Abstract
Abasic sites and single base bulges are thermodynamically destabilizing DNA defects that can lead to cancerous transformations if left unrepaired by the cell. Here we discuss the binding properties with abasic sites and single base bulges of Rh(bpy)2(chrysi)3+, a complex previously shown to bind thermodynamically destabilized mismatch sites via metalloinsertion. Photocleavage experiments show that Rh(bpy)2(chrysi)3+ selectively binds abasic sites with affinities of 1-4 × 106 M-1; specific binding is independent of unpaired base identity but is somewhat contingent on sequence context. Single base bulges are also selectively bound and cleaved, but in this case, the association constants are significantly lower (~105 M-1), and the binding is dependent both on unpaired base identity and bulge sequence context. A wide variety of evidence, including strand scission asymmetry, binding enantiospecificity, and MALDI-TOF cleavage fragment analysis, suggests that Rh(bpy)2(chrysi)3+ binds abasic sites, like mismatches, through insertion of the bulky chrysi ligand into the base pair stack from the minor groove side and ejection of the unpaired base. At single base bulge sites, a similar, though not identical, metalloinsertion mode is suggested. The recognition of abasic sites and single base bulges with bulky metalloinsertors holds promise for diagnostic and therapeutic applications.
Genomic integrity is of paramount importance to cellular survival and replication. However, a wide variety of agents, ranging from genotoxic chemicals to error-prone cellular processes, render DNA dangerously susceptible to damage and mutation.1 The types of DNA defects are as varied as their causative agents, yet the most common forms are single base mismatches, abasic sites, single base bulges, and oxidized bases. Left unrepaired, all of these defects can lead to deleterious mutations, often in the form of single nucleotide polymorphisms. To counter these threats, the cell has evolved complex DNA repair machineries, most notably the mismatch repair (MMR) and base excision repair (BER) pathways.2,3 Under normal conditions, the MMR (mismatches and single base bulges) and BER (abasic sites and oxidized bases) machineries will quickly and efficiently repair their target defects, thereby preventing any lasting damage to the cell or its genome. However, the suppression or disabling of these pathways is often met with dire consequences: mismatch repair deficiency, for example, has been implicated in 80% of hereditary non-polyposis colon cancers in addition to significant percentages of breast, ovarian, and skin cancers.4-6 It thus becomes clear that the synthesis and study of molecules able to specifically target these defects may aid in the development of new cancer diagnostics and therapeutics.7
The design and application of metal complexes capable of specifically targeting one such defect, single base mismatches, have been focuses of our laboratory for over a decade.8 These metal complexes, most notably Rh(bpy)2(chrysi)3+ (chrysi = chrysene-5,6-quinone diimine) and Rh(bpy)2(phzi)3+ (phzi = benzo[a]phenazine-5,6-quinone diimine) (Figure 1), bear sterically bulky ligands that are too wide to fit between matched base pairs and thus instead preferentially target thermodynamically destabilized mismatched sites.9,10 The compounds are highly specific (≥ 1000-fold) for mispaired sites over matched base pairs and recognize over 80% of mismatches in all possible sequence contexts, with only thermodynamically stable, G-containing mismatches escaping binding altogether.11 Furthermore, the complexes can, upon irradiation with ultraviolet light, promote direct cleavage of the DNA backbone at the binding site. More recently, crystallography and NMR studies have revealed that these complexes do not bind via classical intercalation, in which the complex binds from the major groove and increases the base pair rise by stacking an aromatic ligand between intact base pairs. Rather, they employ a unique binding mode that we have termed metalloinsertion: the complex binds the DNA from the minor groove, ejecting the mismatched bases into the major groove and replacing them in the base stack with the sterically expansive aromatic ligand.12,13 These structural data make quite clear the origin of the correlation between recognition and thermodynamic destabilization: the less stable the mismatch, the easier the ejection of the mismatched bases.
Figure 1.
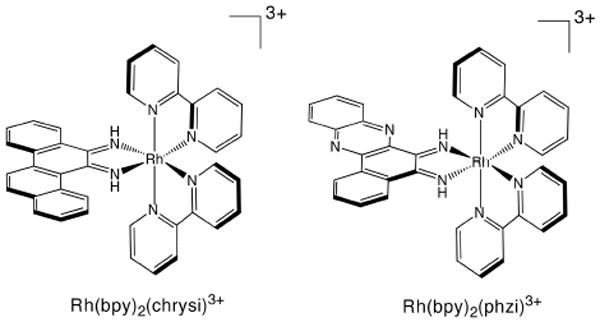
Δ-Rh(bpy)2(chrysi)3+ and Δ-Rh(bpy)2(phzi)3+
Yet mismatches are not the only destabilizing DNA defect. Indeed, the relationship between thermodynamic instability and specific metalloinsertor binding has led our laboratory to investigate the recognition of two different DNA defects: abasic sites and single base bulges. Abasic sites arise from the cleavage of the glycosidic bond between the ribose and the nucleobase; this can occur spontaneously, as a result of exogenous agents, or as an intermediate in the BER pathway (Figure 2).14 In the cell, abasic sites exist primarily in equilibrium between two hemiacetal anomers; just as important to the structure of the site, however, is the unpaired base complementary to the abasic site. Numerous structural studies have shown that the conformation of this unpaired base can be extra- or intrahelical depending upon its identity and the surrounding bases.15-17 Unpaired purines are almost always intrahelical, whereas unpaired pyrimidines likely exist in equilibrium between extrahelical and intrahelical forms, with the extrahelical form favored when the base is flanked by other pyrimidines. Relative to intact duplex DNA, abasic sites are thermodynamically destabilized by 3-11 kcal/mol.18,19 Both the sequence context and the identity of the unpaired base play roles in the magnitude of the destabilization: sites in which the abasic ribose is flanked by purines are more stable than those flanked by pyrimidines, and, to a lesser degree, sites with unpaired purines are more stable than those with unpaired pyrimidines.
Figure 2.
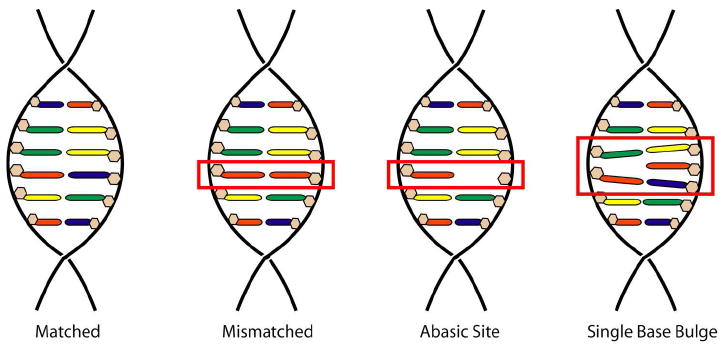
Schematic illustration of structural relationship between matched, mismatched, abasic, and single base bulge sites
Single base bulges are defects in which a base is inserted in one strand of an otherwise well matched duplex. Caused by errors in recombination and replication, these sites are more thermodynamically stable than abasic sites, with destabilizations ranging from 0-3 kcal/mol.20 Recent computational and spectroscopic studies have shown that while bulged base identity and sequence context certainly influence the destabilization of the site, reliable patterns such as those for abasic sites do not exist.21 Several structural studies have shown that the unpaired base may be intra- or extrahelical.22-24 Similar to the case for abasic sites, unpaired purines are almost always intrahelical, whereas an equilibrium between intra- and extrahelical conformations is likely for unpaired pyrimidines. Unpaired bases flanked by purines are more likely to remain intrahelical than those surrounded by pyrimidines. Regardless of unpaired base helicity, all duplexes with single base bulges are bent relative to well-matched DNA.
