount an effective immune response to the bacterial challenge experienced after calving and also delay the general repair process within the endometrium, thus prolonging the time required for the recovery phase. The array data were analyzed and have been presented according to the original experimental design, with six cows in the SNEB group and five in the MNEB group. A cluster analysis based on the 274 differentially expressed genes/probes for all cows indicated that two of the SNEB cows occupied an intermediate position between the two main treatment groups, suggesting that these two animals may have reached a different stage of uterine recovery. They could not, however, be distinguished from the rest of the SNEB group in terms of the blood parameters measured (data not shown). Further study of a larger number of cows will be therefore be required to identify in more detail additional factors that may affect recovery rates in individual animals.
Effects of treatment on metabolism, acute phase response, and oxidative stress.
The SNEB cows had clear evidence of liver damage, associated with lipid infiltration, high circulating NEFAs and BHB, and reduced glucose and IGF-I (17). Although the differential feeding and milking treatments did not begin until after calving, adaptive changes are initiated prepartum. In late pregnancy falling insulin and elevated placental lactogen stimulate adipose mobilization, providing nutrients for fetal growth (2). The main control of the insulin signaling pathway occurs downstream of the insulin receptor. Insulin receptor activation results in tyrosine phosphorylation of IRS proteins-1 and -2 (74). This step is inhibited by lipid accumulation in muscle, contributing to the pregnant mother developing peripheral insulin resistance (34). Insulin signaling was restored within a few days of giving birth in normal weight women, but this took up to 15 wk in obese women (68). Acute infections are also accompanied by tissue insulin resistance (15). AHSG, a natural inhibitor of insulin receptor signaling (46), is a plasma protein, produced primarily by the liver, whose circulating concentration is positively associated with insulin resistance and hepatic fat accumulation (69). In our study uterine expression of AHSG was significantly increased in cows in SNEB, implying that local insulin receptor signaling was likely to be impaired.
Acute-phase proteins are primarily produced by hepatocytes and can directly neutralize inflammatory agents, minimizing the extent of tissue damage. Our results confirmed previous studies in showing a peak in circulating SAA between 1 and 3 days postpartum (64). SAA can affect a variety of cellular functions including adhesion, migration, and proliferation (71). Cows are also under oxidative stress around calving, as indicated by an increase in reactive oxygen metabolites, decreased CuZn-superoxide dismutase (SOD), and raised plasma Se-glutathione peroxidase (GSH-Px) (3). Analysis of livers from the cows in the present study using Affymetrix arrays showed elevated stress response genes (e.g., GPX and HSP70) (47). In the SNEB cows there was altered expression in uterine endometrium of a number of key genes involved in both the acute phase response and Nrf-2-mediated oxidative stress. For example, C3 and SOD2 were increased, whereas expression of PIK3R1, GSTA1, and CRABP1 was decreased. PI3-kinase controls Nrf-2 in response to oxidative stress, and GSTA1 contains functional antioxidant response elements (31). CRABP1 alters cellular responses to retinol. Overall the evidence suggests that there is a complex interrelationship in postpartum cows between acute infection and tissue insulin resistance. Together with the elevated circulating NEFA concentration this may predispose the animals to peroxidative damage of lipids and other macromolecules, chronic inflammation, and a reduced capacity for tissue repair. In addition, previous studies have shown that NEFAs can directly impair the functional capacity of mononuclear cells from sheep (36).
Hematology.
Previous work in cattle has indicated that immune function is suppressed over the periparturient period (7, 32, 43). Blood leukocyte counts decrease in the first 2 wk postpartum then recover over the following 3 wk (67). Increased liver triacylglycerol content in the first 2 wk postcalving is also associated with decreased functional capacities of polymorphonuclear leucocytes derived from both blood and uterus (77, 24). We showed here that the circulating WBC, in particular lymphocyte number, was reduced in SNEB cows. In addition the red blood cell distribution width (RDW) was higher, and the platelet crit and mean platelet volume were lower. In humans low platelet counts and greater RDW are both associated with liver disease (26, 44). In our study the platelet crit was positively associated with circulating IGF-I (r = 0.696, P < 0.02) and negatively with circulating SAA (r = −0.690, P < 0.04), supporting an involvement with liver function in early postpartum cows.
