Abstract
Objectives
To advance our biological understanding of pediatric septic shock, we measured the genome-level expression profiles of critically ill children representing the systemic inflammatory response syndrome (SIRS), sepsis, and septic shock spectrum.
Design
Prospective observational study involving microarray-based bioinformatics.
Setting
Multiple pediatric intensive care units in the United States.
Patients
Children ≤10 years of age: 18 normal controls, 22 meeting criteria for SIRS, 32 meeting criteria for sepsis, and 67 meeting criteria for septic shock on day 1. The available day 3 samples included 20 patients still meeting sepsis criteria, 39 patients still meeting septic shock criteria, and 24 patients meeting the exclusive day 3 category, SIRS resolved.
Interventions
None other than standard care.
Measurements and Main Results
Longitudinal analyses were focused on gene expression relative to control samples and patients having paired day 1 and day 3 samples. The longitudinal analysis focused on up-regulated genes revealed common patterns of up-regulated gene expression, primarily corresponding to inflammation and innate immunity, across all patient groups on day 1. These patterns of up-regulated gene expression persisted on day 3 in patients with septic shock, but not to the same degree in the other patient classes. The longitudinal analysis focused on down-regulated genes demonstrated gene repression corresponding to adaptive immunity-specific signaling pathways and was most prominent in patients with septic shock on days 1 and 3. Gene network analyses based on direct comparisons across the SIRS, sepsis, and septic shock spectrum, and all available patients in the database, demonstrated unique repression of gene networks in patients with septic shock corresponding to major histocompatibility complex antigen presentation. Finally, analyses focused on repression of genes corresponding to zinc-related biology demonstrated that this pattern of gene repression is unique to patients with septic shock.
Conclusions
Although some common patterns of gene expression exist across the pediatric SIRS, sepsis, and septic shock spectrum, septic shock is particularly characterized by repression of genes corresponding to adaptive immunity and zinc-related biology. (Crit Care Med 2009; 37:1558–1566)
Keywords: microarray, T cell, antigen presentation, children, inflammation, zinc
The systemic inflammatory response syndrome (SIRS) is common in critically ill children (1). SIRS is not a specific disease and recognition of SIRS does not necessarily drive clinical decision processes (2). SIRS is defined by alterations of three fundamental physiologic variables and one basic laboratory variable (3). Although the clinical utility of SIRS is debatable (4, 5), at a minimum, the SIRS concept serves to broadly describe the clinical and heterogeneous systemic inflammatory response of critically ill patients to a variety of insults, including trauma, surgery, and pancreatitis (2).
When critically ill patients meet criteria for SIRS, in the context of infection, they are said to have “sepsis.” Critically ill patients with sepsis, who progress to the development of cardiovascular and other organ failure, are said to have “septic shock” or “severe sepsis.” Although sepsis and septic shock are also highly heterogeneous clinical syndromes, the recognition of sepsis and septic shock does drive clinical decision processes and dictates specific therapeutic strategies (2, 6). Thus, in critically ill children, there exists a clinically relevant, conceptual spectrum of increasing illness severity and specificity ranging from SIRS to sepsis to septic shock.
In an ongoing translational research program based on genome-wide expression profiling, we have generated novel observations in children with septic shock (7–9). An important caveat to our observations surrounds the issue of specificity. That is, all of our previous genome-wide expression studies have focused exclusively on children with septic shock, while ignoring the categories of SIRS and sepsis. Thus, an important question to address is: are the genome-wide expression signatures that we have derived for pediatric septic shock, and the resulting hypotheses, specific to septic shock, or are they generalizable to other populations of critically ill children? We recently addressed this question, in part, by formally validating our previous observations in a separate validation septic shock cohort (10). In the current study, we have addressed this question by directly comparing the genome-wide expression signatures of children with SIRS, sepsis, or septic shock.
METHODS
Patients
The study protocol was approved by the individual institutional review boards of each participating institution. Children ≤10 years of age admitted to the pediatric intensive care units (PICUs) and meeting published, pediatric-specific criteria for SIRS, sepsis, or septic shock were eligible (3). All patients were assigned to one of these three classifications at study entry (day 1), and when appropriate re-classified on day 3 based on the same criteria. An additional day 3 classification, “SIRS resolved,” was used for patients not meeting criteria for at least SIRS on day 3. Control patients were recruited from the ambulatory departments of participating institutions using previously published exclusion criteria (7, 8, 10).
Sample and Data Collection
After obtaining informed consent from parents or legal guardians, blood samples were obtained on day 1 of the study and when possible on day 3 of the study. Day 1 of the study was defined as within the first 24 hours of meeting criteria for a given study category in any PICU patient, whether at admission to the PICU or after initial admission to the PICU with a nonstudy-related classification. Day 3 of the study was defined as 48 hours after drawing day 1 samples, regardless of the PICU status. Severity of illness was calculated using the Pediatric Risk of Mortality III Score (11). Organ failure was defined using pediatric-specific criteria (1, 12). Annotated clinical and laboratory data were collected daily while in the PICU using a web-based database developed locally.
RNA Extraction and Microarray Hybridization
The data and protocols described in this article are Minimum Information About a Microarray Experiment (MIAME) compliant and can found under the Gene Expression Ommibus accession number GSE13904 (http://www.ncbi.nlm.nih.gov/geo/). Sub-groups of all the controls and the patients in the day 1 septic shock category have been previously reported in subcohort analyses addressing different questions than that addressed in the current study (7, 8, 10), but have not been analyzed or reported as one single cohort. None of the patients with SIRS or sepsis have been previously reported.
Total RNA was isolated from whole blood samples using the PaxGene Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA) according the manufacturer’s specifications. Microarray hybridization was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children’s Hospital Research Foundation as previously described using the Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA) (7, 8).
