Summary
The centromere is responsible for accurate chromosome segregation. Mammalian centromeres are specified epigenetically, with all active centromeres containing centromere specific chromatin in which CENP-A replaces histone H3 within the nucleosome. The proteins responsible for assembly of human CENP-A into centromeric nucleosomes during the G1 phase of the cell cycle are now identified to be distinct from the chromatin assembly factors that load other histone H3 variants. Prenucleosomal CENP-A is complexed with histone H4, nucleophosmin 1 and HJURP. Recruitment of new CENP-A into nucleosomes at replicated centromeres is dependent on HJURP. Recognition by HJURP is mediated through the centromere targeting domain (CATD) of CENP-A, a region that induces a unique conformational rigidity to both the subnucleosomal CENP-A heterotetramer and the corresponding assembled nucleosome. We propose HJURP to be a cell cycle regulated CENP-A specific histone chaperone required for centromeric chromatin assembly.
Introduction
The ability of cells to properly apportion a complete set of chromosomes to each daughter cell during mitosis is dependent on a unique chromatin domain known as the centromere. It is this locus on the chromosome, through its recruitment of a large macromolecular protein complex, that mediates the attachment of chromosomes to spindle microtubules as well as the transient recruitment of proteins involved in the mitotic or spindle assembly checkpoint (Cleveland et al., 2003; Musacchio and Salmon, 2007), the major cell cycle control pathway in mitosis. Centromeric chromatin incorporates a unique centromeric nucleosome containing Centromere Protein-A (CENP-A). In humans, CENP-A assembles into centromeric nucleosomes that recruit a CENP-A nucleosome associated complex (CENP-ANAC) present throughout the cell cycle (Foltz et al., 2006) as part of a larger group of proteins that make up a constitutive centromere complex (Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006). Distinct from the CENP-ANAC, the centromeric CENP-A nucleosome also interacts with three additional components, HJURP (Holliday Junction Recognition Protein, previously known as hFLEG1) and Nucleophosmin1 (NPM1) as well as the FACT complex (Foltz et al., 2006; Obuse et al., 2004). The consequence of the interaction of the CENP-A nucleosome with HJURP is explored below.
Human centromeric DNA is primarily comprised of 171 base pair alpha satellite elements arranged in tandem repeats (Manuelidis and Wu, 1978; Willard, 1985). However centromere identity in mammals is primarily defined epigenetically, with the underlying DNA sequence neither necessary nor sufficient (Marshall et al., 2008; Vafa and Sullivan, 1997; Warburton et al., 1997). The 0.5-5 megabases of alpha satellite DNA that are present within human centromeres (Cleveland et al., 2003) are packaged into chromatin by the assembly of centromere specific nucleosomes in which CENP-A replaces histone H3 (Palmer et al., 1987; Sullivan et al., 1994; Yoda et al., 2000). The centromere-specific nucleosomes are interspersed with canonical histone H3 containing nucleosomes (Blower et al., 2002). It is this unique CENP-A containing chromatin that is the most likely candidate to constitute the epigenetic mark of the centromeres. Obviously, each round of DNA synthesis presents a challenge for the stable propagation of a centromeric epigenetic mark, including deposition at replicated centromeres of new CENP-A nucleosomes.
Various compositions of CENP-A nucleosomes (or nucleosome-like complexes) have been suggested including tetrameric and hexameric complexes that could distinguish it from the canonical H3.1 containing octameric nucleosome (Dalal et al., 2007; Mizuguchi et al., 2007). The predominant form of the CENP-A in chromatin in vertebrate cells (Blower et al., 2002; Foltz et al., 2006), as well as in Drosophila (Blower et al., 2002), is a nucleosome containing both H2A and H2B in addition to H4 and CENP-A. Recombinant CENP-A combines with histone H4 to spontaneously form a heterotetramer containing two copies each of CENP-A and histone H4 (Black et al., 2004), similar to the subnucleosomal (H3:H4)2 heterotetramer. Further, in the presence of a DNA template CENP-A nucleosomes are formed in vitro into octameric nucleosomes with equal stoichiometries of CENP-A, H4, H2A and H2B (Black et al., 2007; Yoda et al., 2000), containing a conformationally more rigid core (Black et al., 2007), and accompanied by a steady-state unwrapping of 7 base pairs at the DNA entry/exit site, relative to H3-containing nucleosomes (Conde e Silva et al., 2007). On the other hand, in budding yeast, a hexameric nucleosome-like structure containing Cse4, the CENP-A homolog, and H4 (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007) in which Scm3 replaces histones H2A and H2B has been proposed.
Assembly of histone H3.1-containing nucleosomes is coincident with DNA replication and is accomplished through a stepwise mechanism (Jackson, 1990; Smith and Stillman, 1989; Smith and Stillman, 1991). Soluble, prenucleosomal histones H3.1 and H4 associate with the chromatin assembly factor-1 (CAF-1) complex consisting of CAF1p150, CAF1p60, and CAF1p46/48, and as a dimer with the anti-silencing factor 1 chaperone (ASF1) (English et al., 2006; English et al., 2005; Groth et al., 2007; Natsume et al., 2007). Assembly of the H3 and H4 heterotetramer along with two H2A:H2B dimers into the nucleosome is facilitated through its interaction with the CAF-1 complex (Kaufman et al., 1995; Smith and Stillman, 1989; Verreault et al., 1996). In contrast, while the histone H3.3 variant also interacts with ASF1 outside of S phase, it is incorporated into chromatin independent of DNA synthesis through the action of a distinct prenucleosomal complex that includes HIRA and CAF1p48, but is devoid of CAF1p150 and CAF1p60 (Ahmad and Henikoff, 2002; Tagami et al., 2004).
Very surprisingly, recruitment of new CENP-A to centromeric chromatin is not contemporaneous with replication of centromere DNA. Rather, it is restricted to a brief interval in G1 immediately following mitosis in human cells (Hemmerich et al., 2008; Jansen et al., 2007) and slightly earlier in anaphase in the rapidly dividing Drosophila syncytial embryo (Schuh et al., 2007). The assembly of new CENP-A nucleosomes in early G1 is coincident with the accumulation of the Mis18 complex (Mis18α, Mis18β and Mis18BP1/hsKNL2) at the centromere (Fujita et al., 2007; Hayashi et al., 2004; Maddox et al., 2007). CENP-A loading is dependent on this complex for assembly, although no direct interaction has been observed between CENP-A and Mis18.
Although members of the CAF-1 complex have been implicated in CENP-A nucleosome assembly in yeast, flies and humans (Furuyama et al., 2006; Hayashi et al., 2004; Sharp et al., 2002), no direct interaction has been demonstrated between human CENP-A and members of the CAF-1 complex. To this preceding work, we now use affinity tagging to identify prenucleosomal complexes containing human CENP-A. One component, HJURP is shown to be a CENP-A selective histone chaperone required for assembly of CENP-A nucleosomes.