Effects of NEB on the innate and mucosal immune system.
In both the gastrointestinal and urogenital tracts the epithelium normally forms a critical physical and chemical defense barrier that separates the underlying mucosa from the luminal contents and transmits signals generated in response to microbial infection to cells of the innate and acquired immune systems (8, 30, 33, 76). Epithelia prevent adhesion of bacteria and subsequent colonization in part by the release of antimicrobial substances (e.g., lactoferrin, lipocalin) into the overlying mucosal fluid. This system is now known to include antimicrobial peptides such as α- and β-defensins and cathelicidins (8, 76). β-Defensins are found in both macrophages and epithelial cells, have a broad range of antimicrobial activity and are rapidly inducible as an initial part of the innate immune system (12, 58).
Following parturition in the cow there is substantial damage to the surface epithelium (21, 39). In the ewe regeneration of luminal epithelium did not commence until after day 8 and was completed by days 28–31 postpartum (23). There is thus a lengthy period after calving when the normal mucosal defense system is jeopardized. In a previous study, BBD119, BBD123, BBD124, LAP, DEFB7, BNBD4, and BNBD5 were all identified in bovine uterus by qPCR (10). We have here confirmed these findings in the postpartum uterus. Furthermore, we show that expression of DEFB1, TAP, and LAP (significant by qPCR only) was increased in the SNEB cows. Although expression of several members of the cathelicidin family were also readily detectable by the array analysis, none of these were altered by EB, although in ewes the cathelicid bactenecin-1 was upregulated eightfold in cervico-vaginal fluid in association with labor (75).
S100A8, S100A9, and S100A12 are all members of the S100 family of Ca2+ binding proteins. S100A8 and S100A9 together form a heterodimer called calprotectin, which has antimicrobial properties and plays an important role in innate immunity. S100A8 and S100A9 can be produced by a variety of cell types including neutrophils, activated macrophages, keratinocytes, and fibroblasts, and their presence acts as a marker of inflammation in oral mucosa, skin disorders, and wounds (49, 54, 59). Expression in vitro can be upregulated by a variety of proinflammatory cytokines including IFN-γ, TNF-α, IL-1, or FGF2 but is suppressed by TGF-β or retinoic acid (27, 49, 54). Proteomic analysis of human amniotic fluid has identified S100A8 and S100A9 as markers of inflammation associated with intrauterine infection, a major cause of preterm labor in human pregnancies (52, 61). Our data indicate that the members of this family are significantly elevated in postpartum cows in association with uterine infection, and they may thus represent a useful marker of inflammation in this situation.
Effects of NEB on inflammatory responses in uterus.
Bacteria that overwhelm the early mucosal defenses activate an immune response by signaling through receptors such as Toll-like receptor (TLR) 4, which recognize bacterial LPS (11). TLR4 signaling can increase synthesis of β-defensins, whose role is discussed above (20). Some previous studies have also suggested a link between TLR4 signaling and metabolism. For example, LPS can inhibit GHR expression (11, 72). The TLR4 signaling cascade involves MyD88, CD14, IRAK-1, IRAK-4, IRAK-M, and TRAF-6, activating NF-κB and MAPK, leading to cytokine production (20). LPS binds to circulating LPS-binding protein, and this complex binds to cell membrane-bound CD14. With persistence of increasing numbers of microbes, chemokines such as IL-8 and proinflammatory cytokines such as IL-1 and TNF-α upregulate expression of vascular adhesion proteins that attract and activate large numbers of neutrophils, macrophages, and T-lymphocytes (25). Both IL1 and IL8 were upregulated in SNEB cows. IL-1 is a key player in both Network 1 and Network 2. CCL2 and CXCL5 are strong chemoattractants for neutrophils and monocytes and CXCL5 was the second most highly upregulated gene detected (see Table 3) (25). A neutrophilic influx into the superficial endometrium characterizes the early response of the uterus to surface infection, and macrophages, lymphocytes, eosinophils, and mast cells may subsequently be mobilized (4). This is associated with vascular congestion and stromal edema. In this study expression of many chemokines, proinflammatory cytokines and their receptors, and vascular adhesion molecules were all higher in the SNEB cows at 14 days postpartum (see Table 6 and Network 4).