Under normal conditions, abasic sites and single base bulges are repaired through the BER and MMR pathways, respectively. However, if left unrepaired, both lesions represent significant threats to cell viability: abasic sites can lead to single nucleotide polymorphisms, block transcription and replication, and act as a potent topoisomerase poison,25 while single base bulges are a common source of frame-shift mutations.26 Indeed, deficiency in the repair of both types of lesions has been associated with several different cancers.27-30
Given the links to cancer, it is not surprising that agents that recognize these lesions have already been designed and studied. Methodologies for the targeting of abasic sites include organic bis-intercalators31 and nucleophilic amines32 that react with the minor aldehylic form of the natural abasic site. Organic agents have also been designed for single base bulge recognition.33,34 Other recognition agents resemble metalloinsertors; dinuclear ruthenium35 and octahedral cobalt36 compounds have been shown to bind multiple base bulges along with DNA hairpins. Yet despite some successes, almost all of these recognition agents exhibit affinities, specificities, or reactivities that are less than ideal for diagnostic or therapeutic applications.
Our investigation of metalloinsertors for abasic site and single base bulge recognition is thus motivated both by the desire to augment our understanding of DNA lesion recognition by metal complexes and by the opportunity to create a useful diagnostic reagent for the detection of these deleterious defects. We have previously communicated our initial finding that bulky metalloinsertors specifically bind and photocleave abasic sites and single base bulges.37 Here, we present a more comprehensive study geared at elucidating the scope and means of recognition at both types of defects.
EXPERIMENTAL
Reagents, instrumentation, and general methods
All reagents were the highest purity commercially available and, unless otherwise noted, were used as obtained without further purification. Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+ were synthesized and purified as previously reported.38 The enantiomers of Rh(bpy)2(chrysi)3+ were likewise resolved as described earlier. Standard oligonucleotides were synthesized from phosphoramidites on an ABI 3400 DNA synthesizer (reagents from Glen Research). Given the instability of the natural hemiacetal abasic lesion, the often employed tetrahydrofuranyl abasic site analogue was used instead.39 In all text, the symbol Φ denotes the abasic site. Abasic site-containing oligonucleotides were ordered from Integrated DNA Technologies. Following synthesis or delivery, the oligonucleotides were purified both with and without dimethoxytrityl (DMT) protecting groups via reverse phase HPLC [HP1100 HPLC system with Varian DynaMax™ C18 semi-preparative column, gradient of 5:95 to 45:55 MeCN:50 mM NH4OAc (aq) over 30 min for DMT-on purification and 2:98 to 17:83 MeCN:50 mM NH4OAc (aq) over 30 min for DMT-off purification]. UV-Vis absorption spectra were taken on a Beckman DU 7400 spectrophotometer.
Metal complex concentrations were determined using UV-visible spectrophotometry with extinction coefficients of ε302 = 57,000 cm-1M-1 and ε315 = 52,200 cm-1M-1 for Rh(bpy)2(chrysi)3+ and ε304 = 65,800 cm-1M-1 and ε314 = 67,300 cm-1M-1 for Rh(bpy)2(phzi)3+. DNA strand concentrations were also determined spectrophotometrically using base extinction coefficients of ε260 = 15,400 cm-1M-1 (A), ε260 = 7,400 cm-1M-1 (C), ε260 = 11,500 cm-1M-1 (G), and ε260 = 8,700 cm-1M-1 (T). DNA concentrations are presented per strand. Duplex melting temperatures were determined by following hypochromicity at 260 nm for 1 μM duplex in a buffer of 50 mM NaCl, 10 mM NaPi, pH 7.1, via variable temperature UV-Vis.
All oligonucleotid es were 5’-radioactively labeled with 32P using [γ-32P]ATP (MP Biomedicals) and polynucleotide kinase (Roche) employing standard methodologies and purified via 20% polyacrylamide gel electrophoresis (National Diagnostics).38 All photocleavage experiments were performed using end-labeled DNA with identical sequence, unlabeled carrier DNA in a buffer of 50 mM NaCl, 10 mM NaPi, pH 7.1. Duplexes were annealed by incubation at 90 °C for 15 min followed by slow cooling to room temperature.
Recognition and photocleavage experiments
Solutions of Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+ were incubated with 5’-32P-labeled oligonucleotides either containing or lacking a central DNA lesion (see Results section for sequence details). Unless otherwise noted, final solutions were prepared 20 minutes prior to irradiation, contained 1 μM duplex and 1 μM metalloinsertor, and were 20 μL in volume. Dark and light control samples, of course, lacked the appropriate solution components. Because metalloinsertor photocleavage is single-stranded, each duplex was interrogated twice, once with each of the two strands radioactively labeled. Samples were irradiated with an Oriel Instruments 1000 W solar simulator (320-440 nm). Irradiations were performed in open, horizontally oriented 1.7 mL microcentrifuge tubes. After irradiation, samples were incubated at 60 °C for 30 min and then dried under vacuum. Dried samples were redissolved in denaturing formamide loading dye and electrophoresed on 20% denaturing polyacrylamide gels. Images of the gels were obtained via phosphorimagery (Molecular Dynamics) and quantified using ImageQuant software.
Determination of defect-specific binding constants
Photocleavage titrations were performed to determine the thermodynamic binding constants for Rh(bpy)2(chrysi)3+ with lesion sites of interest. Solutions of DNA (1 μM) were incubated with variable concentrations of Rh(bpy)2(chrysi)3+ (0-20 μM) and subsequently irradiated on an Oriel Instruments solar simulator for 10 min. After irradiation, the samples were incubated at 60 °C for 30 min and then dried under vacuum. Dried samples were redissolved in denaturing formamide loading dye and electrophoresed on 20% denaturing polyacrylamide gels. Images of the gels were obtained via phosphorimagery (Molecular Dynamics). The fraction cleaved at the lesion site was quantitated using ImageQuant software, expressed as a fraction of the total parent DNA, and fit to a single site, one parameter binding model.
MALDI-TOF cleavage product analysis
For mass spectrometry analysis of photocleavage products, 2 μM solutions of duplex were incubated with 2 μM Rh(bpy)2(chrysi)3+ and irradiated as described above. After irradiation and incubation, the samples were dried under vacuum, resuspended in 10 μL water, and desalted using 10 μL OMIX C18 tips (Varian). Light and dark controls were also prepared. Mass spectrometry was performed using a Voyager DE-PRO MALDI-TOF instrument with a 337 nm nitrogen laser source (Applied Biosystems). A 4-hydroxypicolinic acid matrix was employed. All mass spectra were internally calibrated using the mass of the parent oligonucleotide.
RESULTS
Sequence design and melting temperature analysis
A series of oligonucleotides was synthesized and purified to allow for the interrogation of abasic sites and single base bulges in variable sequence contexts and with all possible unpaired bases. The 27-mer single strands are identical except for a central six base region in which the sequence variation occurs. Four different oligonucleotides containing abasic sites were designed, each placing the abasic site in a different sequence context: 5’-GΦT-3’ (AB1), 5’-GΦA-3’ (AB2), 5’-AΦG-3’ (AB3), and 5’-TΦC-3’ (AB4) (Table 1). For each abasic strand, four complements were prepared. Each positions a different base complementary to the abasic site: for example, 3’-CAA-5’ (AB1-A), 3’-CCT-5’ (AB2-C), 3’-TGC-5’ (AB3-G), and 3’-ATG-5’ (AB4-T). These oligonucleotides, taken together, allow us to examine the recognition of abasic sites in the three major sequence context types (Pur/Pur, Pyr/Pur, Pyr/Pyr) with all possible opposing unpaired bases. For purposes of comparison, matched and mismatched strands were also created for each sequence context; complementary in each case to the AB#-C strand, these oligonucleotides create either a fully matched duplex or one containing a central CC mismatch.
Table 1.
Sequence and recognition information for abasic assemblies.