Data Analysis
Analyses were performed using one patient sample per chip. Image files were captured using an Affymetrix GeneChip Scanner 3000. CEL files were subsequently preprocessed using robust multiple-array average normalization using GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA) (13). All signal intensity–based data were used after robust multiple-array average normalization, which specifically suppresses all but significant variation among lower intensity probe sets. All chips representing patient samples were then normalized to the respective median values of controls.
Differences in messenger RNA abundance between the individual patient classifications and controls were measured by sequential statistical and expression filters using GeneSpring GX 7.3. The statistical filter consisted of analysis of variance (Welch Student’s t test) using the respective patient categories and controls as the comparison groups, and corrections for multiple comparisons using the Benjamini-Hochberg false discovery rate (10). The false discovery rate used for patients with SIRS, sepsis, or SIRS resolved was 1%. Because the septic shock group had a substantially larger number of patients, we adjusted for this discrepancy by applying a more stringent false discovery rate of 0.1% to this patient group. The expression filter was applied after the statistical filter and selected only the genes having at least two-fold expression difference between the medians of the respective patient categories and the controls.
Gene lists of differentially expressed genes were primarily analyzed using the Ingenuity Pathways Analysis application (Ingenuity Systems, Redwood City, CA) that provides a tool for the discovery of signaling pathways and gene networks within the uploaded gene lists as previously described (8, 14). Adjunct analyses of gene lists were performed using three distinct, public, relational databases of functional gene annotations: D.A.V.I.D. (Database for Annotation, Visualization and Integrated Discovery) (15), the PANTHER classification system (protein analysis through evolutionary relationships) (16, 17), and ToppGene (18). These applications are all based on the established biomedical literature and use specific approaches to estimate significance (p values) based on nonredundant representations of the microarray chip and to convert the uploaded gene lists to gene lists containing a single value per gene. The p values provide an estimate of the probability that a given enrichment is present by chance alone and are derived using corrections for multiple comparisons (see Table footnotes).
RESULTS
Gene Expression Relative to Controls
Longitudinal analyses were conducted using 18 control subjects, and a total of 84 patients having SIRS, sepsis, or septic shock at study entry (day 1) and having available day 3 data if still alive. Table 1 provides the number of patients in each category at study entry, and other demographic and clinical data. As expected, the patients with septic shock had a significantly higher illness severity (Pediatric Risk of Mortality Scores), higher mortality, and a greater degree of organ failure, compared with patients with SIRS or sepsis. None of the other variables provided in Table 1 were significantly different. The median absolute neutrophil, lymphocyte, and monocyte counts were not significantly different between the respective patient categories (data not shown), thus, indicating that the patterns of differential gene expression among the patient groups (provided below) are not simply an artifact of differential white blood cell counts.
Table 1.
Patient demographics for longitudinal analysis
| Controls | SIRS | Sepsis | Septic Shock | |
|---|---|---|---|---|
| Number of patients | 18 | 14 | 14 | 56 |
| Median age in years (IQR) | 1.4 (0.2–3.8) | 3.1 (1.8–7.1) | 2.2 (1.2–5.5) | 2.3 (0.8–6.1) |
| Median PRISM score (IQR) | n/a | 12 (10–17) | 10 (5–12) | 18 (12–23)a |
| % Mortality (28 d) | n/a | 0 | 7 | 27b |
| Median number of organ failure (IQR)c | n/a | 1 (0–1) | 1 (0–1.2) | 2 (2–3)a |
| Number of males/females | 10/8 | 8/6 | 10/4 | 27/29 |
| Number with documented infection | n/a | 0 | 32d | 43e |
| % With comorbidityf | n/a | 38 | 37 | 40 |
| % Immunosuppressedg | n/a | 33 | 22 | 37 |
| Day 3 classification | ||||
| SIRS | n/a | 4 | 0 | 1 |
| SIRS resolved | n/a | 9 | 5 | 8 |
| Sepsis | n/a | 1 | 8 | 5 |
| Septic shock | n/a | 0 | 1 | 33 |
| Death before day 3 | n/a | 0 | 0 | 9 |
SIRS, systemic inflammatory response syndrome; IQR, interquartile range; PRISM, Pediatric Risk of Mortality Score; ANOVA, analysis of variance; PICU, pediatric intensive care unit; n/a, not applicable.
p < 0.05 vs. SIRS and sepsis (ANOVA Kruskal-Wallis);
p < 0.05 vs. SIRS and sepsis (chi-square);
refers to the maximum number of organ failures over the initial 7 d in the PICU;
Gram-negative bacteria (38%); Gram-positive bacteria (36%); and other (26%);
Gram-negative bacteria (22%); Gram-positive bacteria (54%); and other (24%);
refers to any comorbidity or chronic condition other than the primary diagnosis leading to PICU admission;
refers to the presence of an intrinsic immunodeficiency, or a patient receiving immunosuppressive medications (including steroids).
Seventy-five of the 84 patients had a paired day 3 sample available for analysis. The nine patients not having a paired day 3 sample were those with septic shock who died before the day 3 sampling time point. These nine patients were included in the longitudinal analysis because of the clinical importance of this phenotype. The respective day 3 categories for the 84 patients are also shown in the table.