Several genes associated with interferons were upregulated in SNEB cows: MX1, MX2, IFIH1, ISG20, GBP1, and Loc512486. The type 1 interferons IFN-α and IFN-β increase in response to many viral infections, have potent antiviral activities, and can also promote expression of IFN-γ in T cells. They increase cellular responsiveness to other stimuli, including LPS, but can also produce excessive responses leading to tissue damage (27). MX1 and guanylate-binding protein (GBP) 1 both have antiviral activity, and GBP1 also has potent antiangiogenic activity in endothelial cells (50). Loc512486 on the array is identified as being similar to GBP1. Viral causes of uterine disease are rarely considered in cattle, but there is some evidence to associate bovine herpesvirus 4 with bovine metritis (13). Antimicrobial polypeptides such as defensins provide initial mucosal protection against viruses. In humans, impairment of this system can increase susceptibility to HIV-1 infection in the cervix and vagina (9).
Tissue repair.
Many of the other genes showing greatest differential expression between the two groups of cows have previously been associated with tissue remodeling and inflammatory responses (see Networks 3 and 4). MMP1, MMP3, MMP9, and MMP13 were all highly upregulated in the SNEB group. In human uterus MMP1, MMP3, MMP7, and MMP9 mRNAs all increase at menstruation when progesterone withdrawal is an important stimulus (63). In this situation MMP7 is produced by epithelial cells, MMP1 and MMP3 by stromal cells, and MMP9 by migratory immune cells, attracted by chemokines. Mast cell activation releases the proinflammatory cytokines IL-1 and TNF-α (which increase stromal cell MMP1 and MMP3 production) and proteases (which releases active MMPs from pro-MMPs), leading to breakdown of the extracellular matrix (63). Plasmin is one such proteolytic enzyme, and the increased expression of PLAUR is indicative of activation of this system.
MMP expression is also related to that of AHSG, another highly upregulated gene in SNEB cows. In addition to its role in insulin signaling discussed above, AHSG is found on the cell surface where it functions to anchor other molecules to the plasma membrane (38). It interacts with a variety of MMPs, both activating them and protecting them from autolytic cleavage (35, 55). In bone MMP2 is an important activator of IGFBPs and its production is stimulated by IGF-II and TGF-β (48). IGFBP1 expression was increased in SNEB cows and we have previously shown that IGF-II is highly expressed in endometrial stroma (41), although expression was not affected by EB status. Together these data suggest that IL-1 signaling in the postpartum uterus induces MMP expression and activation. The much higher levels of MMP expression in the SNEB cows suggests that the necessary remodeling of the uterus following calving, which requires extensive tissue breakdown, was proceeding more slowly in SNEB than in MNEB cows.
Conclusions
In conclusion, available evidence suggests that there is a complex interrelationship in the postpartum uterus between acute infection, a predisposition to chronic inflammation, and a reduced capacity for tissue repair. The ability to clear the infection is impaired in cows in SNEB, thus also delaying the repair processes. Such effects may be mediated directly by altered concentrations of metabolic hormones and metabolites acting on the uterine cellular mechanisms. Indirect effects are also likely, as cows in SNEB are predisposed to liver damage and increased peripheral insulin resistance, both of which can have a negative impact on the immune system. Our work thus supports previous suggestions that uterine involution and elimination of contaminant bacteria will be delayed in animals in NEB postcalving (40). An increased rate of uterine involution is associated with earlier resumption of ovarian activity (45). Conversely, endometrial damage associated with subclinical endometritis delays cervical involution, disrupts the preovulatory LH surge, and perturbs embryo survival, leading to prolonged intervals to conception with many cows failing to conceive at all (5, 22, 51, 65).