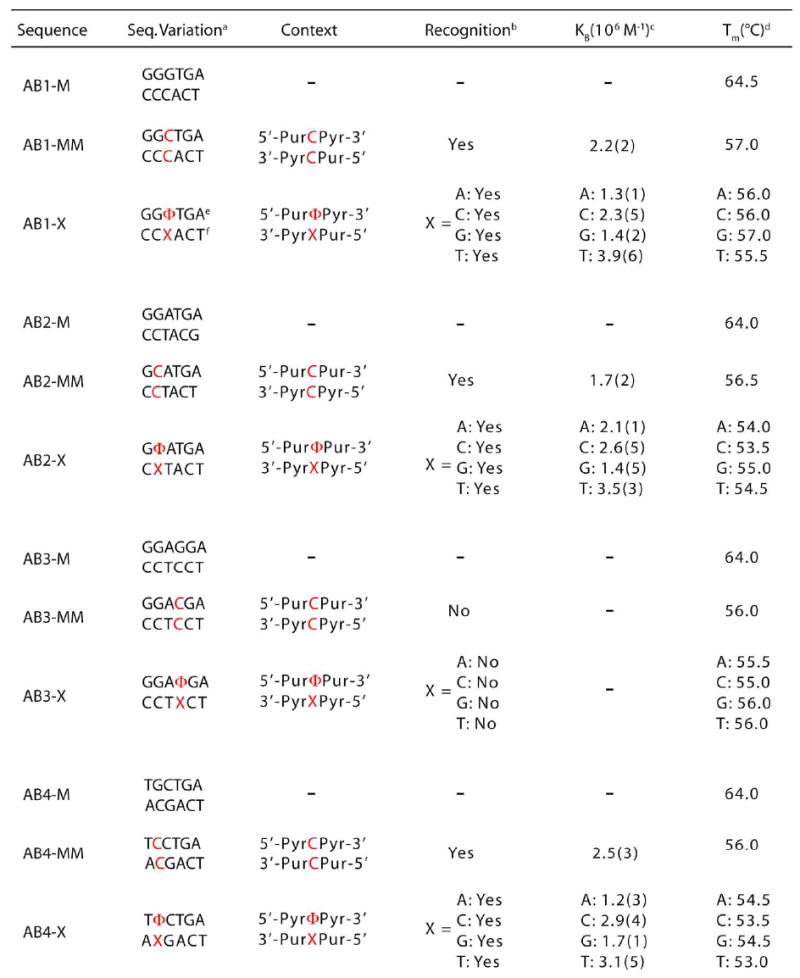 |
Sequence within variable region of 5’-GACTTATCT AGW NNNNNT AAG CTG GTC-3’ (top) and complement (bottom).
Determined by photocleavage assay employing 1 μM Rh(bpy)2(chrysi)3+and 1 μM duplex DNA in buffer (50 mM NaCI, 10 mM NaPi,pH 7.1).
Measured via binding titration experiment using 1 μM duplex DNA and variable concentrations (0-20 μM) of Rh(bpy)2(chrysi)3+5’ in buffer.
Determined with UV-Visible spectrophotometry employing 1 μM duplex DNA in buffer. Accurate within 1 °C.
Φ denotes tetrdhydrofuranyl abasic site.
X denotes base complementary to abasic site.
Four additional oligonucleotides were synthesized to facilitate the study of single base bulges (Table 2). These, termed B1-B4, are identical to the AB# strands in all respects except that they lack the tetrahydrofuranyl abasic site. Thus, when these 26mers are annealed to the complements of the abasic oligonucleotides, duplexes with single base bulges are formed. In each case, the nucleotide formerly complementary to the abasic site is now the bulged base: for example, 3’-CTA-5’ (B1-T), 3’-CAT-5’ (B2-A), 3’-TCC-5’ (B3-C), and 3’-AGG-5’ (B4-G). The same sets of matched and mismatched duplexes were employed as controls. In all, 32 oligonucleotides forming 28 unique duplexes were created.
Table 2.
Sequence and recognition information for single base bulge assemblies.
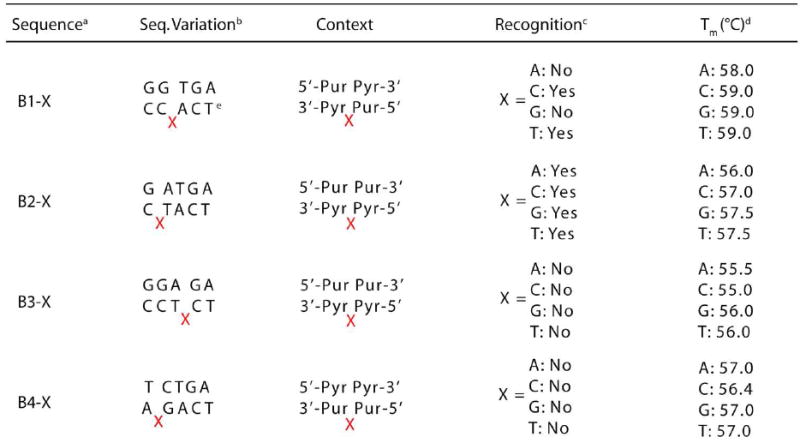 |
Data for the corresponding matched and mismatched strands (e.g.ABI-M and AB1-MM for B1-X) can be found in Table 1.
Sequence within variable region of 5’-GACTTATCT AGA/WA/W/WT AAG CTG GTC-3’(top) and complement (bottom). The complementcontains the bulged base.
Determined by photocleavage assay employing 5 |iM Rh(bpy)2(chrysi}3+and luM duplex DNAin buffer (50 mM NaCI, 10 mM NaPi.pH 7.1).
Determined with UV-Visible spectrophotometry employing 1 (.nVl duplex DNA in buffer. Accurate within 1 °C.
X denotes the bulged base.
Melting temperature analysis of the DNA allows us to determine the relative thermodynamic destabilization created by the lesions. All four matched duplexes have melting temperatures around 64 °C. Relative to these, the mismatched duplexes are destabilized by 7-8 °C. Duplexes containing single base bulges are similarly destabilized, if not slightly more stable, with melting temperatures 6-8 °C lower than that of the corresponding matched duplex. In contrast, abasic site duplexes are even less stable than their mismatched counterparts with melting temperatures reduced by 8-11 °C. Taken together, these ΔTm values are in agreement with the published literature.18,19 It is somewhat surprising, however, that in the family of abasic duplexes we do not see significant variation in ΔTm based upon sequence context or unpaired base identity. This result is more likely a product of instrument sensitivity rather than the absence of such influences on site stability. Nonetheless, these measurements plainly illustrate the relative stabilities of the duplexes at hand: abasic site < mismatched base pair < single base bulge ≪ matched base pair.
Recognition and photocleavage of abasic sites by Rh(bpy)2(chrysi)3+
Polyacrylamide gel assays clearly indicate that Rh(bpy)2(chrysi)3+ specifically recognizes and photocleaves abasic sites in DNA (Figure 3). Indeed, the metalloinsertor binds and promotes strand scission at lesion sites in all sequence context types (5’-PurΦPur-3’, 5’-PurΦPyr-3’, and 5’-PyrΦPyr-3’) and with all possible unpaired bases. No photocleavage is observed in the absence of metalloinsertor or with well matched DNA. In total, twelve of the sixteen abasic sites are bound and cleaved. Specifically, duplexes AB1, AB2, and AB4 are recognized and cleaved regardless of unpaired base identity; surprisingly, however, no photocleavage is observed for the AB3 duplexes. This pattern corresponds precisely to that observed for the strands bearing a central CC mismatch: AB1-MM, AB2-MM and AB4-MM are all recognized and cleaved, while AB3-MM escapes binding and scission. That the AB3 duplexes are not bound and cleaved is certainly not a consequence of the sequence context type, for AB2, like AB3, places the abasic site in a 5’-PurΦPur-3’ sequence context and is, in fact, cleaved quite readily. The answer likely lies in the sensitivity of metalloinsertor mismatch recognition to specific sequence contexts. Similar effects of sequence context have been seen previously for the family of mismatched duplexes.11 Indeed, experiments employing higher rhodium concentrations and longer irradiation times suggest that Rh(bpy)2(chrysi)3+ does bind and cleave the AB3 abasic sites, just not nearly as strongly or efficiently as those in the other sequence contexts.