Results
Identification of a CENP-A associated prenucleosomal complex
Prenucleosomal CENP-A or histone H3.1 and their associated proteins were purified from chromatin-depleted extracts of cells stably expressing tandem affinity purification (TAP) tagged versions of CENP-A or histone H3.1 (Fig. 1A, B). Since the chromatin bound CENP-A-TAP in these cells had been demonstrated previously to directly bind a collection of centromere proteins that comprise the CENP-ANAC (Foltz et al., 2006) and localize properly to centromeres (Fig. 1A), we reasoned that it must participate in the appropriate protein-protein interactions required for targeting and assembling new CENP-A nucleosomes at centromeres. Comparable proportions of endogenous CENP-A, TAP-tagged CENP-A, or histone H3 were in the initial chromatin free extracts (Fig. 1C). Affinity purification of CENP-TAP or H3.1-TAP (using IgG coupled beads) yielded complexes that were devoid of histone H2B (Fig. 1D), consistent with complexes representing prenucleosomal forms. This is in contrast to the purification of histone H3.1 and CENP-A from nucleosome-containing chromatin extracts where stoichiometric amounts of histone H2A and H2B were present (Foltz et al., 2006).
Figure 1. Affinity purification of the CENP-A prenucleosomal complex.
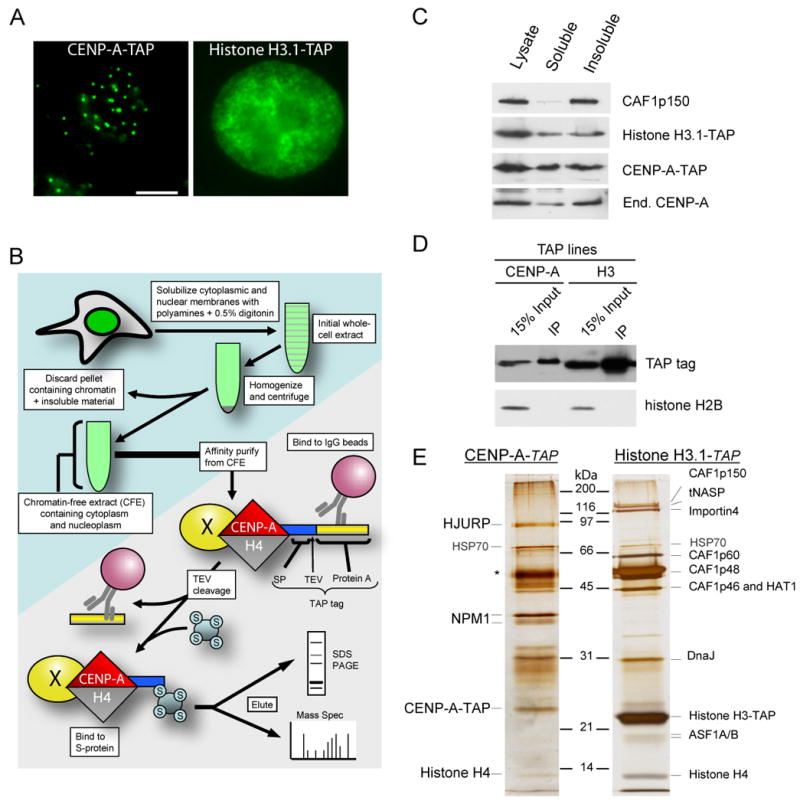
(A) Localization of CENP-A-TAP and histone H3.1-TAP in stable cell lines to centromeres and chromatin, respectively. The scale bar equals 5μm. (B) Purification scheme for identification of a soluble CENP-A prenucleosomal complex by production of a chromatin-free extract followed by tandem affinity purification. (C) Immunoblots of chromatin-free extract extracts derived from parental HeLa cells and stable cell lines expressing TAP-tagged CENP-A or histone H3.1 demonstrate the presence of tagged and endogenous histones as well as chromatin assembly factors. (D) Single-step affinity purifications of TAP-tagged CENP-A and histone H3.1 immunoblotted for the affinity tag as well as histone H2A. (E) Tandem affinity purified CENP-A and histone H3.1 and the associated complexes from chromatin free extracts were visualized by silver stain. Asterisk indicates contaminant present in both preparations. Proteins associated with the soluble complexes were identified in solution by MudPIT mass spectrometry.
A combination of silver staining (Fig. 1E) and mass spectrometry (Supp. Table 1) was used to demonstrate that the CENP-A and histone H3.1 prenucleosomal complexes obtained were almost completely distinct. Only histone H4, part of the final nucleosome, and HSP70 were common to both CENP-A and histone H3.1 prenucleosomal complexes. Consistent with previous reports (Kaufman et al., 1995; Smith and Stillman, 1989; Tagami et al., 2004), prenucleosomal TAP-tagged histone H3.1 was associated with members of the chromatin assembly factor 1 (CAF-1) complex including CAF1p150, CAF1p60, CAF1p48 and CAF1p46, as well as importin 4, histone acetyl transferase-1 (HAT1) and ASF1 (Fig. 1 E; Supp. Table 1).
The most prominent proteins uniquely associated with prenucleosomal CENP-A were the 32kD phosphoprotein nucleophosmin 1 (NPM1) and the 83 kD protein HJURP [Holliday Junction Recognizing Protein (Kato et al, 2007)]. (Fig. 1E).] Homologues of HJURP were identified in several mammals (human and mouse share only 40% sequence identity; Supp. Fig. 1B). Both NPM1 and HJURP were completely absent from prenucleosomal histone H3.1-TAP complexes. Both were also previously found to be associated with CENP-A nucleosomes present within centromeric chromatin, albeit at substantially lower levels as judged by silver staining and mass spectrometry (Foltz et al., 2006).
The CENP-A prenucleosomal complex did not contain any of the known constitutive centromere components. Inspection of the HJURP sequences revealed five highly conserved tryptophan residues (Supp. Fig. 1A,C) resembling the tryptophan – aspartate (WD40) repeats found in chromatin assembly factors such as CAF1p60, RbAp46, RbAp48 and HIRA (Supp. Fig. 1D). Two additional proteins, RuvB like-1 (RuvBL1) and replication protein A1 (RPA1) were identified in association with prenucleosomal CENP-A by solution mass spectrometry, although with low sequence counts (Supp. Table 1). Neither were observable by silver staining (Fig. 1E) and mass spectrometry of isolated silver stained bands failed to re-identify these proteins, indicating that these proteins are at best, substoichiometric components of the CENP-A prenucleosomal complex.
Direct association of HJURP and CENP-A
Sucrose gradient sedimentation of chromatin-free extracts from untransfected HeLa cells was used to characterize the prenucleosomal complexes of endogenous CENP-A. The majority of CENP-A and HJURP migrated together with a 10 S sedimentation coefficient (Fig. 2A). No significant pool of free CENP-A:H4 heterotetramer (3.2 S; Black et al., 2004) or dimer was present and no histone H2B sedimented with CENP-A in this prenucleosomal fraction. A minority of NPM1 cofractionated with prenucleosomal CENP-A, consistent with only a small proportion of total NPM1 stably associated with CENP-A. The partial overlap of NPM1 and HJURP with each other supports their formation of distinct prenucleosomal complexes with CENP-A. The peak of soluble CENP-A fractions did not contain an enrichment of RPA1 or RuvBL1, further suggesting that these proteins may be associated with only in a small subset of CENP-A prenucleosomal complex.
