Abstract
Background & Aims
Vesicular glutamate transporter (VGLUT) has been reported to be involved in glucose-induced insulin secretion. It has been shown that glucose stimulates the expression of VGLUT isoform 2 (VGLUT2) in β cells via transcriptional mechanism. In this study, we identified the mouse VGLUT2 (mVGLUT2) promoter and characterized the transcriptional mechanism of glucose-stimulated mVGLUT2 expression in β-cells.
Methods
A promoter region of mVGLUT2 was cloned by genomic PCR. The mechanism of Sp1 in glucose-induced transactivation of mVGLUT2 was investigated by luciferase assay, electrophoretic mobility shift assay, Chromatin immunopreciptation assay and Western blot analysis.
Results
A promoter containing 2133-base pair of upstream sequence of the 5’-flanking region of mVGLUT2 cDNA was cloned. Transient transfection of various 5'-end deletion constructs of the mVGLUT2 promoter/luciferase reporter indicated that the region between − 96 to + 68 base pair contains the basal promoter for mVGLUT2. Mutational analysis and electromobility shift assay demonstrated an important role for the transcription factor Sp1 in both basal and glucose-induced mVGLUT2 transcription. The interaction between Sp1 and mVGLUT2 was confirmed by chromatin immunoprecipitation (ChIP) assays. Glucose stimulates the phosphorylation of Sp1 via mitogen activated kinase (MAPK) P38 and P44/42. This leads to increase binding activity of Sp1 to the mVGLUT2 promoter and results in activation of the gene.
Conclusion
We cloned the mouse VGLUT2 promoter and demonstrated a novel molecular mechanism of glucose-induced mVGLUT2 transcription.
Keywords: Glutamate, Pancreatic cells, Glucose response gene, VGLUT2, Transcription factor Sp1
Introduction
Glucose-induced insulin secretion involves two major signaling pathways leading to the production of triggering and amplifying signals respectively 1. In a consensus model of glucose-induced insulin secretion, the metabolism of glucose by oxidative glycolysis increases the ATP/ADP ratio, and subsequently promotes the closure of ATP-sensitive K+ channels and depolarization of the plasma membrane. As a consequence, the concentration of free cytoplasmic Ca2+ is raised by the opening of voltage-dependent Ca2+ channel and the increased cytoplasmic Ca2+ triggers the exocytosis of insulin containing granules2–4. However, the triggering action of cytoplasmic Ca2+ does not completely explain the stimulation of insulin secretion by glucose. A growing body of evidence has suggested that glutamate, the major excitatory neurotransmitter in the central nervous system, may act as an intracellular messenger coupling glucose metabolism to insulin secretion in β cells5–7. It was noticed that total cellular glutamate levels were elevated in human, rat and mouse islets as well as clonal β cells upon glucose stimulation8–11. Under permissive condition, glutamate directly stimulates insulin exocytosis from INS-1 cells, an insulinoma cell line, at permissive cytosolic Ca2+ concentration5. In addition, overexpression of glutamate decarboxylating enzyme GAD65 in INS-1E cells and rat islets resulted in a significant reduction of the intracellular glutamate levels and impairment of glucose-induced insulin secretion7. These results support the role of glutamate in glucose-induced insulin secretion.
The glutamatergic system consists of glutamate receptor, vesicular glutamate transporter (VGLUT) and plasma glutamate transporter12. Multiple glutamate receptors have been found in the pancreas13–16. By functional and immunohistochemical studies, VGLUT, the protein responsible for the accumulation of glutamate from cytoplasm into the vesicles, has also been identified in islets of Langerhans11,13.