Figure 3.
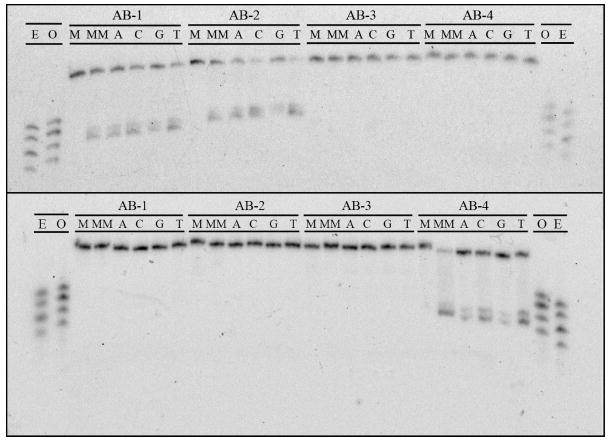
PAGE assay illustrating the recognition and photocleavage of mismatch and abasic site recognition. Sequence contexts are listed along top line of each gel, and individual duplexes are indicated in the second line (M = matched, MM = mismatched, A = unpaired adenine, C = cytosine, G = guanine, and T = thymine). In the top gel, the single strand beginning 5’-GAC CAG … (that containing the unpaired base in the abasic assemblies) is 5’-32P-labeled. In the bottom gel, the single strand beginning 5’-GAC TTA … (that containing the abasic site) is 5’-32P-labeled. In both experiments, 1 μM duplex was incubated with Rh(bpy)2(chrysi)3+ in 50 mM NaCl, 10 mM NaPi, pH 7.1. Samples were irradiated for 10 min on an Oriel Instruments solar simulator (320-440 nm emission) and incubated for 30 min at 60 °C prior to electrophoresis. “E” and “O” denote lanes containing even (10, 12, 14, 16) and odd (11, 13, 15, 17) standardization fragments.
Photocleavage experiments also reveal interesting patterns in the strand asymmetry of scission. Regardless of unpaired base identity, duplexes AB1 and AB2 are cleaved on the strand containing the unpaired nucleotide. Interestingly, however, duplex AB4 is cleaved instead on the strand containing the tetrahydrofuranyl abasic site, again irrespective of unpaired base. This behavior exactly mirrors mismatch photocleavage. While, of course, the mismatched duplexes contain no unpaired bases or abasic sites, the AB1-MM and AB2-MM assemblies are cleaved on the strand corresponding to that containing an unpaired base in the abasic duplexes, and the AB4-MM assembly is cleaved on the strand corresponding to that containing the abasic site in the abasic duplex. This observation must reflect the binding architecture of the complex in the abasic site (see Discussion).
Another important similarity between mismatch and abasic photocleavage is the length of the scission products. Regardless of unpaired base identity, AB1 cleavage fragments are 14 base pairs long, AB2 fragments 15 base pairs long, and AB4 fragments 13 base pairs long. These fragments correspond to cleavage at the ribose 3’ to the unpaired base in duplexes AB1 and AB2 and at the ribose 3’ to the abasic site in the AB4 duplexes. Importantly, photocleavage at the CC mismatch in each duplex produces fragments of identical length.
Recognition and photocleavage of abasic sites by Rh(bpy)2(phzi)3+
In order to probe the generality of metalloinsertor recognition of abasic sites, photocleavage experiments were also performed using Rh(bpy)2(phzi)3+, a second generation complex with a heterocyclic bulky ligand (Figure 4).9 Rh(bpy)2(phzi)3+ is clearly able both to recognize and, upon irradiation, to cleave the representative abasic sites. Again, no recognition or photocleavage is observed in the absence of metalloinsertor or DNA lesion. Significantly, photocleavage with Rh(bpy)2(phzi)3+ is observed at much lower concentrations (100 nM) than with Rh(bpy)2(chrysi)3+, a characteristic also observed for mismatch photocleavage and attributed to the added π-stacking capabilities of the heterocyclic inserting ligand.
Figure 4.
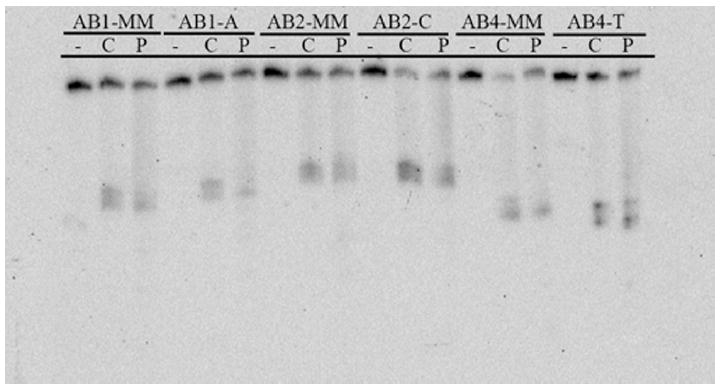
PAGE assay illustrating the recognition and photocleavage of mismatches and abasic sites by Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+. 1 μM duplex was incubated without metal complex (lanes marked “-”) , with 1 μM Rh(bpy)2(chrysi)3+ (lanes marked “C”), or with 100 nM Rh(bpy)2(phzi)3+ (lanes marked “P”) in 50 mM NaCl, 10 mM NaPi, pH 7.1. Duplex identity is indicated at the top of the gel. Samples were irradiated for 10 min on an Oriel Instruments solar simulator (320-440 nm emission) and incubated for 30 min at 60 °C prior to electrophoresis.
Binding affinities of Rh(bpy)2(chrysi)3+ for abasic sites
Photocleavage titration experiments were employed to determine site-specific binding constants for the twelve abasic sites and three mismatches for which photocleavage was observed (Figure 5 shows a representative titration, Table 1). The binding constants for the mismatched sites, 2.2(2) × 106 M-1 (AB1-MM), 1.7(2) × 106 M-1 (AB2-MM), 2.5(3) × 106 M-1 (AB4-MM), are comparable to those previously reported for CC mismatches.11 Since metalloinsertor binding affinity correlates directly to site destabilization, it is not surprising that the binding constants of Rh(bpy)2(chrysi)3+ for abasic sites are similar to if not somewhat greater than those for the most destabilizing (e.g., CC) mismatches.
Figure 5.
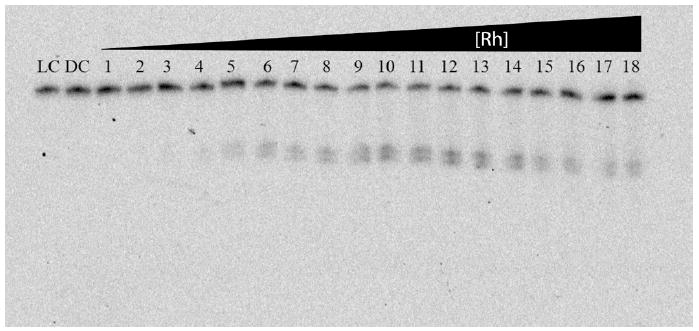
PAGE assay illustrating a typical photocleavage assay for binding constant determination. 1 μM duplex was incubated with increasing concentrations of Rh(bpy)2(chrysi)3+ in 50 mM NaCl, 10 mM NaPi, pH 7.1. The AB1-C duplex was employed for this titration. Samples were irradiated for 10 min on an Oriel Instruments solar simulator (320-440 nm emission) and incubated for 30 min at 60 °C prior to electrophoresis. LC and DC represent light (no rhodium, 10 min irradiation) and dark (1 μM Rh, no irradiation) controls. Lanes 1-18 contain 50 nM, 100 nM, 200 nM, 300 nM, 400 nM, 500 nM, 600 nM, 700 nM, 800 nM, 1 μM, 2 μM, 3 μM, 4 μM, 5 μM, 7 μM, 9 μM, 13 μM, 15μM, 17.5 μM, 20 μM Rh(bpy)2(chrysi)3+.