Figure 2. Identification of an endogenous CENP-A prenucleosomal complex.
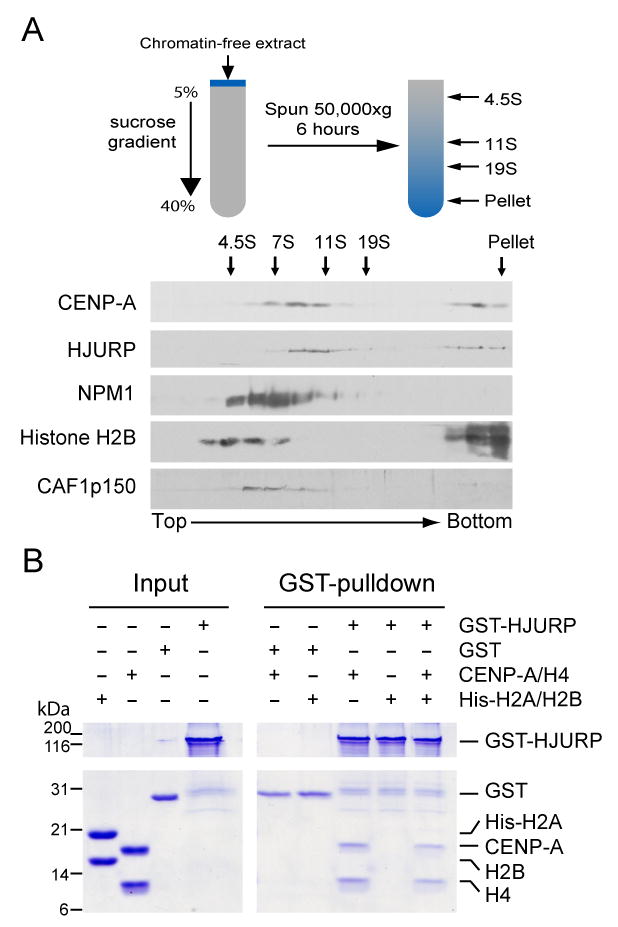
(A) Chromatin-free extract derived from HeLa cells was subjected to sucrose gradient sedimentation. The bottom of the gradient appears on the right. The migration of sedimentation coefficient standards bovine serum albumin, aldolase, catalase and thyroglobulin are indicated by arrows at the top. (B) A direct interaction was observed between recombinant GST-HJURP and recombinant untagged CENP-A:H4 but not recombinant His-H2A:H2B dimer. Following purification on glutathione agarose, proteins associated with GST-HJURP were separated by SDS-PAGE and stained with Coomassie blue.
To determine whether HJURP directly binds CENP-A, GST-HJURP, CENP-A and histone H4 were expressed and purified from E. coli. In assays where the CENP-A and histone H4 heterotetramer was combined with GST-HJURP or with GST, a complex with equimolar levels of CENP-A and H4 was selectively recovered with GST-HJURP (Fig. 2B). When added to these binding assays recombinant his-tagged H2A:H2B dimers did not interact with HJURP either alone or as part of the CENP-A:H4-HJURP complex. Thus, association of CENP-A:H4 and HJURP reflects a direct interaction that is independent of H2A:H2B and can form spontaneously in the absence of other cellular factors.
HJURP is required for CENP-A centromeric localization
To test if HJURP is required for CENP-A localization to centromeres, HJURP levels were reduced by transfection of siRNA targeting HJURP mRNA. Within 72 hours, HJURP protein levels were reduced to below 5% of their initial levels (Fig. 3A). Cell cycle distribution of the resulting cell population was not affected (Supp. Fig. 2). By three cell cycles after initiating HJURP depletion, the majority of cells had substantially reduced levels of endogenous CENP-A (Fig. 3B,D) or YFP-CENP-A (Fig. 3C,E) at individual centromeres. Since expression of YFP-CENP-A is controlled by the 5′ LTR of the virus used to produce the stable lines, this latter finding demonstrates that CENP-A loss is not due to cell cycle dependent transcriptional regulation of CENP-A. Cells reduced in HJURP developed a higher proportion (accumulating to more than a third of the total by 72 hours) of misshapen, multi-lobed nuclei or contained micronuclei (Fig. 3F). Both morphological abnormalities were phenocopies of siRNA mediated reduction in CENP-A itself (Black et al., 2007; Goshima et al., 2003) that drives chromosome missegregation events underlying the interphase nuclear defects. Our attempts to alter CENP-A nucleosome assembly by reducing NPM1 protein levels by siRNA showed no effect on overall levels of CENP-A at the centromere following a 72 hour treatment (data not shown). As we were only able to obtain modest suppression of NPM1 (70% reduction), we cannot rule out that the degree of NPM1 knockdown is insufficient to alter CENP-A assembly, especially given that NPM1 is a highly expressed protein. However, it is also possible that NPM1 plays a non essential role in the assembly of CENP-A nucleosomes or the nucleophosmin paralogues NPM2 and NPM3 may compensate for the absence of NPM1.
Figure 3. Loss of CENP-A recruitment in HJURP depleted cells.
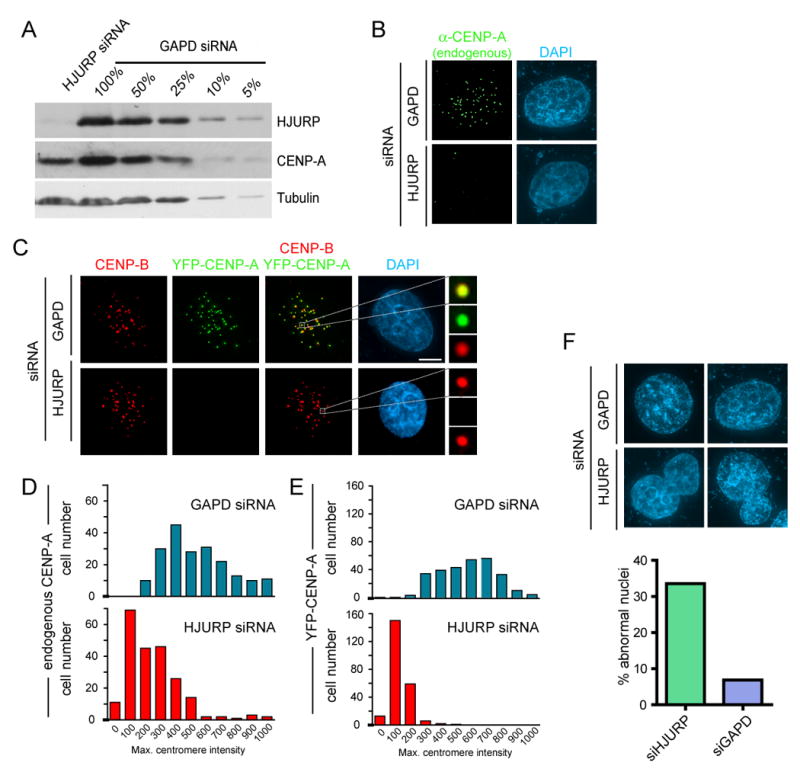
(A) Extracts from HeLa cells depleted of endogenous HJURP by treatment with siRNA pools for 72 hours were subjected to immunoblot. Serial dilution of GAPD control treated cell extracts was used to determine the degree of HJURP knockdown. Parental HeLa cells (B,D) or HeLa cells stably expressing YFP-CENP-A (C and E) treated with HJURP siRNA pools. Cells were pre-extracted and fixed 72 hours after the initiation of siRNA treatment and CENP-A and YFP-CENP-A were detected by immunofluoresence. Centromeres were identified by using anti-CENP-B monoclonal antibody in (C). Reduction of endogenous (B, n=200) and YFP-CENP-A (C, n=250) in response to HJURP siRNA was assessed by measuring the maximum pixel intensity per nucleus. All values are background corrected. Scale bar equals 5μm. (F) Abnormally shaped nuclei were observed by DAPI stain in HJURP siRNA treated cells at a greater frequency than controls. Abnormal nuclei included those that were multi lobed or contained micronuclei.