Vesicular glutamate transporter plays an essential role in glutamate signal output through vesicular storage of glutamate17. We have recently cloned a neuronal vesicular glutamate transporter isoform 2 (VGLUT2)18 and have shown that VGLUT2 is expressed in clonal β cells11. Vesicular glutamate transporter is directly involved in insulin secretion from pancreatic β cells, as inhibitors of vesicular glutamate transport suppress the glutamate-evoked insulin exocytosis5. Moreover, high glucose concentration increases the cellular glutamate level in rat insulinoma INS-1 cells and this increase can be abolished by carbonyl cyanide p-trifluoromethoxyphenlhydrazone, an inhibitor of vesicular glutamate transporter19. These previous studies suggested a potential regulatory role of VGLUTs in glucose-induced insulin secretion of β cell. In support of these findings, we have recently found that high glucose concentration stimulates vesicular glutamate uptake in clonal β cells. The increases of vesicular glutamate transport in β cells are due to glucose-induced up-regulation of VGLUT2 mRNA via a transcriptional mechanism11. These findings suggest that high glucose stimulates glutamate uptake into secretory vesicles in β cells and favors the glutamate-evoked insulin release. In the present study, we identified the VGLUT2 promoter and characterized the transcriptional mechanism of glucose-stimulated VGLUT2 expression in β-cells that involves activation of the transcriptional factor Sp1.
Methods
Reagents
MIN6 cell, a mouse insulin-secreting pancreatic beta cell line was kindly provided by Dr. Morris F. White with permission of Dr. Junichi Miyazaki20. Drosophila SL2 cells and cell culture medium were obtained from American Type Culture Collection (Manassas, VA). The inhibitors of MAPK P38 and P44/42 were purchased from Calbiochem (San Diego, CA). All other reagents were obtained from Sigma.
Cell Culture
MIN6 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 mg/ml glucose and 0.39 mmol/L mercaptoethanol and supplemented with 10% fetal bovine serum, 100 U/ml penicillin sulfate and 70µmol/L streptomycin at 37°C in a humidified atmosphere of 5% CO2/95% air20. Drosophila SL2 cells were maintained under the manufacture’s protocol.
Cloning and Analysis of the Mouse VGLUT2 Promoter
The first 100 bp at the 5′ end of mVGLUT2 cDNA (GenBank accession number AF324864) was selected to BLAST search the mouse genome. This 100-bp sequence corresponded to a region on mouse chromosome 7 with 100% identity. A pair of primers (sequences: 5'-ggagccccttggtaccatggaggttg-3’ and 5’-ggtgctctcgagtccctgttctgga-3′) was designed to amplify a 2133-bp fragment of the mouse genomic DNA, which contains 251 bp of mVGLUT2 cDNA sequence and 1912 bp of upstream sequence of the 5′-flanking region of mVGLUT2 cDNA. Genomic PCR was performed under standard protocol21. The isolated 2133-bp fragment was confirmed to have 100% identity with the mouse genome, shown by double-strand DNA sequencing (GenBank accession number DQ812098).
The genomic PCR fragments were then subcloned into pGL3-basic vector (Promega, Madison, WI). The serial deletions of the mVGLUT2 promoter were created by PCR amplification22. Site-directed mutagenesis of the Sp1 and GATA3 binding sites was conducted using the QuikChange® II Site-Directed Mutagenesis Kit’s (Stratagene, La Jolla, CA). The sequences of the top primers were −40/−8, 5’-acaattacgccttcgcgatgaggagaccatg-3’ for Sp1 and −111/−72, 5’-ctctatcgatagggtaccaaatatcagcaacattctcagtc-3’ for GATA3. The altered nucleotides are underlined. All deletions and site-directed mutants were confirmed by DNA sequencing.
Identification of the Transcriptional Start Site by Primer Extension and 5’- Rapid Amplification of cDNA Ends (5’-RACE)
Transcription start site of mVGLUT2 was independently determined by primer extension and 5’-RACE. Primer extension was performed with a standard protocol as described before22. Mouse brain mRNA was used for 5’-RACE with the FirstChoice™ RACE-Ready cDNA kit (Ambion). The following mouse VGLUT2 gene-specific primers were used to identify the 5' end of the mRNA. The outer primer was 5’- cttctcagcagaggtgagctttcc- 3’ and the inner primer was 5’- tgagtccctattctggaagtcacct -3’.