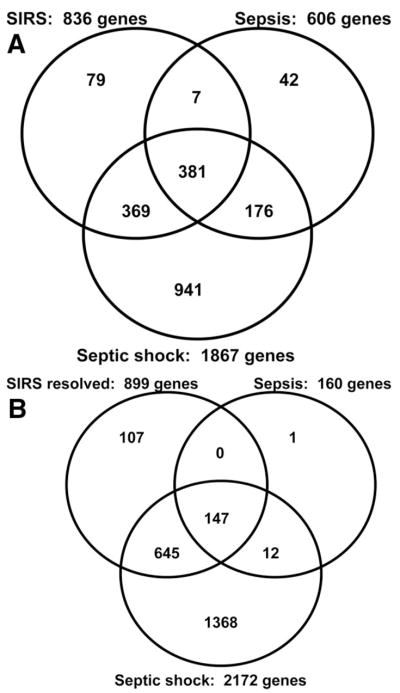
Lists of differentially regulated genes between each of the respective study categories and controls were generated as described in the Methods section (days 1 and 3), and as summarized in Table 2 (see Supplemental Digital Content 1, http://links.lww.com/A1100; Supplemental DigitalContent2, http://links.lww.com/A1101; Supplemental Digital Content 3, http://links.lww.com/A1102; Supplemental Digital Content 4, http://links.lww.com/A1103 for complete gene lists). As shown in Figure 1A, Venn analysis demonstrated that 381 genes were common to all three respective day 1 gene lists. The majority of genes differentially regulated in the SIRS (90%) and the sepsis categories (92%), respectively, were also differentially regulated in the septic shock category.
Table 2.
Number of differentially regulated genes between the respective patient categories and control subjectsa
| SIRSb | Sepsis | Septic Shock | SIRS Resolvedc | |
|---|---|---|---|---|
| Day 1 | ||||
| Total | 836 | 606 | 1867 | n/a |
| Up-regulated (%) | 537 (64) | 448 (74) | 995 (53) | n/a |
| Down-regulated (%) | 299 (36) | 158 (26) | 872 (47) | n/a |
| Day 3 | ||||
| Total | n/a | 160 | 2172 | 899 |
| Up-regulated (%) | n/a | 150 (94) | 1224 (56) | 686 (76) |
| Down-regulated (%) | n/a | 10 (6) | 948 (44) | 213 (24) |
SIRS, systemic inflammatory response syndrome.
See Methods section for description of gene list derivations;
day 3 analysis not conducted because there were only five patients still meeting SIRS criteria on day 3;
day 1 analysis not conducted because “SIRS resolved” was an exclusive day 3 category.
Figure 1.
A, Venn analysis comparing the day 1 genes differentially regulated between the respective patient categories and the control subjects (see Table 2). B, Venn analysis comparing the day 3 genes differentially regulated between the respective patient categories and the control subjects (see Table 2). SIRS, systemic inflammatory response syndrome.
Because there were only five patients meeting SIRS criteria on day 3, the day 3 analysis was focused on patients with SIRS resolved (n = 22), sepsis (n = 14), and septic shock (n = 34). As shown in Figure 1B, Venn analysis demonstrated that 147 genes were common to all three respective day 3 gene lists. Compared with the day 1 data, the day 3 data demonstrate that by the third day of illness, patients with sepsis had a much smaller number of genes differentially regulated relative to control subjects. Also, the majority of genes differentially regulated in the SIRS resolved (88%) and the sepsis categories (99%), respectively, were also differentially regulated in the septic shock category. Finally, the data demonstrate that only patients in the septic shock category continued to have a large unique number of differentially regulated genes on day 3.
The lists of differentially regulated day 1 genes were uploaded to the Ingenuity Pathways Analysis application (see Methods section), and the analytic output was focused on coordinated expression or repression of genes corresponding to signaling pathways (data not shown). The longitudinal analysis focused on up-regulated genes revealed common patterns of up-regulated gene expression, primarily corresponding to inflammation and innate immunity, across all patient groups on day 1. These patterns of up-regulated gene expression persisted on day 3 in patients with septic shock, but not to the same degree in the other patient classes. The longitudinal analysis focused on down-regulated genes demonstrated gene repression corresponding to adaptive immunity-specific signaling pathways and was most prominent in patients with septic shock on days 1 and 3.
In previous studies, we have made the novel observation that pediatric septic shock is characterized by large scale and persistent repression of genes having functional annotations related to zinc and metal binding (7–10). When the down-regulated genes were uploaded to the D.A.V.I.D. database, the lists from patients with SIRS, sepsis, or SIRS resolved were not significantly enriched, on days 1 and 3, for any functional annotations related to zinc and heavy metal binding (data not shown), thus, indicating that our previous observations are specific to septic shock.
Differential Gene Expression Across Patient Groups—Day 1
The longitudinal analyses described above were based on differential gene expression between the respective patient categories and controls, and were restricted only to patients having paired day 1 and day 3 data. Herein, we took an alternative analytic approach by directly comparing gene expression across the patient groups on days 1 and 3. To increase the statistical power of these cross-patient group comparisons, we included all patients in the database, thus, removing the restriction of having paired day 1 and day 3 samples.
For the day 1 analysis, we conducted a three-group analysis of variance (Welch test with a Benjamini false discovery rate of 5%) using patients with SIRS (n = 22), sepsis (n = 32), and septic shock (n = 67) as the comparison groups. The starting gene list for this three-group analysis consisted of the 1995 genes that were found in any of the three gene lists depicted in Figure 1A. This direct statistical analysis yielded 136 genes that were differentially regulated between patients with SIRS, sepsis, or septic shock.