Long-term reduction of HJURP resulted in a reduction of the level of CENP-A protein overall (Fig. 3A, Supp. Fig. 3B), consistent with failure to load new CENP-A and/or loss from centromeres and suggesting instability of the pool of CENP-A that is not associated with the prenucleosomal complex or incorporated into centromeric chromatin. To test if putative histone chaperone activity of HJURP was limited to stabilizing prenucleosomal CENP-A, but not directly involved in its centromeric loading, CENP-A was expressed at high levels. If HJURP was required for stability but not loading, CENP-A should be incorporated into centromeres in the absence of HJURP. However, this was not the case. When cells were treated with siRNA against HJURP for 24 hours and subsequently transfected with YFP-CENP-A for the following 48 hours (Supp. Fig. 3B), few cells were still able to load YFP-CENP-A at centromeres (Supp. Fig. 3C). Indeed, most cells with reduced HJURP along with a sustained high accumulation of CENP-A showed a pattern of YFP-CENP-A staining consistent with its inclusion into general chromatin.
Cell-cycle regulated accumulation of HJURP at centromeres
Chromatin-free extracts derived from synchronized HeLa cells were immunoblotted to determine when in the cell cycle the CENP-A prenucleosomal complex was present (Fig. 4A). Levels of non-chromosomal CENP-A and HJURP were at their lowest in S-phase and early G2-phase, but rose together to peak levels in mitosis and early G1-phase. This is consistent with previous reports on CENP-A mRNA levels which rise during G2 (Shelby et al., 1997). Comparison of asynchronous cells with those blocked in mitosis (by treatment with nocodazole), confirmed that mitotic cells had a 2.5 fold higher level of non-chromatin bound CENP-A and HJURP than asynchronous cells and a 4.5 fold high level than cells at the G1/S boundary. Single step affinity purifications of CENP-A-TAP from these different points in the cell cycle demonstrated that HJURP and CENP-A remained associated at all times (Fig. 4A), even though loading of CENP-A is restricted to early G1 (Jansen et al., 2007).
Figure 4. Cell cycle regulation of HJURP accumulation and recruitment.
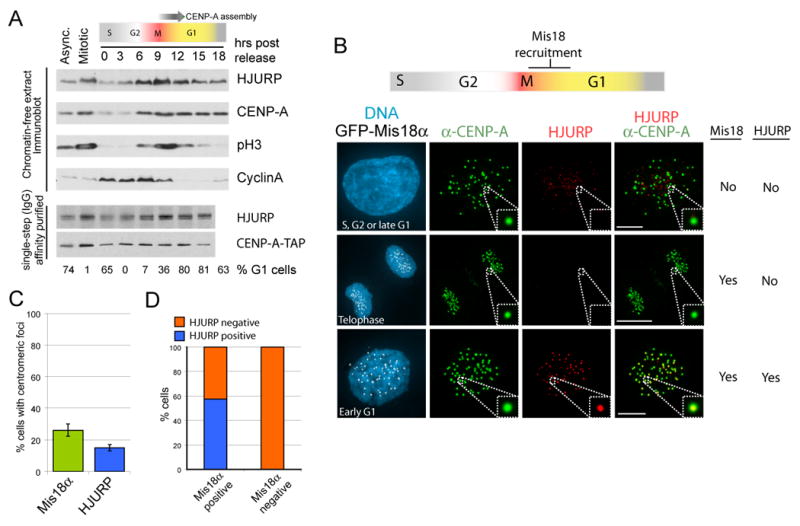
(A) The levels of soluble CENP-A and HJURP across the cell cycle were assessed in chromatin-free extracts from synchronized cells by immunoblot. Each lane contains extract from 1 X 105 cells. The percent of cells in G1 was determined by FACS analysis. (B) Cells stably expressing GFP-Mis18α were pre-extracted and fixed prior to immunostaining for HJURP and CENP-A. (C,D) Cells were scored for the accumulation of HJURP and GFP-Mis18α at CENP-A foci (n=280 cells). Data are represented as the mean (+/- S.D.), Scale bar equals 5μm.
HJURP localization was undertaken in cells stably expressing GFP-Mis18 α. The Mis18 complex is recruited to centromeres beginning in late anaphase and persists for approximately 3 hours after metaphase, which overlaps with the time during which new CENP-A is recruited to centromeres (Fujita et al., 2007; Jansen et al., 2007). HJURP was found to be strongly localized to centromeres during the period in early G1 when CENP-A nucleosomes are being assembled (Figure 4B). HJURP co-localized to centromeres with CENP-A in 15% of cells (Fig. 4B,C) In all cells where HJURP was present at centromeres, GFP-Mis18α was also centromere-bound (Fig. 4D). In contrast, only a subset, fifty-seven %, of cells with GFP-Mis18α present at centromeres also recruited HJURP. During anaphase and telophase HJURP was never visible at centromeres, demonstrating HJURP is recruited to centromeres during early G1, slightly later than Mis18. In the 85% of interphase cells where HJURP was not present at centromeres (Fig. 4B, C) it was distributed throughout the nucleus and accumulated to varying levels consistent with the cell-cycle regulation shown in Fig. 4A. The non-centromeric localization of HJURP was often punctate and may represent a pool of protein involved in a DNA damage response (as characterized by Kato et al. (2007)).
Centromeres cluster around nucleoli throughout the cell cycle (Suppl Fig. 4A,B); however, during G1, NPM1 was also found in extra-nucleolar foci following the disassembly of the nucleoli during mitosis (Boisvert et al., 2007). During G1, a subset of NPM1 foci was found in close association with centromeres (Supp. Fig. 4A,B), although these foci were usually larger and more diffuse than the centromere itself.