Transfections and Reporter Gene Assays
MIN6 cells (passage 25) were transfected in 24 well plates at 90–95% confluence with 0.5µg mVGLUT2 promoter constructs or PGL3-basic vector (Promega, Madison, WI) with 0.05µg PRL-SV40 vector (Promega, Madison, WI) containing Renilla luciferase reporter gene as an internal control. For Sp1 overexpression studies, 0.5µg pPAC vector, pPAC/Sp1, pPAC/Sp2, pPAC/Sp3 or pPAC/Sp4 (generously provided by Dr. R. Tjian) were co-transfected along with mVGLUT2 promoter constructs into Drosophila SL2 cells. All transfections were performed with lipofectamine 2000 (Invitrogen) in an OptiMEM medium. Luciferase activity was measured 48 h after transfection by a standard method. Each experiment was repeated a minimum of three times with different populations of cells on different days, and two wells were averaged from each experiment to obtain n = 1.
Nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts from MIN6 cells were prepared as described previously22. Gel mobility shift assay was carried out using the DIG Gel Shift kit (Roche, Indianapolis, IN).
Semi-quantitative RT-PCR
Total RNA was isolated from MIN6 cells using TRIZOl LS Reagent (Invitrogen). RT-PCR was performed with mVGLUT2 primers (upstream: 5’-atctgctaggtgcaatggaaaaattt-3’ and downstream: 5’-gaataatcatctcggtccttataggtg-3), Sp1-specific primers (upstream: 5’-cacctgatagacgaccagttcag-3’ and downstream: 5’-gtctctggaattgttgtaggtg-3’), and β-actin primers (upstream: 5’-tcatgaagtgtgacgttgacatccgt-3’ and downstream: 5’-cttagaagcatttgcggtgcacgatg-3’). Subsaturation levels of cDNA templates that were needed to produce a dose-dependent amount of PCR products were defined in initial experiments by testing a range of template concentrations. Subsequent PCR was carried out with subsaturation levels of RT reaction with mVGLUT2, Sp1, and β-actin primers in separate PCR reactions with identical parameters. The PCR products were analyzed by agarose gel electrophoresis.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP was performed under the manual of Chromatin Immunopreciptiation Assay Kit (Upstate Temecula CA). The cells were treated with different glucose concentrations for 48 hours before harvested for SDS-PAGE. The PCR products were analyzed on 2% Nusieve 3:1-agarose and visualized by with MultiImage Light Cabinet with Quantity One software.
Western Blotting
Western blotting was performed with standard protocols of our lab. Anti-Sp1 rabbit polyclonal antibody was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), anti-PhosphoThreonine and PhosphoSerine antibodies from Qiagen(Qiagen CA), anti-P38, P44/42, p-P38, p-P44/42, β-actin polyclonal or monoclonal antibodies from Cell Signalling (Cell Signalling MA), anti-mVGLUT2 antibody form Sigma (Sigma,MO).
Sp1 Gene Silencing With siRNA
Sp1 siRNA targeted to AATGAGAACAGCAACAACTCC was designed under the standard principles with GC ratio about 50% by Qiagen and was transfected to the cells with TransMessenger Reagent according to the manufacturer’s instructions (Qiagen). A scramble siRNA with same GC content was used as control. The sequences of these siRNA were digitally searched and no similar sequences were found in the current genomic database.