GRANTS
This work was supported by the Wellcome Trust UK and the Irish National Development Plan.
Supplementary Material
Acknowledgments
The authors are grateful to the following for assistance: Alison Downing, ARK-Genomics, Edinburgh, UK for performing the array hybridizations; Dr. Erin Williams, Royal Veterinary College for assistance with the SAA ELISAs; Andrew Clemson, Royal Veterinary College for assistance with some of the qPCR; Prof. Martin Sheldon, University of Swansea for helpful discussions; and Dr. Kieran Meade, Trinity College, Dublin for providing information on the β-defensin genes.
Current addresses: M. A. Fenwick, Inst. of Reproductive and Developmental Biology, Div. of Surgery, Oncology, Reproductive Biology and Anaesthetics (SORA), Faculty of Medicine, Imperial College London, Hammersmith Hospital, Du Cane Rd., London W12 0NN, UK; R. Fitzpatrick, Boston Scientific Galway, Ballycleary, Kinvara, Co. Galway, Ireland.
Address for reprint requests and other correspondence: D. C. Wathes, Reproduction Group, Dept. of Veterinary Basic Sciences, Royal Veterinary College, London, UK (e-mail: dcwathes@rvc.ac.uk).
Footnotes
The online version of this article contains supplemental data.
REFERENCES
- 1.Bauman DE, Currie WB. Partitioning of nutrients during pregnancy and lactation: a review of mechanisms involving homeostasis and homeorhesis. J Dairy Sci 63: 1514–1529, 1980. [DOI] [PubMed] [Google Scholar]
- 2.Bell AW. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci 73: 2804–2819, 1995. [DOI] [PubMed] [Google Scholar]
- 3.Bernabucci U, Ronchi B, Lacetera N, Nardone A. Influence of body condition score on relationships between metabolic status and oxidative stress in periparturient dairy cows. J Dairy Sci 88: 2017–2026, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Bondurant RH. Inflammation in the bovine female reproductive tract. J Anim Sci 77, Suppl 2: 101–110, 1999. [DOI] [PubMed] [Google Scholar]
- 5.Borsberry S, Dobson H. Periparturient diseases and their effect on reproductive performance in five dairy herds. Vet Rec 124: 17–219, 1989. [DOI] [PubMed] [Google Scholar]
- 6.Butler WR. Energy balance relationships with follicular development, ovulation and fertility in postpartum dairy cows. Livest Prod Sci 83: 211–221, 2003. [Google Scholar]
- 7.Cai TQ, Weston PG, Lund LA, Brodie B, McKenna DJ, Wagner WC. Association between neutrophil functions and periparturient disorders in cows. Am J Vet Res 55: 934–943, 1994. [PubMed] [Google Scholar]
- 8.Cole AM. Innate host defence of human vaginal and cervical mucosae. Curr Top Microbiol Immunol 306: 199–230, 2006. [PubMed] [Google Scholar]
- 9.Cole AM, Cole AL. Antimicrobial polypeptides are key anti-HIV-1 effector molecules of cervicovaginal host defense. Am J Reprod Immunol 59: 27–34, 2008. [DOI] [PubMed] [Google Scholar]
- 10.Cormican P, Meade KG, Cahalane S, Narciandi F, Chapwanya A, Lloyd AT, O'Farrelly C. Evolution, expression and effectiveness in a cluster of novel bovine beta-defensins. Immunogenetics 60: 147–156, 2008. [DOI] [PubMed] [Google Scholar]