HJURP is required for loading of new CENP-A nucleosomes
To determine if HJURP is required for cell cycle dependent incorporation of CENP-A into centromeric nucleosomes after mitotic exit into G1, as opposed to HJURP acting only to maintain already assembled CENP-A nucleosomes, loading of newly synthesized CENP-A was tested during the first cell cycle after depletion of HJURP by siRNA. We have previously developed a pulse-chase labeling technique by fusing a SNAP tag to CENP-A (Jansen et al., 2007). Cell lines in which a CENP-A-SNAP fusion protein is localized to centromeres (Jansen et al., 2007) were partially synchronized with a first thymidine arrest and then released, transfected with siRNA to HJURP or to a control (GAPD), and then arrested at the next G1/S boundary (Fig. 5A). Under these conditions, HJURP is reduced to below 10% of initial levels (data not shown). Visualization of all existing CENP-A-SNAP was blocked with non-fluorescent benzylguanine Newly synthesized CENP-A-SNAP was labeled with TMR-Star (the fluorescent benzyl guanine) during the subsequent G2 phase and assembly of new labeled CENP-A nucleosomes was then assessed at the subsequent G1/S boundary. Post-mitotic loading of new CENP-A into centromeres was severely diminished in cells with reduced HJURP levels compared with the GAPD siRNA treated control cells where TMR-Star labeled CENP-A-SNAP was easily apparent (Fig. 5B,C).
Figure 5. HJURP is required for recruitment of new CENP-A nucleosomes.
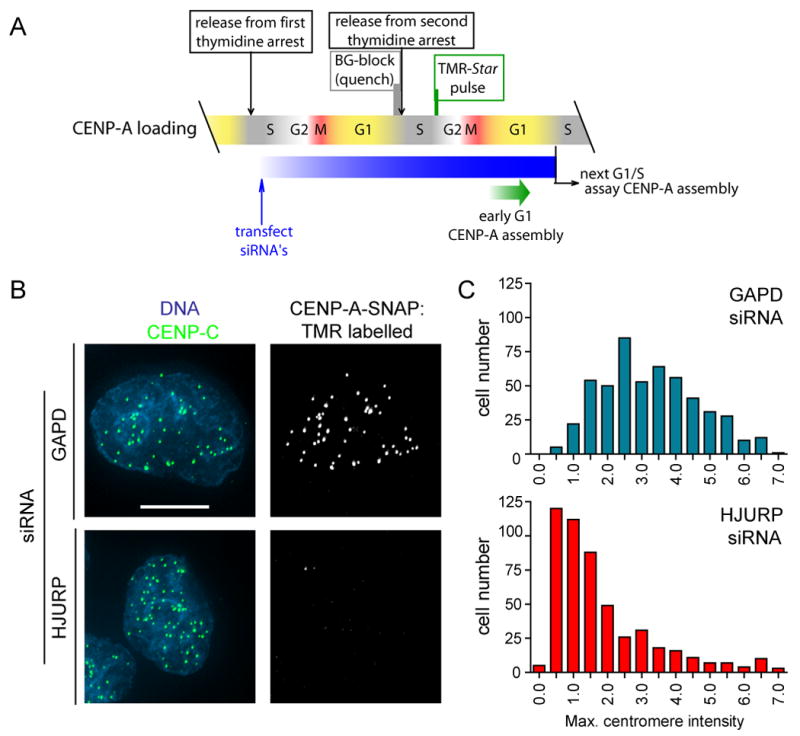
(A) Scheme for siRNA treatment and SNAP labeling to visualize loading of newly synthesized CENP-A-SNAP in the absence of HJURP. (B) Loading of newly synthesized CENP-A was assessed by the ability of cells to recruit TMR-Star labeled (i.e. newly synthesized) CENP-A to centromeres in cells treated with HJURP or GAPD siRNA and SNAP labeled as described in (A). Centromeres are shown by immunostaining for CENP-C. Scale bar equals 5μm. (C) The degree of TMR labeled CENP-A-SNAP loading into centromeres in siRNA treated cells was quantified by measuring the maximum pixel intensity per nucleus in >500 cells per condition.
The CENP-A targeting domain mediates the interaction with HJURP
We have previously established (Black et al., 2004; Black et al., 2007) that the cis-acting element within CENP-A required for its assembly at centromeres is the CENP-A targeting domain (CATD) comprised of loop 1 and α2 helix of the histone fold domain (Fig. 6A). Swapping the 22 amino acids of CENP-A that differ between the two variants within the CATD into histone H3.1 is sufficient to convert the corresponding heterotetramers (Black et al., 2004) or corresponding nucleosomes (Black et al., 2007) into structures that have an increased conformational rigidity relative to the corresponding complexes assembled with histone H3. Moreover, H3CATD not only assembles at centromeres but also provides an essential role of CENP-A in centromere maintenance (Black et al., 2007).
Figure 6. Histone H3CATD chimeric protein recruits HJURP.
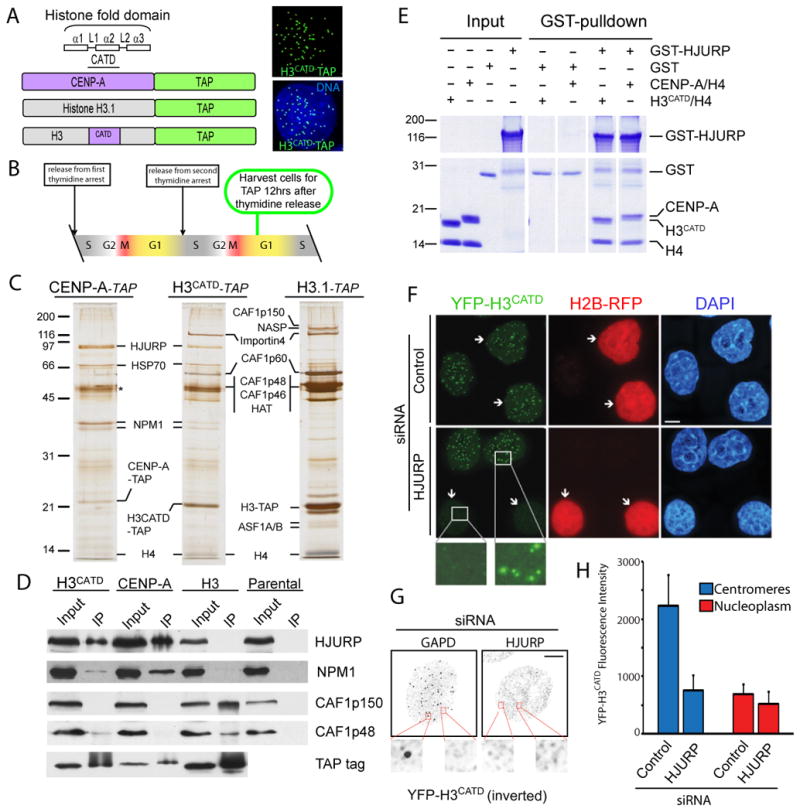
(A) Stable cell lines expressing a chimeric H3CATD in which the loop1 and α2 helix of CENP-A were swapped into histone H3 and fused to the TAP tag. H3CATD-TAP localizes to discreet centromeric foci. (B) Cells were harvested for tandem affinity purification (TAP) 12 hours after release from double thymidine arrest when the majority of cells are in the G1 phase. (C) Tandem affinity purifications conducted from stable cell lines during the G1 phase of the cell cycle. Proteins were identified by in-gel trypsin digestion and mass spectrometry, (D) Immunoblots of single-step affinity purifications. (E) Recombinant co-expressed chimeric H3CATD and histone H4 directly interacts with GST-HJURP in vitro. (F) A cell line that stably expresses YFP-H3CATD was treated with control or HJURP-directed siRNA and processed for immunofluorescence 72 hours following transfection. Arrowheads indicate cells co-transfected with the indicated siRNA plasmids and a plasmid encoding H2B-RFP. (G) Inverted grayscale images of YFP-H3CATD in which soluble protein was pre-extracted prior to fixation. Scale bars equal 5μm. (H) Quantitation of the effect of HJURP siRNA or control treatment on YFP-H3CATD fluorescent intensity. Centromeres and nucleoplasm were distinguished based on ACA immunostaining (n=12).