Despite likely differences in site destabilization, little variation is observed between the values for the three different sequence contexts, a result that suggests a threshold behavior in the relationship between destabilization and binding affinity. Such behavior has previously been suggested for mismatch binding.11 Small differences, however, do appear based on unpaired base identity within a single sequence context. For example, the values for AB2 are 1.4(5) × 106 M-1 (G), 2.1(1) × 106 M-1 (A), 2.6(5) × 106 M-1 (C), and 3.5(3) × 106 M-1 (T). The metalloinsertor seems to bind abasic sites with unpaired pyrimidines slightly tighter than sites with unpaired purines. These differences are admittedly minor; however, the trend is consistent among the three sequence contexts. An explanation based on the kinetics and helicity of the unpaired base in each case is perhaps most likely.
Enantiospecificity of Rh(bpy)2(chrysi)3+ for abasic sites
Photocleavage assays employing Δ-Rh(bpy)2(chrysi)3+ and Λ-Rh(bpy)2(chrysi)3+ clearly indicate that abasic recognition is enantiospecific for the right-handed isomer of the metalloinsertor (Figure 6). PAGE experiments reveal that concentrations of 1 μM Δ-Rh(bpy)2(chrysi)3+ bind and cleave all abasic sites interrogated while incubation and irradiation with 1 μM Λ-Rh(bpy)2(chrysi)3+ produce no photocleavage bands. This chiral specificity has been well-documented for metalloinsertor recognition of mismatched sites.40 Recent structural studies of Rh(bpy)2(chrysi)3+ bound to a CA mismatch have shed light on the question; because the metalloinsertor binds the mismatch site from the narrow, sterically constrictive minor groove, the chirality of complex must match that of the helix to prevent steric clash between the ancillary ligands and the DNA backbone. In short, the right-handed helix can only accommodate the right-handed enantiomer. The observation that metalloinsertor recognition of abasic sites is also enantiospecific argues strongly for site binding via the minor groove.
Figure 6.
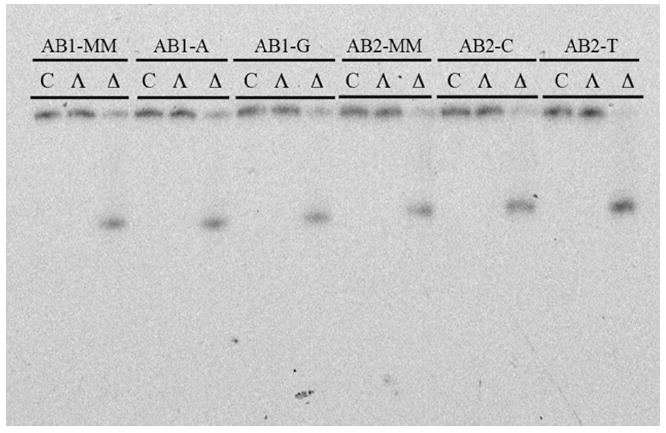
PAGE assay illustrating the enantioselectivity of mismatch and abasic site recognition. 1 μM duplex was incubated with either Δ-Rh(bpy)2(chrysi)3+, Λ-Rh(bpy)2(chrysi)3+, or no Rh complex at all in 50 mM NaCl, 10 mM NaPi, pH 7.1. Samples were irradiated for 10 min on an Oriel Instruments solar simulator (320-440 nm emission) and incubated for 30 min at 60 °C prior to electrophoresis.
MALDI-TOF analysis of abasic site photocleavage products
While we have predominantly employed gel electrophoresis in our study of abasic site recognition, MALDI-TOF mass spectrometry affords a unique opportunity to investigate not only the site specificity of recognition but also the identity of the individual photocleavage products. A similar investigation has been previously reported for metalloinsertor mismatch recognition.41 Here the photocleavage of all 12 cleaved abasic duplexes and their mismatched analogues was investigated. The MALDI-TOF analysis of AB1-C photocleavage provides a suitable example (Figure 7). In light (no Rh, with irradiation) and dark (Rh, no irradiation) controls, only peaks corresponding to the singly (DNA1+) and doubly (DNA2+) charged parent single strands are observed, m/z = 8198.7 and 4100.3 for AB1 and 8213.2 and 4106.9 for AB1-C (Supp. Info.). Photocleavage samples reveal three new masses in addition to the parent strands at m/z = 3733.7, 4286.8, and 4475.9. These fragments are consistent with the DNA only being cleaved on the AB1-C strand. We assign the cleavage fragment at m/z = 3733.7 as a 12-mer with a 5’-phosphate group and the product at m/z = 4286.8 as a 14-mer with a 3’-phosphate group. These fragments correspond to common DNA oxidation products and clearly indicate scission on the 3’-side of the unpaired base. The final cleavage fragment, appearing at m/z = 4475.9, corresponds to the above 14-mer with a 3’-2,3-dehydronucleotide rather than a phosphate. Upon sample incubation for 24 h at 20 °C, however, complete conversion of the dehydronucleotide product to the 3’-phosphate fragment is observed, suggesting that the former is a metastable intermediate.
Figure 7.
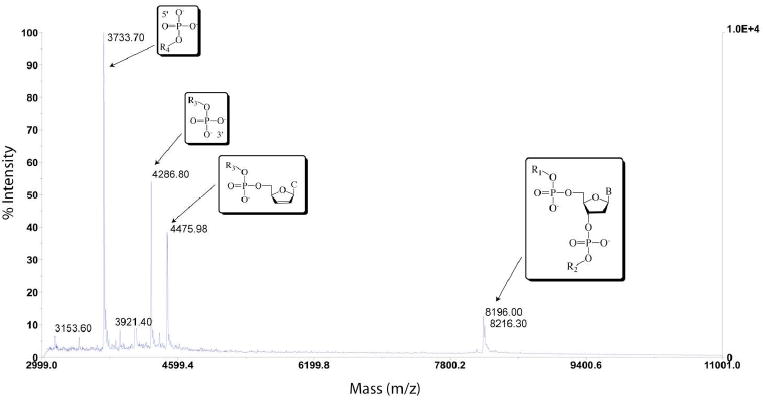
MALDI-TOF mass spectrograph of photocleavage products of duplex AB1-C, 5’- GAC CAG CTT ATC ACC CCT AGA TAA GCG -3’ in which the underlined, italicized cytosine is the unpaired complement of an abasic site. The rightmost peaks correspond to the full, uncleaved parent strands. Assigned scission products can be viewed on the left-hand side of the plot and correspond to 5’-PO4-CCT AGA TAA GCG-3’, 5’-GAC CAG CCT ATC AC-PO4-3’, and 5’-GAC CAG CCT ATC AC-dehydroC-3’. R1 = GAC CAG CTT ATC A; R2 = CCC TAG ATA AGC G; R3 = GAC CAG CCT ATC AC; R4 = CCT AGA TAA GCG; B = cytosine.
Analogous results are obtained for all abasic sites that are cleaved on the strand containing the unpaired base. The situation changes only slightly for the AB4 sequence context, in which scission occurs on the strand containing the abasic site; for these duplexes, all of the same cleavage products are observed, but strand scission occurs on the 3’ side of the abasic site. Importantly, analogous products are also seen for photocleavage of the mismatched strands. Indeed, exactly the same products are seen for AB1-C and AB1-MM: strand scission occurs on the 3’-sides of the unpaired cytosine in AB1-C and the corresponding mispaired cytosine in AB1-MM, resulting in identical fragments (Supp. Info.).