- 11.Dejkhamron P, Thimmarayappa J, Kotlyarevska K, Sun J, Lu C, Bonkowski EL, Denson LA, Menon RK. Lipopolysaccharide (LPS) directly suppresses growth hormone receptor (GHR) expression through MyD88-dependent and -independent Toll-like receptor-4/MD2 complex signaling pathways. Mol Cell Endocrinol 274: 35–42, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond G, Kaiser V, Rhodes J, Russell JP, Bevins CL. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect Immun 68: 113–119, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donofrio G, Herath S, Sartori C, Cavirani S, Flammini CF, Sheldon IM. Bovine herpesvirus 4 is tropic for bovine endometrial cells and modulates endocrine function. Reproduction 134: 183–197, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drackley JK. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci 82: 2259–2273, 1999. [DOI] [PubMed] [Google Scholar]
- 15.Drobny EC, Abramson EC, Baumann G. Insulin receptors in acute infection: a study of factors conferring insulin resistance. J Clin Endocrinol Metab 58: 710–716, 1984. [DOI] [PubMed] [Google Scholar]
- 16.Dunbar ME, Wysolmerski JJ. Parathyroid hormone-related protein: a developmental regulatory molecule necessary for mammary gland development. J Mammary Gland Biol Neoplasia 4: 21–23, 1999. [DOI] [PubMed] [Google Scholar]
- 17.Fenwick MA, Fitzpatrick R, Kenny DA, Diskin MG, Patton J, Murphy JJ, Wathes DC. Interrelationships between negative EB (NEB) and IGF regulation in liver of lactating dairy cows. Domest Anim Endocrinol 34: 31–44, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Foldi J, Kulcsar M, Pecsi A, Huyghe B, de Sa C, Lohuis JA, Cox P, Huszenicza G. Bacterial complications of postpartum uterine involution in cattle. Anim Reprod Sci 96: 265–281, 2006. [DOI] [PubMed] [Google Scholar]
- 19.Fourichon C, Seegers H, Malher X. Effects of disease on reproduction in the dairy cow. A meta-analysis. Theriogenology 53: 1729–1759, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Froy O, Hananel A, Chapnik N, Madar Z. Differential effect of insulin treatment on decreased levels of beta-defensins and Toll-like receptors in diabetic rats. Mol Immunol 44: 796–802, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Gier HT, Marion GB. Uterus of the cow after parturition: involutional changes. Am J Vet Res 29: 83–96, 1968. [PubMed] [Google Scholar]
- 22.Gilbert RO, Shin ST, Guard CL, Erb HN, Frajblat M. Prevalence of endometritis and its effects on reproductive performance of dairy cows. Theriogenology 64: 1879–1888, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Gray CA, Stewart D M, Johnson GA, Spencer TE. Postpartum uterine involution in sheep: histoarchitecture and changes in endometrial gene expression. Reproduction 125: 185–198, 2003. [PubMed] [Google Scholar]
- 24.Hammon DS, Evjen IM, Dhiman TR, Goff JP, Walters JL. Neutrophil function and energy status in Holstein cows with uterine health disorders. Vet Immunol Immunopathol 113: 21–29, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Harada A, Sekido N, Akahoshi T, Wada T, Mukaida N, Matsushima K. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J Leukoc Biol 56: 559–564, 1994. [PubMed] [Google Scholar]
- 26.Hay JE. Liver disease in pregnancy. Hepatology 47: 1067–1076, 2008. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Chavravarty SD, Ivashiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunologic Rev 226: 41–56, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iles KE, Forman HJ. Macrophage signaling and respiratory burst. Immunologic Res 26: 95–105, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Jarrige J. INRAtion V2.7: Microsoft computer program of ration formulation for ruminant livestock. In: Logiciel de Rationnement des Ruminants, edited by Agabriel J, Champciaux P, Espinasse C. Dijon, France: CNERTA, 1989.