Since these observations suggested that proteins important for targeting CENP-A must also associate with H3CATD, we isolated proteins associated with H3CATD in cell lines stably expressing a TAP-tagged version of this chimeric histone. H3CATD-TAP localized to centromeres as expected (Fig. 6A). After double thymidine arrest and release to produce cells synchronized to be in the G1 cell cycle phase (Fig. 6B), proteins associated with prenucleosomal CENP-A, H3CATD and histone H3.1 were purified by tandem affinity (Fig. 6C). H3CATD bound a dual set of proteins that reflected both its histone H3 and CENP-A characteristics. CAF1p60, CAF1p46 and CAF1p48 were bound to prenucleosomal H3CATD, albeit the levels were clearly reduced relative to authentic H3.1 and the CAF1p150 subunit was absent. More importantly, despite its complete absence from histone H3.1 purifications done in parallel, swapping of the CATD region into histone H3 was sufficient for recognition and binding by HJURP, producing sufficient levels of bound HJURP to be clearly visible by silver stain (Fig. 6C) or immunoblotting (Fig. 6D). Inclusion of the CATD also recruited NPM1 to the prenucleosomal H3CATD, although not as efficiently as authentic CENP-A (Fig. 6D). The inclusion of the CATD within histone H3 is sufficient to mediate a direct interaction between between H3CATD and HJURP, as the recombinant GST-HJURP was able pulldown H3CATD:H4 in an equimolar ratio to a comparable degree as CENP-A:H4 (Fig. 6E).
Consistent with dual CENP-A and histone H3 characteristics, when YFP-H3CATD was stably expressed in a monoclonal cell line (Black et al., 2007) at a level comparable to endogenous CENP-A (2.4 X 106 copies per cell versus 2.0 X 106 copies per cell, respectively), it localized primarily to centromeres (as seen previously with ∼85% of the selectivity of bona fide CENP-A; Black et al., 2004) (Supp. Fig 5). In addition, YFP-H3CATD also incorporated at low level into general chromatin (Fig. 6F,G,H). After siRNA-mediated depletion of HJURP in these YFP-H3CATD-expressing cells, a large majority (84%) lost YFP-H3CATD from centromeres (Fig. 6F,H). In contrast, treatment with HJURP siRNA did not reduce the incorporation of the H3CATD into general chromatin, and this pool was stable in cells that were pre-extracted to remove soluble nuclear proteins (Fig. 6G, H). Therefore, although its incorporation into general chromatin persisted, most probably through its association with CAF1 components, H3CATD was unable to be loaded onto, or maintained at, centromeres in the absence of HJURP.
Discussion
Epigenetic inheritance of the centromere requires that CENP-A nucleosomes are incorporated into centromeric chromatin preferentially over the other histone H3 variants, H3.1 and H3.3. We have identified HJURP as a unique CENP-A histone chaperone required for new CENP-A nucleosome assembly at centromeres. With other chaperones known for the other H3 variants, this demonstrates that all histone H3 variants depend on different assembly factors to achieve distinct temporal and spatial patterns of deposition within chromatin. The complexes responsible for assembling histone H3.1 and H3.3 include CAF1p150 and CAF1p60 for H3.1 and HIRA for H3.3. The CENP-A prenucleosomal complex does not include the canonical CAFs. The interaction of prenucleosomal CENP-A with the HJURP histone chaperone that is unique for it and the absence of canonical histone chaperones supports a model where CENP-A is specifically targeted to centromeres by HJURP dependent deposition (Fig.7). Other proteins may facilitate the process of CENP-A nucleosome assembly including co-factors in the nucleosome assembly process and proteins that dictate the specific targeting of HJURP mediated CENP-A nucleosome deposition.
Figure 7. Model of CENP- A nucleosome deposition at centromeres.
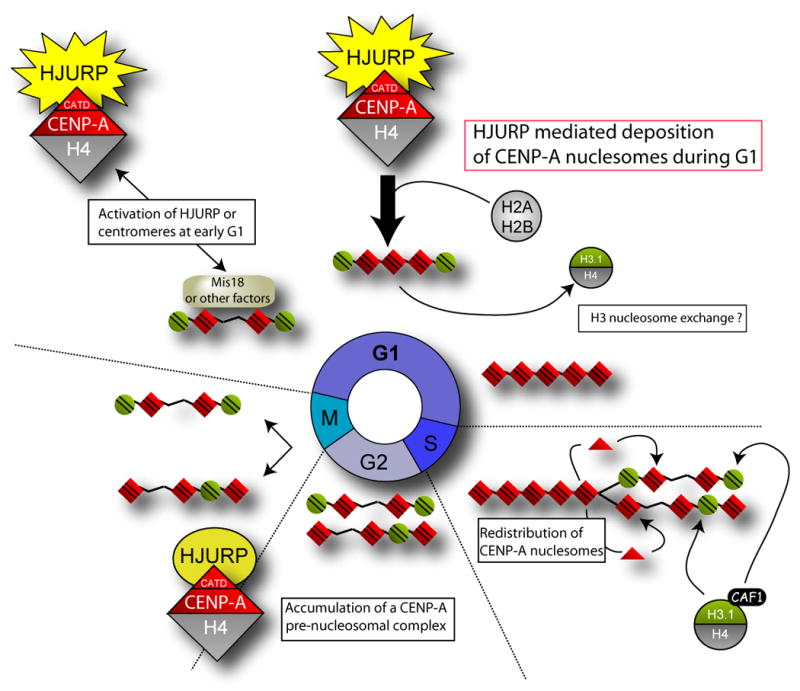
Replication of sister chromatids requires that new CENP-A be deposited specifically at centromeric loci in order to maintain the epigenetic mark of the centromere. Existing CENP-A nucleosomes are distributed to sister chromosomes during DNA replication. Cells progress through G2 and mitosis with half the maximal centromeric complement of CENP-A nucleosomes. CENP-A nucleosomes are assembled in a HJURP dependent manner during the G1 phase of the cell cycle. HJURP and CENP-A levels accumulate during G2 and peak during M even though active assembly of CENP-A is restricted to G1. Restriction of CENP-A assembly may be achieved by modification of the CENP-A prenucleosomal complex or through the recruitment of other proteins recruited to the centromere during G1, such as the Mis18 complex, that may serve to prime the centromere and restrict CENP-A nucleosomes assembly.