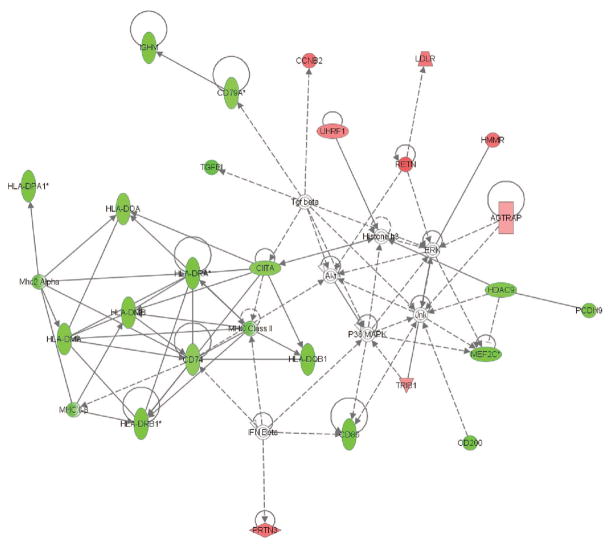
We next uploaded these 136 genes to the Ingenuity Pathways Analysis application and focused the analytic output on the presence of gene networks, with gene nodes being colored based on the degree of increased (red color intensity) or decreased (green color intensity) expression in the patients with septic shock relative to patients with SIRS or sepsis. As shown in Figure 2, these 136 genes corresponded to a network of genes containing multiple repressed genes from the human leukocyte antigen family. To derive more objective biological meaning from this gene network, we uploaded the network genes to three, distinct, independent ontology databases (PANTHER, ToppGene, and D.A.V.I.D., see Methods section) and focused the analytic output on the most significant (based on p values) functional annotations derived from each database. As shown in Table 3, this gene network was enriched for functional annotations related to major histocompatibility complex (MHC) class II–mediated antigen function and T-cell function. Thus, when differential gene expression is directly compared across the day 1 patient groups, the differentially regulated genes correspond to a gene network related to repression of antigen processing and T-cell function in patients with septic shock.
Figure 2.
Gene network derived from the 136 genes differentially regulated on day 1 between patients with systemic inflammatory response syndrome (SIRS), sepsis, and septic shock (see text for derivation of the 136 genes and for network derivation). Red intensity within a gene node corresponds to increased expression in the patients with septic shock, relative to the patients with SIRS or sepsis, and green intensity within a gene node corresponds to decreased expression in the patients with septic shock, relative to the patients with SIRS or sepsis. This network has a score of 54, which is equivalent to a p value of 1.0E–54. The p value provides an estimate of the probability that the network genes are present in the uploaded gene list by chance alone. A network legend and a complete list of the network genes are provided in Supplemental Digital Content 5, http://links.lww.com/A1104. HLA, human leukocyte antigen; MHC, major histocompatibility complex; IFN, interferon; IL, interleukin; IGHM, immunoglobulin heavy constant mu; UHRF, ubiquitin-like with PHD and ring finger domains 1; LDLR, low density lipoprotein receptor; AGTRAP, angiotensin II receptor-associated protein; CCNB, cyclin B2; RETN, resistin; HMMR, hyaluronan-mediated motility receptor (RHAMM); HDAC, histone deacetylase 9; MAPK, mitogen activated protein kinase; ERK, extracellular regulated MAP kinase; JNK, jun n-terminal kinase; TRIB, tribbles homolog 1; MEF, myocyte enhancer factor.
Table 3.
Functional annotations corresponding to the differentially expressed gene network shown in Figure 2
| Database | Functional Annotations (p) |
|---|---|
| PANTHERa | Pathway: T-cell activation (5.1E–15) |
| Biological process: MHC II-mediated immunity (1.8E–17) | |
| Molecular function: major histocompatibility complex antigen (5.5E–14) | |
| ToppGenea | Molecular function: MHC class II receptor activity (<1.0E–6) |
| Biological process: antigen processing and presentation (<1.0E–6) | |
| Mouse phenotype: abnormal antigen processing via MHC class II (<1.0E–6) | |
| Pathway: antigen processing and presentation (<1.0E–6) | |
| D.A.V.I.D.b | GOTERM_BP_ALL: MHC class II receptor activity (1.6E–9) |
PANTHER, protein analysis through evolutionary relationships; D.A.V.I.D., Database for Annotation, Visualization and Integrated Discovery; MHC, major histocompatibility complex.
Based on default parameters and Bonferroni correction for multiple comparisons;
based on default parameters and Benjamini-Hochberg false discovery rate of 5%.
Differential Gene Expression Across Patient Groups—Day 3
An identical analysis was conducted using all available day 3 samples for patients with SIRS resolved (n = 24), sepsis (n = 20), or septic shock (n = 39) and the 2280 genes that were found in any of the three gene lists depicted in Figure 1B. This analysis yielded 535 genes that were differentially regulated on day 3 between patients with SIRS resolved, sepsis, or septic shock.
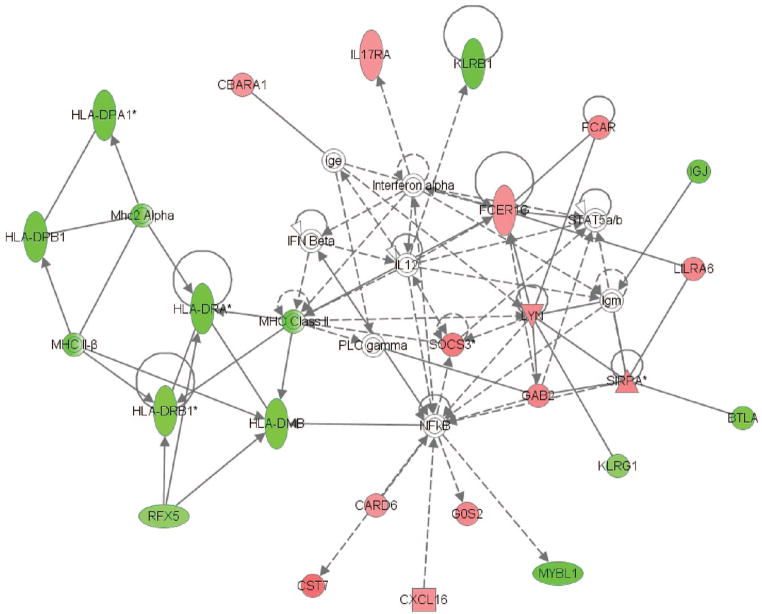
These 535 genes corresponded to two gene networks depicted in Figures 3 and 4, respectively. Similar to the day 1 network, the day 3 network shown in Figure 3 contains multiple genes from the human leukocyte antigen family that were repressed in the patients with septic shock, relative to the patients with SIRS resolved or sepsis. As shown in Table 4, this day 3 gene network was also enriched for functional annotations related to MHC class II–mediated antigen presentation and T-cell function.