Taken together, these mass spectrometry experiments provide a number of key insights. First, the data confirm observations made via gel electrophoresis regarding site specificity, strand asymmetry of scission, and cleavage product length. More important, however, is light shed on the relationship between abasic site and mismatch recognition and photocleavage. As stated above, analogous, and in some cases indistinguishable, products are observed for mismatch and abasic site photocleavage. This result strongly suggests a similar, if not identical, binding mode for metalloinsertors at abasic sites. Furthermore, based on cleavage product analysis and structural information, it has been posited that mismatch photocleavage proceeds via an H1-abstraction mechanism;41 based on these results, it is almost certain that abasic site strand scission occurs via the same pathway.
Recognition and photocleavage of single base bulges by Rh(bpy)2(chrysi)3+
Compared to abasic sites, single base bulges are recognized less effectively and, when bound, cleaved less efficiently. In fact, out of the sixteen possible single base bulges in this investigation, only seven were recognized and cleaved: B1-C, B1-G, B1-T, B2-A, B2-C, B2-G, and B2-T (Figure 8). Furthermore, in some cases, even faint bulge photocleavage bands required longer irradiation times (20-30 min, compared to 10 min for substantial abasic site cleavage). Based on comparison to shorter labeled oligonucleotides, the bulge photocleavage fragments appear to be 14 bases long for the B1 duplexes and 15 bases long for the B2 duplexes, indicating strand scission on the 3’-side of the bulged base. However, the low photocleavage efficiency at single base bulges precludes the accurate determination of binding affinities. Based on photocleavage titrations and qualitative observations, however, it is evident that in each case the metalloinsertor binding affinity is ~105 M-1.
Figure 8.
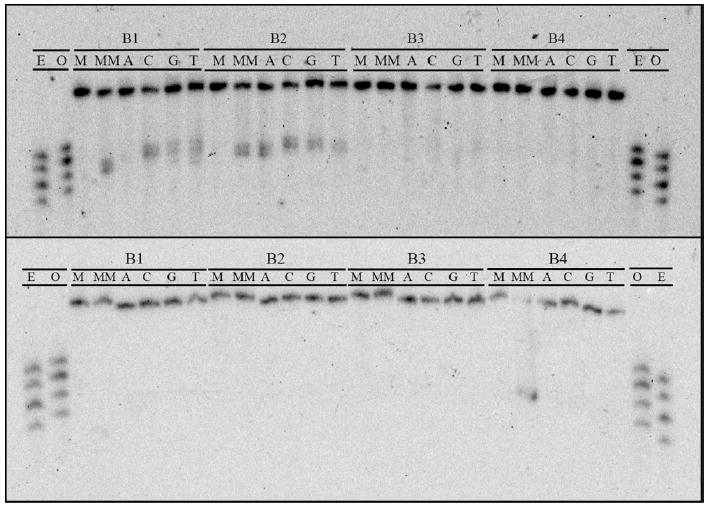
PAGE assay illustrating the recognition and photocleavage of mismatch and single base bulge recognition. Sequence contexts are listed along top line of each gel, and individual duplexes are indicated in the second line (M = matched, MM = mismatched, A = bulged adenine, C = cytosine, G = guanine, and T = thymine). In the top gel, the single strand beginning 5’-GAC CAG … (that containing the bulged base in SBB assemblies) is 5’-32P-labeled. In the bottom gel, the single strand beginning 5’-GAC TTA … is 5’-32P-labeled. In both experiments, 1 μM duplex was incubated with Rh(bpy)2(chrysi)3+ in 50 mM NaCl, 10 mM NaPi, pH 7.1. Samples were irradiated for 30 min on an Oriel Instruments solar simulator (320-440 nm emission) and incubated for 30 min at 60 °C prior to electrophoresis. “E” and “O” denote lanes containing even (10, 12, 14, 16) and odd (11, 13, 15, 17) standardization fragments.
Both sequence context and bulged base identity appear to play a role in recognition. Single base bulges in the B3 and B4 sequence contexts escape binding and photocleavage in toto, whereas all of the bulges in the B2 sequence context are recognized and cleaved to some extent. The recognition of single base bulges in the B1 sequence context seems to be dependent upon bulged base identity; the bulged cytosine, guanine, and thymine are cleaved, whereas the bulged adenine is not. Proffering an explanation for this behavior proves difficult, especially without the aid of simple bulge site destabilization trends (see Discussion).
Despite the lack of generality in single base bulge recognition, the initial photocleavage assay and subsequent experimentation do provide some insight into how the metalloinsertor may bind single base bulges. First, the strand asymmetry and cleavage product length of single base bulge scission match those of mismatch photocleavage. Second, photocleavage assays employing Δ- and Λ-Rh(bpy)2(chrysi)3+ clearly indicate that bulge recognition is enantiospecific for the right-handed isomer of the metalloinsertors. Third, MALDI-TOF analysis of bulge photocleavage products reveal fragments analogous to those produced in mismatch and abasic site recognition and scission (Supp. Info.).42 For example, Rh(bpy)2(chrysi)3+ photocleavage of the B2 -A duplex produces fragments of m/z = 7999.9, 8251.1, 3442.7, 4614.8, and 4802.3. The first two values correspond to the parent single strands of the duplex. The peak at m/z = 3442.7 corresponds to an 11-mer fragment with a 5’-phosphate, that fragment at m/z = 4614.8 to a 15-mer with a 3’-phosphate, and that at m/z = 4798.7 to the same 15-mer fragment but with a 3’-2,3-dehydronucleotide instead of a 3’-phosphate. These products are, in fact, almost identical to those produced via cleavage of the AB2-A abasic site. Thus the data clearly suggest that even though Rh(bpy)2(chrysi)3+ only recognizes single base bulges in a minority of cases, lesion binding, when it does happen, likely occurs in a mode analogous to that of the metalloinsertor at mismatches and abasic sites.
DISCUSSION
Based upon the data described, Rh(bpy)2(chrysi)3+ recognizes abasic sites with high affinity and specificity with little regard for sequence context or the opposing unpaired base. The targeting of single base bulges, however, appears to be more complicated, with only seven of sixteen possible abasic sites bound and cleaved by the metal complex. Yet, now that we have shown that Rh(bpy)2(chrysi)3+ can, indeed, bind both lesions, two simple questions follow: (1) how does the complex bind each lesion and (2) what are the constraints upon the recognition of these defects?
Rh(bpy)2(chrysi)3+ binds abasic sites in DNA via metalloinsertion
NMR and X-ray crystallographic evidence has revealed that Rh(bpy)2(chrysi)3+ binds mismatched sites not by classical major groove intercalation but rather via a previously unseen binding mode: insertion. The complex approaches the DNA from the minor groove, ejects the mismatched bases into the major groove, and replaces the extruded bases in the π-stack with its own aromatic ligand.