While CENP-A nucleosomes are quantitatively redistributed to daughter centromeres contemporaneous with DNA replication, as shown by pulse-chase labeling of CENP-A with SNAP tagging (Jansen et al., 2007), new CENP-A incorporation is delayed until the subsequent telophase or early G1 and we have shown this to require HJURP. Consequently, cells must progress through G2 and mitosis with only 50% of the maximal CENP-A nucleosome complement. It is not known whether histone H3 nucleosomes are assembled in place of CENP-A nucleosomes following their redistribution during DNA synthesis or whether these sites remain unoccupied through G2. Certainly H3.1 nucleosomes are able to occupy alpha-satellite DNA and can be found interspersed with CENP-A nucleosomes (Blower et al., 2002). If H3.1 nucleosomes are assembled with the centromeres during S-phase, HJURP-dependent assembly of new CENP-A nucleosomes in late M/early G1 occurs via a reaction in which histone H3 nucleosomes are exchanged for CENP-A nucleosomes.
A previous report suggested a role for HJURP in a double strand DNA damage break response and in vitro it can interact with a synthetic Holliday junction-like structure (Kato et al., 2007), an observation upon which the HJURP name was proposed. HJURP was also reported to interact with the mismatch repair protein hMSH5 and the MRN complex component NBS1 involved in double stranded break processing, consistent with a role for HJURP mediated CENP-A nucleosome assembly in chromatin remodeling accompanying DNA repair. In addition, HJURP was independently identified as a 14-3-3 interacting protein by yeast-two hybrid screen (Luhn et al., 2007). That the serine threonine kinase Akt/PKB is able to phosphorylate HJURP in vitro (leading to the additional proposed name FAKTS [Fourteen-three-three Akt substrate]) and may regulate its binding to 14-3-3 proteins has suggested a possible mechanism of HJURP regulation.
NPM1 is a highly abundant phosphoprotein which acts as a histone chaperone for both H3:H4 and H2A:H2B, in addition to playing many other cellular roles (Frehlick et al., 2007; Grisendi et al., 2006). NPM1 can bind ATP and the Drosophila homologue functions as an ATP-dependent chromatin remodeler suggesting that NPM1 may provide ATPase actitivty in the CENP-A histone deposition/exchange reaction (Chang et al., 1998; Ito et al., 1996). The requirement for ATP hydrolysis may be an important aspect of histone variant exchange. Deposition of Drosophila histone H3.3 is also dependent on both HIRA and the ATPase CHD1 (Konev et al., 2007). Two other potential ATPases with roles in CENP-A nucleosome assembly include RuvBL1, identified in this study as a substiochiometric pre-nucleosomal CENP-A associated component, and the hSNF2H component of the remodeling and spacing factor (RSF) found associated with CENP-A containing chromatin, but not the prenucleosome (Obuse et al., 2004).
CENP-A assembly is a tightly regulated process. Levels of HJURP and CENP-A protein are cell cycle regulated, accumulating in G2 and showing maximal levels during mitosis (Fig. 2C). The co-regulation of CENP-A and HJURP levels and the loss of CENP-A protein in the absence of HJURP suggests an important role for HJURP in CENP-A chromatin assembly is to stabilize prenucleosomal CENP-A. Recruitment of HJURP to centromeres and the subsequent assembly of new CENP-A nucleosomes only occurs during G1 following mitosis, suggesting an additional telophase/early G1-dependent activation event. G1 phase initiation of CENP-A deposition may be regulated by modification of the CENP-A prenucleosomal complex or centromeric chromatin, or by the activation and recruitment of chromatin-bound, G1 specific assembly promoting factors. The Mis18 complex is a good candidate to fulfill this role in centromere assembly as it is localized to centromeres only during early G1 (Fujita et al., 2007; Maddox et al., 2007) coincident with the packaging of new CENP-A into nucleosomes. Mutations and reductions of components of the Mis18 complex (Mis18a or Mis18BP1/hsKNL2) profoundly reduce the accumulation of CENP-A nucleosomes at the centromere (Fujita et al., 2007; Hayashi et al., 2004; Maddox et al., 2007). The Mis18 complex is not, however, found as part of the CENP-A prenucleosomal complex (Figure 1D) or in association with CENP-A nucleosomes (Foltz et al., 2006; Obuse et al., 2004), nor are CENP-A or HJURP ssociated with the Mis18 complex purified from nuclease-digested chromatin (Fujita et al., 2007). A simple view would be that Mis18 may alter centromeric chromatin structure possibly by promoting priming modifications in late mitosis or early G1 that are required for CENP-A nucleosome incorporation (a possibility supported by complementation of a Mis18 requirement by an inhibitor of histone deacetylation (Fujita et al., 2007)) or by affecting the removal histone H3 from the centromere as an initiating step for HJURP-mediated deposition of new CENP-A nucleosomes. This scenario would be consistent with positioning of the Mis18 complex and CENP-A nucleosomes near to one another along centromeric chromatin (Maddox et al., 2007). Alternatively, the Mis18 complex when bound to chromatin may facilitate the deposition of CENP-A nucleosomes by directly recruiting CENP-A assembly factors such as HJURP.
Results differ between organisms as to the role of the canonical chromatin assembly and remodeling factors in CENP-A deposition. In budding and fission yeast components of the canonical CAF-1 complex have been shown to be involved in CENP-A loading (Hayashi et al., 2004; Sharp et al., 2002). In fission yeast the chromatin remodeler Hrp1 (Walfridsson et al., 2005) and the histone binding protein NASP1-related protein Sim3 (Dunleavy et al., 2007) have been shown to alter spCENP-A assembly at the centromere. Sim3 interacts with non-chromosomal CENP-A, possibly fulfilling a partially overlapping chaperone role with HJURP. In contrast, Sim3 is not found concentrated at centromeres and interacts with histone H3 and human NASP is found only in the H3 prenucleosomal complex suggesting HJURP may play a more inclusive role in CENP-A nucleosome deposition. CAF1p48/RpAb48 (a.k.a. Mis16 in S. pombe) can interact directly with the Drosophila CENP-A homolog CID/CenH3 (Furuyama et al., 2006). While CAF1p46 and p48 have an effect on mammalian CENP-A accumulation (Hayashi et al., 2004), a direct interaction has not been documented, and as we have shown here, it is not found in prenucleosomal CENP-A complexes. While this does not rule out a substoichiometric or transient interaction between CENP-A and CAF1p46 or p48, it seems unlikely especially since interactions between CAF1p46 and p48 and other histone variants are easily seen by similar approaches (Tagami et al., 2004). It is possible that the tagging approach we have employed disrupts interaction of CAF1p46/48 with CENP-A; however, if this is the case it is clear that CAF1p46/48 is not required for the specific recruitment of CENP-A to centromeres.
With our discovery of HJURP as a unique CENP-A histone chaperone, it is now clear that distinct chromatin assembly factor complexes are used for the unique spatial and temporal accumulation of H3 variant nucleosomes in mammals. Assembly of histone H3.1 nucleosomes is coupled to replication through interaction between histone H3 and the MCM replicative helicase (Groth et al., 2007), and between CAF1p150 and PCNA (Moggs et al., 2000; Shibahara and Stillman, 1999). Both MCM and PCNA are major components of the replication machinery, such that new histone H3.1 nucleosome assembly occurs in close proximity to DNA synthesis. Existing CENP-A nucleosomes may direct the incorporation of new CENP-A nucleosomes either directly or through the recruitment of intermediate factors that could include the covalent modification of surrounding centromeric chromatin. In turn HJURP must recognize either the existing CENP-A nucleosome or the intermediate factors or modifications that they induce in order to direct the deposition of new CENP-A nucleosomes only into active centromeres.