- 30.Kagnoff MF. Microbial-epithelial cell crosstalk during inflammation: the host response. Ann NY Acad Sci 1072: 313–320, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Kang KW, Lee SJ, Kim SG. Molecular mechanism of nrf2 activation by oxidative stress. Antioxid Redox Signal 7: 1164–1173, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Kehrli ME, Burton JL, Nonnecke BJ, Lee EK. Effects of stress on leukocyte trafficking and immune responses: implications for vaccination. Adv Vet Med 41: 61–81, 1999. [DOI] [PubMed] [Google Scholar]
- 33.King AE, Critchley HO, Kelly RW. Innate immune defences in the human endometrium. Reprod Biol Endocrinol 1: 116, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirwan JP, Varastehpour A, Jing M, Presley L, Shao J, Friedman JE, Catalano PM. Reversal of insulin resistance postpartum is linked to enhanced skeletal muscle insulin signaling. J Clin Endocrinol Metab 89: 4678–4684, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Kübler D, Gosenca D, Wind M, Heid H, Friedberg I, Jahnen-Dechent W, Lehmann WD. Proteolytic processing by matrix metalloproteinases and phosphorylation by protein kinase CK2 of fetuin-A, the major globulin of fetal calf serum. Biochimie 89: 410–418, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Lacetera N, Franci O, Scalia D, Bernabucci U, Ronchi B, Nardone A. Effects of nonesterified fatty acids and beta-hydroxybutyrate on functions of mononuclear cells obtained from ewes. Am J Vet Res 63: 414–418, 2002. [DOI] [PubMed] [Google Scholar]
- 37.Lacetera N, Scalia D, Bernabucci U, Ronchi B, Pirazzi D, Nardone A. Lymphocyte functions in overconditioned cows around parturition. J Dairy Sci 88: 2010–2016, 2005. [DOI] [PubMed] [Google Scholar]
- 38.Leite-Browning ML, McCawley LJ, Choi OH, Matrisian LM, Ochieng J. Interactions of alpha2-HS-glycoprotein (fetuin) with MMP-3 and murine squamous cell carcinoma cells. Int J Oncol 21: 965–971, 2002. [PubMed] [Google Scholar]
- 39.Leslie KE. The events of normal and abnormal postpartum endocrinology and uterine involution in dairy cows: a review. Can Vet J 24: 67–71, 1983. [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis GS. Uterine health and disorders. J Dairy Sci 80: 984–994, 1997. [DOI] [PubMed] [Google Scholar]
- 41.Llewellyn S, Fitzpatrick R, Kenny DA, Patton J, Wathes DC. Endometrial expression of the insulin-like growth factor system during uterine involution in the postpartum dairy cow. Domest Anim Endocrinol 34: 391–402, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loor JJ, Dann HM, Guretzky NA, Everts RE, Oliveira R, Green CA, Litherland NB, Rodriguez-Zas SL, Lewin HA, Drackley JK. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiol Genomics 27: 29–41, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Mallard BA, Dekkers JC, Ireland MJ, Leslie KE, Sharif S, Vankampen CL, Wagter L, Wilkie BN. Alteration in immune responsiveness during the peripartum period and its ramification on dairy cow and calf health. J Dairy Sci 81: 585–595, 1998. [DOI] [PubMed] [Google Scholar]
- 44.Maruyama S, Hirayama C, Yamamoto S, Koda M, Udagawa A, Kadowaki Y, Inoue M, Sagayama A, Umeki K. Red blood cell status in alcoholic and non-alcoholic liver disease. J Lab Clin Med 138: 332–337, 2001. [DOI] [PubMed] [Google Scholar]
- 45.Mateus L, Lopes da Costa L, Bernardo F, Robalo Silva J. Influence of puerperal uterine infection on uterine involution and postpartum ovarian activity in dairy cows. Reprod Domest Anim 37: 31–35, 2002. [DOI] [PubMed] [Google Scholar]
- 46.Mathews ST, Chellam N, Srinivas PR, Cintron VJ, Leon MA, Goustin AS, Grunberger G. Alpha2-HSG, a specific inhibitor of insulin receptor autophosphorylation, interacts with the insulin receptor. Mol Cell Endocrinol 164: 87–98, 2000. [DOI] [PubMed] [Google Scholar]
- 47.McCarthy SD, Morris DG, Diskin MG, Kenny D, Patton J, Murphy JJ, Fenwick M, Wathes DC, Fitzpatrick R. Negative energy balance alters hepatic gene expression and key biological processes in dairy cows during the early post partum period. In: Proc Irish Agr Res Forum [http://www.agresearchforum.com/publicationsarf/2007/page%20030.pdf].