In the absence of concrete structural information for the abasic site binding, we must rely on comparisons to mismatch recognition when considering how Rh(bpy)2(chrysi)3+ targets abasic sites. The similarities are striking. First, mismatch and abasic site photocleavage exhibit identical strand asymmetry. In the AB1 and AB2 duplexes, the metal complex cleaves the strand containing the unpaired bases; in the AB4 duplexes, the strand containing the abasic site is cut. Mismatch photocleavage mirrors this behavior, with the corresponding strands of the mismatched duplexes being photocleaved. Second, the enantiospecificity of recognition is revealing. While bis(bpy) complexes intercalate into B-DNA with very little enantioselectivity,43 Δ- Rh(bpy)2(chrysi)3+ is able to target and cleave mismatched sites enantiospecifically, a consequence of metalloinsertion occurring from the sterically constrictive minor groove. The same high specificity is observed for abasic site recognition: only the right-handed enantiomer targets and cleaves the abasic lesion. This clearly argues strongly for involvement of the minor groove. Third, mass spectrometry photocleavage product analysis provides still more evidence for similarity. This technique reveals that both abasic sites and mismatches are cleaved on the 3’-side of the lesions, producing three products: (1) a fragment containing a 5’-phosphate, (2) a fragment containing a 3’-phosphate, and (3) a metastable fragment identical to (2) but with a 3’-2,3-dehydronucleotide. Indeed, when the unpaired base in the abasic assembly is a cytosine and thus contains the same sequence as the mismatched assembly, identical photocleavage fragments are formed. These products are consistent with H1’-hydrogen abstraction by the photoactivated ligand, a mechanistic pathway accessible only via the minor groove. Finally, a variety of other, more minor similarities between abasic site and mismatch recognition exist, including the failure of Rh(bpy)2(chrysi)3+ to target either defect in the AB3 sequence context and the similarity of metalloinsertor binding affinity for both types of lesion, and these observations also argue for similar binding modes. In sum, this study clearly indicates that abasic site recognition and photocleavage by metalloinsertors occur in a manner almost, if not precisely, identical to mismatch targeting. Thus, these data are fully consistent with Rh(bpy)2(chrysi)3+ targeting abasic sites via metalloinsertion from the minor groove. It should be noted that this conclusion fits well with an intuitive, and teleological, approach to the system: to a metalloinsertor, an abasic site looks like a mismatch with half the extrusion work already accomplished.
Rh(bpy)2(chrysi)3+ likely binds single base bulges in DNA via metalloinsertion
Single base bulge recognition presents a somewhat more difficult task for the metalloinsertor. Of the sixteen different single base bulge sites interrogated in the study, only seven were bound and cleaved by Rh(bpy)2(chrysi)3+. Again, a comparison to mismatch recognition is useful in exploring the recognition of single base bulges.
Several observations point to a binding mode for single base bulges similar to that for mismatches and abasic sites. First, photocleavage strand asymmetry for single base bulges mirrors that for both other defects. Furthermore, bulge recognition, like that of mismatches and abasic sites, is enantiospecific for the Δ-isomer of the metalloinsertor. Lastly, the DNA fragments produced by bulge photocleavage are analogous to those produced by scission neighboring the other two lesions. Yet two significant differences indicate that the binding mode must be at least somewhat different. Both the binding affinity and photocleavage efficiency at single base bulges are substantially reduced compared to that at the corresponding mismatches and abasic sites. While this certainly does not preclude minor groove insertion, it does suggest that the orientation of the complex within the binding site differs somewhat from that at the other two lesions.
Intuitively, this is not surprising. Like binding at an abasic site, metalloinsertion at a single base bulge requires only the ejection of a single base. However, a key structural difference exists between single base bulges and the other two lesions: in a bulged duplex, the ribose of the abasic site or the second mismatched base is not there. It is thus not possible for Rh(bpy)2(chrysi)3+ to target a bulged site in the same manner it does a mismatch or an abasic site. One half of the ligand may bind in a manner similar to insertion, extruding the bulged base and replacing it in the DNA base stack, but the other half of the ligand, without an abasic ribose or a mismatched base to eject, must bind stacked between adjacent bases in a mode far closer to metallointercalation than metalloinsertion.
Factors affecting metalloinsertor recognition of abasic sites and single base bulges
Certainly the most puzzling aspect of the abasic site recognition investigation is the absence of photocleavage for the abasic AB3 duplexes. Neither sequence context nor thermodynamic stabilization provide explanations; the AB2 duplexes, which also house the abasic site in a 5’-PurΦPur-3’ sequence context, are cleaved, and melting temperature measurements suggest that the AB3 duplexes are as destabilized as the other abasic assemblies. The failure of Rh(bpy)2(chrysi)3+ to cleave the AB3 duplex containing a central CC mismatch is equally, if not more, surprising. Cytosine-cytosine mismatches are among the most destabilizing mispairs and are readily recognized and cleaved by metalloinsertors in almost any sequence context. It follows that the most likely, if slightly unsatisfying, explanation is purely based on sequence: the particular 5’-AΦG-3’ sequence context in the AB3 duplexes simply does not allow for efficient lesion binding and photocleavage. Such anomalies, though poorly understood at present, have been reported for mismatch targeting and constitute only a very small percentage of cases.44
The somewhat sporadic single base bulge cleavage of Rh(bpy)2(chrysi)3+ also merits some attention. As we have noted, only seven of sixteen possible bulges were recognized and cleaved. A thermodynamic rationale is not available, principally due to the lack of reliable, reported patterns between bulge sequence and destabilization. Sequence context surely plays a role, but it cannot be the sole determining factor; both the B2 and B3 assemblies place the bulged base in a 5’-PyrXPyr-3’ context, but one set of duplexes (B2) exhibits cleavage regardless of bulged base identity while the other (B3) escapes recognition entirely. The selective cleavage of three bulged bases in the B1 assemblies suggests bulged base identity as a determining factor, but the recognition of the B2 sequence bulges regardless of base identity suggests a slightly more complicated rationale.
One possible explanation may be found in the likely conformation of the bulged base. In the B2 duplexes, all of which are photocleaved by Rh(bpy)2(chrysi)3+, each bulged base is in a 5’-PyrXPyr-3’ sequence context and is therefore likely to spend at least some time in a extrahelical conformation. In contrast, the B4 duplexes house the bulged base in a 5’-PurXPur-3’ conformation, with the better stacking purines shifting the likely position of the bulged base from extra- to intrahelical; in this case, none of the single base bulges is bound and cleaved. The B1 duplexes provide an intermediate case. Here, the bulged bases are in a 5’-PyrXPur-3’ sequence context. In this case, the bulged bases, likely in an extrahelical conformation, the pyrimidines C and T, are bound and cleaved, while one of those more likely to prefer an intrahelical orientation, the purine A, escapes recognition. In sum, the data suggest that the more likely a base is to exist in an extrahelical conformation, the more easily it will be targeted by our metalloinsertors. It should be noted, however, that this hypothesis fails to explain the targeting of the bulged guanine in the B1-G assembly.
CONCLUSIONS
This investigation clearly illustrates that both abasic sites and single base bulges are targeted by Rh(bpy)2(chrysi)3+, a sterically bulky metalloinsertor. Abasic sites are targeted with high specificity and affinity in all sequence contexts and with all unpaired bases, and a wide variety of evidence points to minor groove metalloinsertion as the binding mode of the complex at these defects. The recognition of single base bulges is less reliable, though the available data suggest an insertion binding mode is likely in this case as well.
The broader implications of this study are threefold. The revelation that specific metalloinsertion is not a phenomenon unique to mismatches certainly is important in the development of recognition agents for DNA lesions. Perhaps this and subsequent investigations will enable us to expand the utility of these complexes beyond mismatch recognition into applications involving the detection of abasic sites or other thermodynamically destabilized DNA defects in vivo. Second, the ability of Rh(bpy)2(chrysi)3+ to specifically target abasic sites represents an exciting diagnostic possibility. A reliable probe for these lesions, especially one with the specificity, affinity, and reactivity of Rh(bpy)(chrysi)3+ and Rh(bpy)2(phzi)3+, could prove an invaluable clinical and diagnostic tool. Surely these results dictate that abasic sites and single base bulges may, in addition to mismatches, be in vivo targets for metalloinsertors. Experiments with mismatch repair proficient and deficient cell lines have illuminated the substantial therapeutic potential of metalloinsertors and, furthermore, have strongly suggested that Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+ target mismatches in the cell. Similar studies employing cells deficient in abasic site repair pathways may further illuminate the potential therapeutic value of these complexes. Looking forward, the discovery that metalloinsertors specifically target and photocleave abasic sites creates a variety of new and exciting opportunities in the study and development of metal complexes that target DNA lesions.