Figure 3.
The first gene network derived from the 535 genes differentially regulated on day 3 between patients with systemic inflammatory response syndrome (SIRS) resolved, sepsis, and septic shock (see text for derivation of the 535 genes and for network derivation). Red intensity within a gene node corresponds to increased expression in the patients with septic shock, relative to the patients with SIRS resolved or sepsis, and green intensity within a gene node corresponds to decreased expression in the patients with septic shock, relative to the patients with SIRS resolved or sepsis. This network has a score of 36, which is equivalent to a p value of 1.0E–36. The p value provides an estimate of the probability that the network genes are present in the uploaded gene list by chance alone. A network legend and a complete list of the network genes are provided in Supplemental Digital Content 6, http://links.lww.com/A1105. HLA, human leukocyte antigen; MHC, major histocompatibility complex; IFN, interferon; IL, interleukin; RFX, regulatory factor X; CST, cystatin F; CARD, caspase recruitment domain family, member 6; CXCL, chemokine (C-X-C motif) ligand 16; GOS, G0/G1 switch 2; MYBL, v-myb myeloblastosis viral oncogene homolog; KLRG, killer cell lectin-like receptor subfamily G, member 1; BTLA, B and T lymphocyte associated; GAB, GRB2-associated binding protein 2; PLC, phospholipase C gamma; SOCS, suppressor of cytokine signaling 3; SIRPA, signal-regulatory protein alpha; LLRA, leukocyte immunoglobulin-like receptor, subfamily A; FCER, Fc fragment of IgE, high affinity I; FCAR, Fc fragment of IgA; KLRB, killer cell lectin-like receptor subfamily B, member 1.
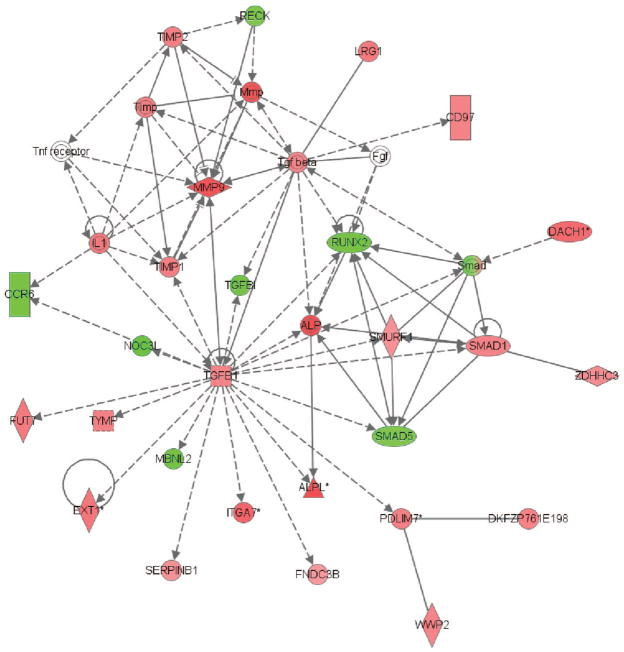
Figure 4.
The second gene network derived from the 535 genes differentially regulated on day 3 between patients with systemic inflammatory response syndrome (SIRS) resolved, sepsis, and septic shock (see text for derivation of the 535 genes and for network derivation). Red intensity within a gene node corresponds to increased expression in the patients with septic shock, relative to the patients with SIRS resolved or sepsis, and green intensity within a gene node corresponds to decreased expression in the patients with septic shock, relative to the patients with SIRS resolved or sepsis. This network has a score of 43, which is equivalent to a p value of 1.0E–43. The p value provides an estimate of the probability that the network genes are present in the uploaded gene list by chance alone. A network legend and a complete list of the network genes are provided in Supplemental Digital Content 7, http://links.lww.com/A1106. TNF, tumor necrosis factor; MMP, matrix metalloproteinase; IL, interleukin; TGF, transforming growth factor; TIMP, TIMP metallopeptidase inhibitor; RECK, reversion-inducing-cysteine-rich protein with kazal motifs; LRG, leucine-rich alpha-2-glycoprotein 1; RUNX, runt-related transcription factor 2; SMURF, SMAD specific E3 ubiquitin protein ligase 1; FUT, fucosyltransferase 7; NOC, nucleolar complex associated 3 homolog; CCR, chemokine (C-C motif) receptor 6; ITGA, integrin, alpha 7; ALPL, alkaline phosphatase, liver/bone/kidney; PDLIM, PDZ and LIM domain 7; MBNL, muscleblind-like 2; DAHC, dachshund homolog 1; WWP, WW domain containing E3 ubiquitin protein ligase 2; FDNC, fibronectin type III domain containing 3B.
Table 4.
Functional annotations corresponding to the differentially expressed gene network shown in Figure 3
| Database | Functional Annotations (p) |
|---|---|
| PANTHERa | Pathway: T-cell activation (4.7E–2) |
| Biological process: T-cell–mediated immunity (4.5E–9) | |
| Molecular function: major histocompatibility complex antigen (3.0E–7) | |
| ToppGenea | Molecular function: MHC class II receptor activity (<1.0E–6) |
| Biological process: antigen processing via MHC class II (<1.0E–6) | |
| Mouse phenotype: abnormal immune system physiology (1.1E–4) | |
| Pathway: antigen processing and presentation (1.0E–6) | |
| D.A.V.I.D.b | GOTERM_MF_ALL: MHC class II receptor activity (4.1E–4) |
PANTHER, protein analysis through evolutionary relationships; D.A.V.I.D., Database for Annotation, Visualization and Integrated Discovery; MHC, major histocompatibility complex.