Experimental procedures
Cell culture, synchronization and transfection
CENP-A-TAP, H3.1-TAP, YFP-CENP-A and YFP-H3CATD stable expressing cells were described previously (Black et al., 2007; Foltz et al., 2006; Kops et al., 2004). Stable H3CATD-TAP cell lines were produced by retroviral infection as described (Foltz et al., 2006). Synchronization was achieved as described by Jansen et al (2007). For siRNA treatment, 1.5×105 cells were plated on glass coverslips in a 6 well plate and duplexed siRNAs were introduced into cells using Oligofectamine (Invitrogen, Carlsbad, CA). siRNA-encoding plasmids were co-transfected with RFP- tagged histone H2B (H2B-RFP; Black et al., 2007, Mol Cell) at a ratio of 20:1 (siRNA:H2B-RFP) using the Effectene transfection reagent (Qiagen, Valencia, CA). Cells were fixed and processed for immunofluorescence 72 hr following transfection. SNAP labeling was conducted as described previously (Jansen et al., 2007).
Affinity purification
Chromatin free-extracts were produced from 5×108 cells expressing histone H3.1-TAP or 1×109 cells expressing either CENP-A.TAP or H3CATD-TAP. Cells where dounce homogenized in buffer A (3.75 mM Tris pH 7.5, 20 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, 0.05 mM spermidine, 0.125 mM spermine, 0.1% digitonin, 1 mM PMSF, 10μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml chymostatin). Homogenized extracts were centrifuged at 300×g for 5 minutes and the pellet was resuspeded in buffer A, homogenized and centrifuged for 5 minutes at 300×g. Supernatants from these two centrifugations were combined and centrifuged at 12000×g for 10 minutes to produce a chromatin free extract. Tandem affinity purifications were conducted as described previously (Foltz et al., 2006). In the case of single step purification, proteins were eluted from the first affinity step (IgG-bound beads) by boiling in SDS sample buffer.
Mass Spectrometry
Total eluates from tandem affinity purifications were analyzed by mass spectrometry using MudPIT analysis as described previously (Foltz et al., 2006) or by excision and in-gell digestion of proteins from silver stained polyacrylamide gels. Gel bands were dehydrated in 50% acetonitrile, rehydrated in 50 mM ammonium bicarbonate pH8 including 1 ug/ul trypsin (Promega, Madison, WI) and incubated overnight at 37°C. Peptides were eluted from the gel in 50% acetonitrile, 5% formic acid, dehydrated, resuspended in 0.1% acetic acid and analyzed by separation on a 10cm C18 column (Michrom BioResources, Auburn, CA) coupled to a Thermo Finnigan LCQ ion trap mass spectrometer (Waltham, MA). Data-dependent collection of MS spectra was conducted by taking a single MS scan followed by MS/MS scans of the top three most intense ions. Peptide identification was conducted using COMET.
Sucrose gradient sedimentation
Chromatin free extracts were prepared as described above from 6×107 randomly cycling HeLa cells and applied to the top of a 2 ml 5-40% sucrose gradient in buffer A buffer. Sucrose gradients were centrifuged at 4°C for 6 hours at 50,000×g in a Beckman TLS55 swinging bucket rotor and the gradient was separated into 150 μl fractions. Proteins were separated by SDS-PAGE and blotted to nitrocellulose and detected by immunoblot in 25 mM Tris, pH 7.4, 150 mM NaCl, 5% dry milk.
Protein purification and in vitro binding
All proteins were expressed in the Rosetta BL21 (DE3) pLysS bacteria strain and purified using either Ni-NTA affinity resin (Qiagen, Valencia, CA) for 6XHis proteins or Glutathione sepharose (GE Healthcare, Piscataway, NJ) for GST fusions. CENP-A:H4 heterotetramers were produced as described (Black et al., 2004). Histone H2A containing amino terminal tandem 6XHis and S tags (His-H2A) and untagged histone H2B were expressed in bacteria and initially purified as monomers. His-H2A:H2B dimers were reconstituted and purified as described (Luger et al., 1999). In vitro binding assays were conducted in 100mM NaH2PO4 pH7.4, 300mM NaCl, 0.05% NP-40, 10% glycerol, and 1mM DTT. Recombinant proteins were combined and incubated at room temperature for 30 minutes and an additional 30 minutes following addition of glutathione sepharose. Bound protein complexes where washed twice in binding buffer, once in low salt buffer (100mM NaH2PO4 pH7.4, 100mM NaCl, 0.05% NP-40, 10% glycerol, and 1mM DTT), eluted from the beads in SDS sample buffer, separated on a 15% SDS-PAGE gel and stained with Coomassie.
Immunocytochemistry
Cells were preextracted using 0.3% TritonX-100 in PBS, fixed in 4% formaldehyde for 10 minutes and quenched in 100 mM Tris pH7.7. Cells were preblocked in PBS containing 2% FBS, 2% BSA, and 0.2% tween 20. Incubations with primary antibodies (see supplemental experimental procedures) were conducted in blocking buffer for one hour at room temperature. DNA was detected using DAPI and cells were mounted in Prolong Antifade (Invitrogen, Carlsbad, CA). Images were collected using a Deltavision microscope (Applied Precision, Issahquah, WA) and deconvolved z-projections were presented. Images of shRNA treated cells were acquired using a Leica DMI 6000 B microscope using Leica LAS software. Quantification of siRNA effect was conducted using Metamorph (Molecular Devices, Sunnyvale, CA) on non deconvolved images collected on the same day with identical exposure times by measuring maximum pixel intensity per nucleus with background subtracted.
Supplementary Material
Acknowledgments
The authors thank Iain Cheeseman, Andrew Holland, Karen Oegema, Arshad Desai and members of Cleveland Lab for helpful discussion and reagents. This work was supported by grants from the National Institutes of Health to B.E.B. and D.W.C. D.R.F. was supported by a Special Fellowship from the Leukemia and Lymphoma Society. B.E.B. was supported by a Career Award in the Biomedical Sciences from the Burroughs Welcome Fund, L.E.T.J. was supported by a postdoctoral fellowship from Philip Morris USA Inc. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Chang JH, Lin JY, Wu MH, Yung BY. Evidence for the ability of nucleophosmin/B23 to bind ATP. Biochem J. 1998;329(Pt 3):539–544. doi: 10.1042/bj3290539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tyler JK, Bulger M, Kobayashi R, Kadonaga JT. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J Biol Chem. 1996;271:25041–25048. doi: 10.1074/jbc.271.40.25041. [DOI] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, Fyodorov DV. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Luhn P, Wang H, Marcus AI, Fu H. Identification of FAKTS as a novel 14-3-3-associated nuclear protein. Proteins. 2007;67:479–489. doi: 10.1002/prot.21288. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Wu JC. Homology between human and simian repeated DNA. Nature. 1978;276:92–94. doi: 10.1038/276092a0. [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. Embo J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Willard HF. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