- 48.Minuto F, Palermo C, Arvigo M, Barreca AM. The IGF system and bone. J Endocrinol Invest 28, Suppl: 8–10, 2005. [PubMed] [Google Scholar]
- 49.Mørk G, Schjerven H, Mangschau L, Søyland E, Brandtzaeg P. Proinflammatory cytokines upregulate expression of calprotectin (L1 protein,MRP-8/MRP-14) in cultured human keratinocytes. Br J Dermatol 149: 484–491, 2003. [DOI] [PubMed] [Google Scholar]
- 50.Naschberger E, Werner T, Vicente AB, Guenzi E, Topolt K, Leubert R, Lubeseder-Martellato C, Nelson PJ, Sturzl M. Nuclear factor-κB motif and interferon-α stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem J 379: 409–420, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Opsomer G, Gröhn YT, Hertl J, Coryn M, Deluyker H, de Kruif A. Risk factors for post partum ovarian dysfunction in high producing dairy cows in Belgium: a field study. Theriogenology 53: 841–857, 2000. [DOI] [PubMed] [Google Scholar]
- 52.Park SJ, Yoon WG, Song JS, Jung HS, Kim CJ, Oh SY, Yoon BH, Jung G, Kim HJ, Nirasawa T. Proteome analysis of human amnion and amniotic fluid by two-dimensional electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proteomics 6: 349–363, 2006. [DOI] [PubMed] [Google Scholar]
- 53.Patton J, Kenny DA, Mee JF, O'Mara FP, Wathes DC, Cook M, Murphy JJ. Effect of milking frequency and diet on milk production, energy balance, and reproduction in dairy cows. J Dairy Sci 89: 1478–1487, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahimi F, Hsu K, Endoh Y, Geczy CL. FGF-2, IL-1beta and TGF-beta regulate fibroblast expression of S100A8. FEBS J 272: 2811–2827, 2005. [DOI] [PubMed] [Google Scholar]
- 55.Ray S, Lukyanov P, Ochieng J. Members of the cystatin superfamily interact with MMP-9 and protect it from autolytic degradation without affecting its gelatinolytic activities. Biochim Biophys Acta 1652: 91–102, 2003. [DOI] [PubMed] [Google Scholar]
- 56.Robinson RS, Pushpakumara PG, Cheng Z, Peters AR, Abayasekara DR, Wathes DC. Effects of dietary polyunsaturated fatty acids on ovarian and uterine function in lactating dairy cows. Reproduction 124: 119–131, 2002. [PubMed] [Google Scholar]
- 57.Roche JF. The effect of nutritional management of the dairy cow on reproductive efficiency. Anim Reprod Sci 96: 282–296, 2006. [DOI] [PubMed] [Google Scholar]
- 58.Roosen S, Exner K, Paul S, Schröder JM, Kalm E, Looft C. Bovine beta-defensins: identification and characterization of novel bovine beta-defensin genes and their expression in mammary gland tissue. Mamm Genome 15: 834–842, 2004. [DOI] [PubMed] [Google Scholar]
- 59.Ross KF, Herzberg MC. Calprotectin expression by gingival epithelial cells. Infect Immun 69: 3248–3254, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowson LE, Lamming GE, Fry RM. Influence of ovarian hormones on uterine infection. Nature 171: 749–750, 1953. [DOI] [PubMed] [Google Scholar]
- 61.Rüetschi U, Rosén A, Karlsson G, Zetterberg H, Rymo L, Hagberg H, Jacobsson B. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. J Proteome Res 4: 2236–2242, 2005. [DOI] [PubMed] [Google Scholar]