Based on default parameters and Bonferroni correction for multiple comparisons;
based on default parameters and Benjamini-Hochberg false discovery rate of 5%.
The other day 3 network, shown in Figure 4, contains transforming growth factor (TGF)-β1 as a highly connected node that was up-regulated in the patients with septic shock, relative to patients with SIRS resolved or sepsis. As shown in Table 5, this day 3 gene network was enriched for functional annotations corresponding to the TGF-β signaling pathway and metalloproteinase inhibition.
Table 5.
Functional annotations corresponding to the differentially expressed gene network shown in Figure 4
| Database | Functional Annotations (p) |
|---|---|
| PANTHERa | Pathway: TGF-β signaling pathway (4.7E–3) |
| Biological process: developmental processes (6.8E–7) | |
| Molecular function: protease inhibitor (1.2E–6) | |
| ToppGenea | Molecular function: metalloendopeptidase inhibitor activity (2.5E–4) |
| Biological process: tissue remodeling (7.7E–5) | |
| Mouse phenotype: increased WBC count (3.9E–4) | |
| Pathway: inhibition of matrix metalloproteinases (1.0E–6) | |
| D.A.V.I.D.b | GOTERM_BP_ALL: organ development (3.8E–5) |
PANTHER, protein analysis through evolutionary relationships; D.A.V.I.D., Database for Annotation, Visualization and Integrated Discovery; TGF, transforming growth factor.
Based on default parameters and Bonferroni correction for multiple comparisons;
based on default parameters and Benjamini-Hochberg false discovery rate of 5%.
Thus, when differential gene expression is directly compared across the day 3 patient groups, the differentially regulated genes correspond to a gene network related to repression of antigen processing and T-cell function in patients with septic shock, and concomitant increased expression of a gene network related to TGF-β signaling in the patients with septic shock.
DISCUSSION
Elucidating the biological basis of disease spectrums affords unique opportunities for the development of more specific therapeutic strategies and novel diagnostic approaches. These opportunities have been most readily leveraged in the field of cancer (19–25), which, similar to septic shock, also involves heterogeneous disease processes that exist across spectrums of disease severity and specificity. We have attempted to elucidate the genome-wide expression profiles that differentiate the SIRS, sepsis, and septic shock spectrum, with the ultimate goal of developing novel knowledge regarding the pathobiology of pediatric septic shock.
Our previous reports involving genome-wide expression profiling in critically ill children were focused exclusively on septic shock relative to normals. These reports demonstrated persistent repression of genes corresponding to the adaptive immune system and zinc biology in children with septic shock (7–9), and were subsequently formally validated (10). However, the specificity of these observations in the broader context of critical illness remained to be formally tested. Accordingly, the primary objective of the current study was to determine whether our previous iterations of the genome-wide expression signatures of pediatric septic shock are specific to septic shock, or are relatively nonspecific epiphenomenon of pediatric critical illness.
The current data represent the most comprehensive genome-wide analysis across the spectrum of pediatric SIRS, sepsis, and septic shock to date. The data demonstrate that there are some common patterns of gene expression and repression across the SIRS, sepsis, and septic shock spectrum. There also exist, however, patterns of gene expression that are relatively specific to each of these three clinical syndromes. In particular, we have demonstrated distinctive patterns of gene expression in septic shock consistent with our previous reports. From the standpoints of morbidity and mortality, septic shock is the most clinically important patient classification used in the current study (2, 26, 27).
The greatest degree of commonality across the SIRS, sepsis, and septic shock spectrum was demonstrated by the day 1 data. The functional relevance of these overlaps was demonstrated by the analysis involving the up-regulated day 1 genes and their correspondence to signaling pathways. The signaling pathways represented by the up-regulated genes in patients with septic shock were broadly related to innate immunity and inflammation. All of these signaling pathways were also significantly expressed in the patients with SIRS or sepsis. These data confirm, at a genomic level, the longstanding concept that SIRS, sepsis, and septic shock share common features related to activation of innate immunity and inflammation (2, 28, 29).
Beyond the above data, the gene expression patterns began to diverge across the various patient groups and the most notable divergence occurred in the patients with septic shock. For example, in the patients with septic shock, the up-regulated signaling pathways related to innate immunity and inflammation persisted on day 3, compared with patients with sepsis or SIRS resolved. These data would indicate that septic shock is characterized by persistent activation of the innate immune and inflammatory systems compared with the relatively milder clinical categories of sepsis and SIRS.
In the context of our previous data, the most biologically important examples of divergence involved the down-regulated genes. When the down-regulated genes in each clinical category were interrogated for enrichment of functional annotations related to zinc and heavy metals, we found this type of enrichment exclusively in the patients with septic shock. As we have stated in our previous reports, the potential significance of this observation is based on the important role that normal zinc homeostasis plays in normal functioning of the innate and adaptive immune systems (30–32).
The analyses involving repression of genes corresponding to signaling pathways also support the specificity of our previous findings. Similar to our previous reports, patients with septic shock were characterized by repression of genes corresponding to the adaptive immune system, and this pattern of repression persisted on day 3 (8, 9). These gene repression patterns do not seem to be an artifact of the white blood cell subpopulation counts as we have previously reported (8). These data indicate that persistent repression of key adaptive immunity genes and pathways are a more prominent feature of septic shock, compared with that of SIRS or sepsis. The network-based analyses, which represent direct statistical comparisons across the patient groups, further support the concept that alterations of the adaptive immune system are particularly characteristic of septic shock, but not of SIRS or sepsis.