Supplementary Material
Selected MALDI-TOF mass spectrographs of photocleavage experiments and relevant controls. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations
- MMR
mismatch repair
- BER
base excision repair
- chrysi
chrysene-5,6-quinone diimine
- phzi
benzo[a]phenazine-5,6-quinone diimine
- DMT
dimethoxyltrityl
Footnotes
Financial support for this work from the National Institutes of Health (GM33309 to J.K.B.) is gratefully acknowledged.
References
- 1.Marnett LJ, Plastaras JP. Endogenous DNA damage and mutation. Trends Genet. 2001;17:214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 2.Iyer RR, Pluciennik A, Burdett V, Modrich P. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.David SS, O’Shea V, Kundu S. Base excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolodner RD. Mismatch repair: mechanisms and relation to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 5.Arzimanoglou II, Gilbert F, Barber HRK. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Nat Acad Sci U S A. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatani K, Sando S, Saito I. Scanning of guanine-guanine mismatches in DNA by synthetic ligands using surface plasmon resonance. Nat Biotech. 2001;19:51–55. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]
- 8.Zeglis BM, Pierre VP, Barton JK. Metallointercalators and metalloinsertors. Chem Comm. 2007;44:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 10.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. A rhodium(III) complex for high affinity DNA base-pair mismatch recognition. Proc Nat Acad Sci U S A. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson BA, Barton JK. Recognition of base mismatches in DNA 5,6-chrysenequinone diimine complexes of rhodium(III): a proposed mechanism for preferential binding in destabilized regions of the double helix. Biochem. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 12.Pierre VP, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc Nat Acad Sci U S A. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordier C, Pierre VP, Barton JK. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: an NMR solution study. J Am Chem Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 2000;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 15.Coppel Y, Berthet N, Coulombeau C, Coulombeau C, Garvia J, Lhomme J. Solution conformtation of an abasic DNA undecamer duplex d(CGCACXCACGC)-d(GCGTGTGTGCG): the unpaired thymine stacks inside the helix. Biochem. 1997;36:4817–4830. doi: 10.1021/bi962677y. [DOI] [PubMed] [Google Scholar]
- 16.Goljer I, Kumar S, Bolton PH. Refined solution structure of a DNA heteroduplex containing an aldehydic abasic site. J Biol Chem. 1995;270:22980–22987. doi: 10.1074/jbc.270.39.22980. [DOI] [PubMed] [Google Scholar]
- 17.Feig M, Zacharias M, Pettitt BM. Conformation of an adenine bulge in a DNA octamer and its influence on DNA structure from molecular dynamics simulations. Biophys J. 2001;81:351–370. doi: 10.1016/S0006-3495(01)75705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vesnaver G, Chang CN, Eisenberg M, Grollman AP, Breslauer KJ. Influence of abasic and a nucleosidic sites on the stability, conformation, and metling behavior of a DNA duplex: correlations of thermodynamic and structural data. Proc Nat Acad Sci U S A. 1989;86:3614–3618. doi: 10.1073/pnas.86.10.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochem. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 20.Alani E, Chi NW, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka F, Kameda A, Tamamoto M, Ohuchi A. Thermodynamic parametres based on a nearest-neighbor model for DNA sequences with a single-bulge loop. Biochem. 2004;43:7143–7150. doi: 10.1021/bi036188r. [DOI] [PubMed] [Google Scholar]
- 22.Nikonowicz EP, Meadows RP, Gorenstein DG. NMR structural refinement of an extrahelical adenosine tridecamer d(CGCAGAATTCGCG)2 via a hybrid relaxation matrix procedure. Biochem. 1990;29:4193–4204. doi: 10.1021/bi00469a024. [DOI] [PubMed] [Google Scholar]
- 23.Hare D, Shapiro L, Patel DJ. Extrahelical adenosine stacks into right-handed DNA: solution conformation of the d(CGCAGAGCTCGCG) duplex deduced from distance geometry analysis of nuclear overhauser effect spectra. Biochem. 1986;25:7456–7464. doi: 10.1021/bi00371a030. [DOI] [PubMed] [Google Scholar]
- 24.Joshua-Tor L, Rabinovich D, Hope H, Frolow F, Appella E, Sussman JL. The three dimensional structure of a DNA duplex containing looped-out bases. Nature. 1988;334:82–84. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- 25.Cline SD, Jones W, Stone MP, Osheroff N. DNA abasic lesions in a different light: solution structure of an endogenous topoisomerase II poison. Biochem. 1999;38:15500–15507. doi: 10.1021/bi991750s. [DOI] [PubMed] [Google Scholar]
- 26.Streisinger G, Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 29.Li GM. DNA mismatch repair and cancer. Front Biosci. 2003;8:D997–U1. doi: 10.2741/1121. [DOI] [PubMed] [Google Scholar]
- 30.Wilson DM, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Fkyerat A, Demeunynck M, Constant JF, Michon P, Lhomme J. A new class of artificial nucleases that recognize and cleave apurinic sites in DNA with great selectivity and efficiency. J Am Chem Soc. 1993;115:9952–9959. [Google Scholar]
- 32.Boturyn D, Boudali A, Constant JF, Defrancq E, Lhomme J. Synthesis of fluorescent probes for the detection of abasic DNA. Tetrahedron. 1997;53:5485–6592. [Google Scholar]
- 33.Kobori A, Murase T, Suda H, Saito I, Nakatani K. 2-Ureidoquinoline: a useful molecular element for stabilizing single cytosine and thymine bulges. Bioorg Med Chem Lett. 2004;14:3431–3433. doi: 10.1016/j.bmcl.2004.04.075. [DOI] [PubMed] [Google Scholar]
- 34.Nakatani K, Sando S, Saito I. Recognition of a single guanine bulge by 2-acylamino-1,8-napthyridine. J Am Chem Soc. 2000;122:2172–2177. [Google Scholar]
- 35.Morgan JL, Buck DP, Turley AG, Collins JG, Keene FR. Selectivity at a three base bulge site in the DNA binding of ΔΔ-[{Ru(phen)2}2(μ-dppm)]2+ J Biol Inorg Chem. 2006;11:824–834. doi: 10.1007/s00775-006-0130-9. [DOI] [PubMed] [Google Scholar]
- 36.Cheng CC, Kuo YN, Chuang KS, Luo CF, Wang WJ. A new CoII complex as a bulge-specific probe for DNA. Ang Chem Int Ed. 1999;38:1255–1257. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1255::AID-ANIE1255>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 37.Zeglis BM, Barton JK. Targeting abasic sites and single base bulges in DNA with metalloinsertors. J Am Chem Soc. 2008;130:7530–7531. doi: 10.1021/ja801479y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeglis BM, Barton JK. DNA base mismatch detection with bulky rhodium intercalators: synthesis and applications. Nat Protocols. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites: model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 40.Barton JK. Metals and DNA: Molecular Left-Handed Complements. Science. 1986;233:727–732. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 41.Brunner J, Barton JK. Site-specific DNA photocleavage by rhodium intercalators analyzed by MALDI-TOF mass spectrometry. J Am Chem Soc. 2006;128:6772–6773. doi: 10.1021/ja0612753. [DOI] [PubMed] [Google Scholar]
- 42.The low photocleavage efficiency associated with single base bulge photocleavage renders MALDI-TOF analysis difficult owing to the low amounts of product fragments produced; however, all relevant peaks can be easily identified above baseline.
- 43.Erkkila KE, Odom DT, Barton JK. Recognition and Reactions of Metallointercalators bound to DNA. Chem Rev. 1999;99:2777–2795. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 44.Jackson BA. Ph. D. Thesis, California Institute of Technology. 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selected MALDI-TOF mass spectrographs of photocleavage experiments and relevant controls. This material is available free of charge via the Internet at http://pubs.acs.org.