- 62.Rutledge RG, Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31: e93, 2003. [DOI] [PMC free article] [PubMed]
- 63.Salamonsen LA, Zhang J, Brasted M. Leukocyte networks and human endometrial remodelling. J Reprod Immunol 57: 95–108, 2002. [DOI] [PubMed] [Google Scholar]
- 64.Sheldon IM. The postpartum uterus. Vet Clin Food Anim 20: 569–591, 2004. [DOI] [PubMed] [Google Scholar]
- 65.Sheldon IM, Dobson H. Postpartum uterine health in cattle. Anim Reprod Sci 82–83: 295–306, 2004. [DOI] [PubMed]
- 66.Sheldon IM, Lewis GS, Leblanc S, Gilbert RO. Defining postpartum uterine disease in cattle. Theriogenology 65: 1516–1530, 2006. [DOI] [PubMed] [Google Scholar]
- 67.Singh J, Murray RD, Mshelia G, Woldehiwet Z. The immune status of the bovine uterus during the peripartum period. Vet J 175: 301–309, 2008. [DOI] [PubMed] [Google Scholar]
- 68.Sivan E, Chen X, Homko CJ, Reece EA, Boden G. Longitudinal study of carbohydrate metabolism in healthy obese pregnant women. Diabetes Care 20: 1470–1475, 1997. [DOI] [PubMed] [Google Scholar]
- 69.Stefan N, Hennige AM, Staiger H, Machann J, Schick F, Krober SM, Machicao F, Fritsche A, Haring HU. Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 29: 853–857, 2006. [DOI] [PubMed] [Google Scholar]
- 70.Taylor VJ, Cheng Z, Pushpakumara PG, Beever DE, Wathes DC. Relationships between the plasma concentrations of insulin-like growth factor-I in dairy cows and their fertility and milk yield. Vet Rec 155: 583–588, 2004. [DOI] [PubMed] [Google Scholar]
- 71.Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol 7: 64–69, 2000. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Jiang J, Warram J, Baumann G, Gan Y, Menon RK, Denson LA, Zinn KR, Frank SJ. Endotoxin-induced proteolytic reduction in hepatic growth hormone receptor: a novel mechanism for GH insensitivity. Mol Endocrinol 22: 1427–1437, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wathes DC, Cheng Z, Bourne N, Taylor VJ, Coffey MP, Brotherstone S. Differences between primiparous and multiparous dairy cows in the inter-relationships between metabolic traits, milk yield and body condition score in the periparturient period. Domest Anim Endocrinol 33: 203–225, 2007. [DOI] [PubMed] [Google Scholar]
- 74.White MF. Regulating insulin signaling and beta-cell function through IRS proteins. Can J Physiol Pharmacol 84: 725–737, 2006. [DOI] [PubMed] [Google Scholar]
- 75.Young IR, Rice GE, Palliser HK, Ayhan M, Dellios NL, Hirst JJ. Identification of bactenecin-1 in cervicovaginal fluid by two-dimensional electrophoresis in an ovine model of preterm labour. Proteomics 7: 281–288, 2007. [DOI] [PubMed] [Google Scholar]
- 76.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol 18: 2810–2816, 2007. [DOI] [PubMed] [Google Scholar]
- 77.Zerbe H, Schneider N, Leibold W, Wensing T, Kruip TA, Schuberth HJ. Altered functional and immunophenotypical properties of neutrophilic granulocytes in postpartum cows associated with fatty liver. Theriogenology 54: 771–786, 2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.