Other aspects of the current data support a distinctive link between alterations of the adaptive immune system and septic shock. First, a large number genes corresponding to the interleukin (IL)-10 signaling pathway were persistently up-regulated in patients with septic shock. IL-10 has pleiotropic effects on the immune/inflammatory system and is generally regarded as an “anti-inflammatory” cytokine in that it counteracts multiple proinflammatory processes (33). In the context of the current data, it is notable that IL-10 negatively modulates MHC class II antigen expression (34) and suppresses T-cell proliferation (35). Second, it is notable that TGF-β was uniquely up-regulated on day 3 in patients with septic shock and served as a highly connected node in one of the gene networks corresponding to the day 3 differentially regulated genes. TGF-β is another pleiotropic cytokine with anti-inflammatory properties (36), and has been demonstrated to decrease expression of MHC-II molecules and class II major histocompatibility complex transactivator (37). The latter (class II major histocompatibility complex transactivator) is one of the down-regulated gene nodes in the day 1 network and is the major transcription factor that positively regulates the expression of human leukocyte antigen genes and MHC-II (38, 39).
Finally, genes corresponding to the IL-4 signaling pathway were also uniquely and persistently repressed in patients with septic shock. Activation of the IL-4 signaling pathway involves the Janus kinase/signal transducer and activator of transcription pathway and the insulin receptor substrate-1/Pi 3-kinase pathway (40). The most well-know functions of IL-4 include the differentiation of naïve T helper cells into Th2 cells and immunoglobulin isotype switching (40). As such, IL-4 is considered to be a key regulator of allergic conditions and immunity against parasites, and has not been a primary focus of investigation in the context of septic shock. Other modulating effects of IL-4 on the adaptive immune system, however, suggest that it could play a role in the pathobiology of septic shock. These include, increased expression of MHC-II molecules, increased expression of CD23, prevention of lymphocyte apoptosis, and modulation of leukocyte adhesion and inflammation (40). Indeed, one study demonstrated increased susceptibility to staphylococcal enterotoxin B–mediated shock in IL-4 null mice (41). Thus, the existing literature surrounding IL-4 biology indicates that further investigation may be warranted regarding the potential role of IL-4 in the pathobiology of septic shock.
This study has two potential limitations. One potential limitation involves the use of whole blood–derived RNA for generating the microarray data. This is a common criticism of our work in that the RNA is derived from a mixed population of white blood cells, thus, raising the possibility that the gene expression patterns we have reported merely reflect differences in the white blood cell populations. However, several aspects of our published and current work well demonstrate that biologically meaningful data can be derived using the current approach based on whole blood–derived RNA, as previously discussed and validated (7, 8, 10, 42).
A second potential limitation of our studies is our use of “day 1” and “day 3” to characterize the longitudinal gene expression patterns. We are cognizant that the “day 1” and “day 3” designations not necessarily reflect actual onset of disease, as could be achieved in the artificial and controlled environment of a research laboratory. Rather, these designations reflect recognition of disease in the PICU setting. The data reflect “real world” expression patterns in the context of the PICU, thus, providing clinically relevant data regarding gene expression patterns in patients with critical illness.
In conclusion, we have generated unprecedented, genome-wide data involving the pediatric spectrum of SIRS, sepsis, and septic shock. Although some biologically important commonalities exist across this spectrum, we have demonstrated that persistent repression of genes corresponding to adaptive immunity is unique to septic shock. These observations are consistent with emerging paradigms involving the pathobiology of septic shock, which have shifted from the traditional focus on innate immunity to that of the adaptive immune system (29, 43– 46). We have also demonstrated that alterations of zinc-related biological processes and IL-4 signaling are specific to septic shock, relative to SIRS and sepsis. The direct link(s) between these two latter observations, the adaptive immune system and septic shock, remain to be elucidated. However, potential links are biologically plausible and readily testable in the laboratory and at the bedside.
Supplementary Material
Acknowledgments
Supported by a grant from the National Institute of General Medical Sciences (RO1 GM064619).
Contributing investigators and centers for genomic database: Paul Checchia (St. Louis Children’s Hospital, St. Louis, MO); Allan Doctor (St. Louis Children’s Hospital, St. Louis, MO); Robert Fitzgerald (Devos Children’s Hospital, Grand Rapids, MI); Meena Kalyanaraman (Newark Beth Israel Medical Center, Newark, NJ); Gary Kohn (Morristown Memorial Hospital, Morristown, NJ); Cheri Landers (Kentucky Children’s Hospital, Lexington, KY); Gwenn McLaughlin (Jackson Memorial Hospital, Miami, FL); Scott Penfil (DuPont Hospital for Children, Wilmington, DE); Neal J. Thomas (Penn State Children’s Hospital, Hershey, PA); Nancy M. Tofil (The University of Alabama at Birmingham, Birmingham, AL); Margaret Winkler (The University of Alabama at Birmingham, Birmingham, AL); and Douglas Willson (University of Virginia, Charlottesville, VA).
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.ccmjournal.org).
The authors have not disclosed any potential conflicts of interest.
References
- 1.Proulx F, Fayon M, Farrell CA, et al. Epidemiology of sepsis and multiple organ dysfunction syndrome in children. Chest. 1996;109:1033–1037. doi: 10.1378/chest.109.4.1033. [DOI] [PubMed] [Google Scholar]
- 2.Shanley TP, Hallstrom C, Wong HR. Sepsis. In: Fuhrman BP, Zimmerman JJ, editors. Pediatric Critical Care Medicine. 3. St. Louis: Mosby; 2006. pp. 1474–1493. [Google Scholar]
- 3.Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6:2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 4.Baue AE. MOF, MODS, and SIRS: What is in a name or an acronym? Shock. 2006;26:438 – 449. doi: 10.1097/01.shk.0000228172.32587.7a. [DOI] [PubMed] [Google Scholar]
- 5.Baue AE. A debate on the subject “Are SIRS and MODS important entities in the clinical evaluation of patients?” The con position. Shock. 2000;14:590–593. doi: 10.1097/00024382-200014060-00003. [DOI] [PubMed] [Google Scholar]
- 6.Carcillo JA, Fields AI. Clinical practice parameters for hemodynamic support of pediatric and neonatal patients in septic shock. Crit Care Med. 2002;30:1365–1378. doi: 10.1097/00003246-200206000-00040. [DOI] [PubMed] [Google Scholar]
- 7.Wong HR, Shanley TP, Sakthivel B, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanley TP, Cvijanovich N, Lin R, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong HR. Pediatric septic shock treatment: New clues from genomic profiling. Pharmacogenomics. 2007;8:1287–1290. doi: 10.2217/14622416.8.10.1287. [DOI] [PubMed] [Google Scholar]
- 10.Cvijanovich N, Shanley TP, Lin R, et al. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34:127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III—Acute Physiology Score (PRISM III-APS): A method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr. 1997;131:575–581. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson JD, Pollack MM, Ruttimann UE, et al. Outcome of pediatric patients with multiple organ system failure. Crit Care Med. 1986;14:271–274. doi: 10.1097/00003246-198604000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 16.Mi H, Guo N, Kejariwal A, et al. PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Xu H, Aronow BJ, et al. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 20.Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr Opin Immunol. 2000;12:219–225. doi: 10.1016/s0952-7915(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 21.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 22.Ebert BL, Golub TR. Genomic approaches to hematologic malignancies. Blood. 2004;104:923–932. doi: 10.1182/blood-2004-01-0274. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 26.Watson RS, Carcillo JA, Linde-Zwirble WT, et al. The epidemiology of severe sepsis in children in the United States. Am J Respir Crit Care Med. 2003;167:695–701. doi: 10.1164/rccm.200207-682OC. [DOI] [PubMed] [Google Scholar]
- 27.Watson RS, Carcillo JA. Scope and epidemiology of pediatric sepsis. Pediatr Crit Care Med. 2005;6:S3–S5. doi: 10.1097/01.PCC.0000161289.22464.C3. [DOI] [PubMed] [Google Scholar]
- 28.Abraham E. Why immunomodulatory therapies have not worked in sepsis. Intensive Care Med. 1999;25:556–566. doi: 10.1007/s001340050903. [DOI] [PubMed] [Google Scholar]
- 29.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 30.Rink L, Haase H. Zinc homeostasis and immunity. Trends Immunol. 2007;28:1–4. doi: 10.1016/j.it.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Fraker PJ, King LE. Reprogramming of the immune system during zinc deficiency. Annu Rev Nutr. 2004;24:277–298. doi: 10.1146/annurev.nutr.24.012003.132454. [DOI] [PubMed] [Google Scholar]
- 32.Ibs KH, Rink L. Zinc-altered immune function. J Nutr. 2003;133:1452S–1456S. doi: 10.1093/jn/133.5.1452S. [DOI] [PubMed] [Google Scholar]
- 33.Moore KW, de Waal Malefyt R, Coffman RL, et al. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 34.de Waal Malefyt R, Abrams J, Bennett B, et al. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: An autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taga K, Mostowski H, Tosato G. Human in-terleukin-10 can directly inhibit T-cell growth. Blood. 1993;81:2964–2971. [PubMed] [Google Scholar]
- 36.Li MO, Wan YY, Sanjabi S, et al. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y, Tang L, Letterio JJ, et al. The smad3 protein is involved in TGF-beta inhibition of class II transactivator and class II MHC expression. J Immunol. 2001;167:311–319. doi: 10.4049/jimmunol.167.1.311. [DOI] [PubMed] [Google Scholar]
- 38.Fontes JD, Kanazawa S, Nekrep N, et al. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect. 1999;1:863–869. doi: 10.1016/s1286-4579(99)00232-4. [DOI] [PubMed] [Google Scholar]
- 39.Masternak K, Muhlethaler-Mottet A, Villard J, et al. Molecular genetics of the Bare lymphocyte syndrome. Rev Immunogenet. 2000;2:267–282. [PubMed] [Google Scholar]
- 40.Nelms K, Keegan AD, Zamorano J, et al. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 41.Aroeira LS, Martinez AC. The role of IL-4 in the staphylococcal enterotoxin B-triggered immune response: Increased susceptibility to shock and deletion of CD8Vbeta8 + T cells in IL-4 knockout mice. Eur J Immunol. 1999;29:1397–1405. doi: 10.1002/(SICI)1521-4141(199904)29:04<1397::AID-IMMU1397>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 42.Wong HR, Cvijanovich N, Wheeler DS, et al. Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med. 2008;178:276–282. doi: 10.1164/rccm.200801-131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: Mediators, macrophages, lymphocytes and apoptosis. Shock. 1996;6:S27–S38. [PubMed] [Google Scholar]
- 44.Hotchkiss RS, Swanson PE, Freeman BD, et al. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit Care Med. 1999;27:1230–1251. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Hotchkiss RS, Tinsley KW, Karl IE. Role of apoptotic cell death in sepsis. Scand J Infect Dis. 2003;35:585–592. doi: 10.1080/00365540310015692. [DOI] [PubMed] [Google Scholar]
- 46.Felmet KA, Hall MW, Clark RS, et al. Prolonged lymphopenia, lymphoid depletion, and hypoprolactinemia in children with nosocomial sepsis and multiple organ failure. J Immunol. 2005;174:3765–3772. doi: 10.4049/jimmunol.174.6.3765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.