Results
Glucose-mediated Regulation of the Mouse VGLUT2 Expression in Clonal β Cells
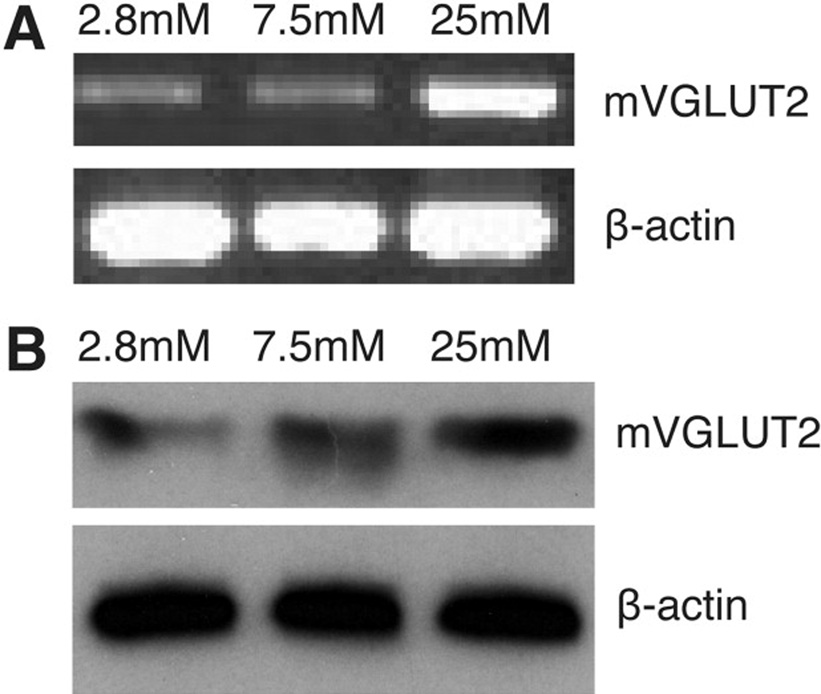
The expressions of VGLUT2 mRNA and protein in MIN6 cells under different glucose concentrations were assessed by semiquantitative RT-PCR and Western blot (Figure 1). Both mRNA and protein expression of VGLUT2 were significantly increased by high glucose concentration in concordance with the previous investigation11.
Figure 1. Glucose increases expression levels of VGLUT2 mRNA and protein in MIN6 cells.
(A) mRNA prepared from MIN6 cells grown in different concentrations of glucose medium for 12 h was used for 1st-strand cDNA synthesis. Semiquantitative RT-PCR was performed with VGLUT2 or β-actin-specific primers in separate reactions. (B) Vesicular protein purified from MIN6 cells grown in different concentrations of glucose medium as shown for 12 h was used for Western blot with VGLUT2 or β-actin-specific antibody.
Identification of the Mouse VGLUT2 Promoter and Gene Structure and Its Regulation by Glucose
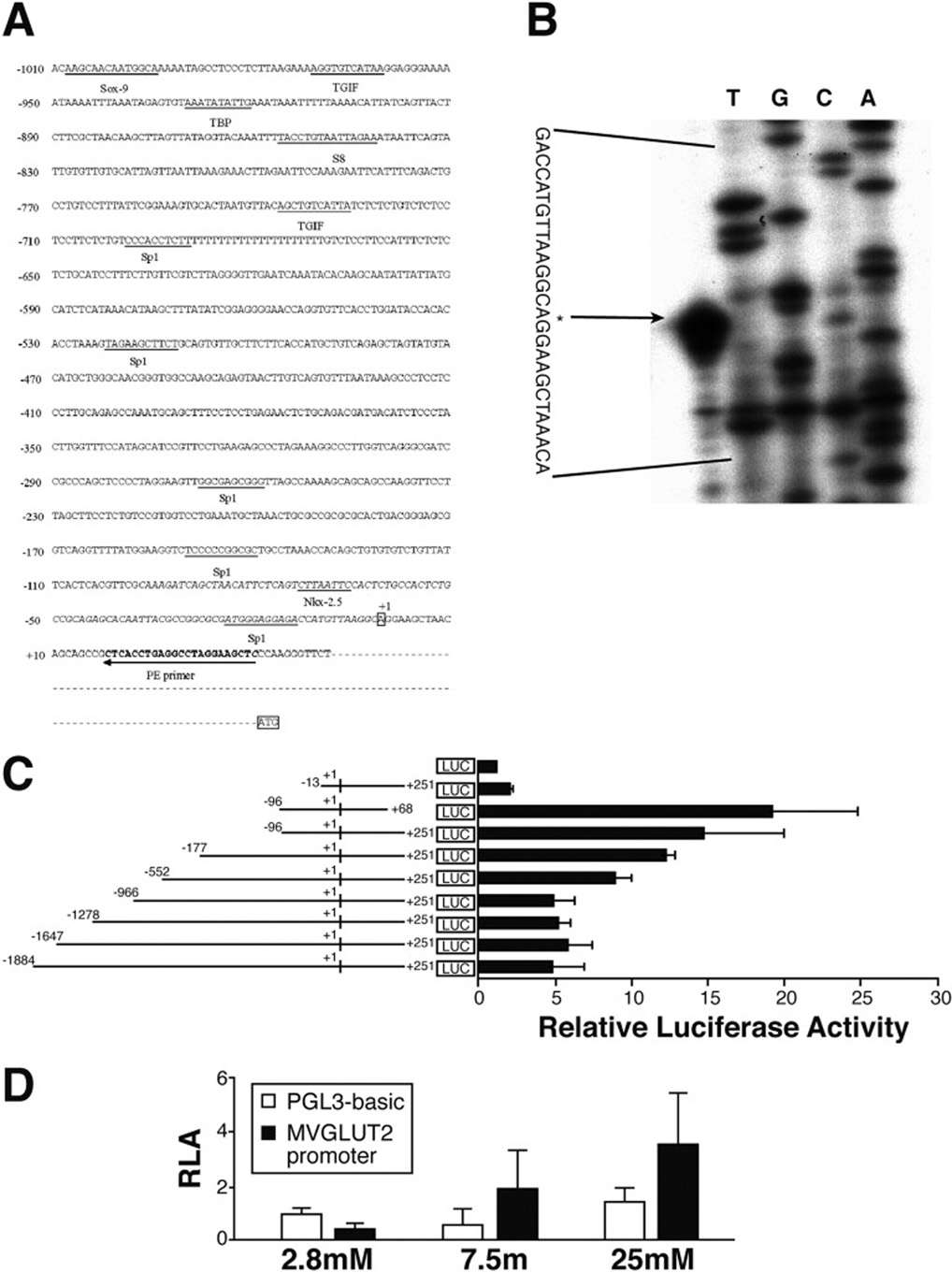
Figure 2A displays the sequence from nt-1010bp to +51 base pair (bp) relative to the transcription start site, which was determined in Figure 2B. Homologues to known gene regulatory elements were identified using the program Mat Inspector (http://www.gene-regulation.com/). Many putative cis-elements were found for ubiquitously expressed transcription factors including five Sp-1 sites, two TGIF sites, one TBP site, one Sox-9 sites, one Nkx-2.5 site, one S8 site, and one CCAAT/enhancer-binding protein α (C/EBPα) site (Figure 2A).
Figure 2. Cloning and functional characterization of mVGLUT2 promoter and its regulation by glucose.
(A) mVGLUT2 upstream genomic sequence and putative transcription factor binding sites. Depicted in this figure is the sequence of the 1010-bp genomic region immediately upstream of the mVGLUT2 transcription start site. The transcription start site is marked as +1. Putative cis-elements are underlined. PE primer is the primer used for primer extension. (B) Identification of the transcription start site of mVGLUT2. The arrow indicates the primer extension product aligned with an adenosine “A” residue as determined by comparison with the sequencing ladder. (C) Identification of the minimal mouse VGLUT2 promoter by deletion analysis. Promoter activity was measured by fold increase over the control promoterless pGL3-Basic vector. (D) Glucose regulation of mVGLUT2 promoter. MIN6 cells expressing the minimal mVGLUT2 promoter were cultured in medium containing different glucose concentration as indicated. Promoter activity was measured by fold increase over the control promoterless pGL3-Basic vector.
Further sequence analyses revealed that the mVGLUT2 gene consists of 11 introns and 12 exons. The methionine initiation codon, ATG, is located within exon 1, and the termination codon, TAA, is within exon 12 (data not shown). The Intron–exon boundaries of the mVGLUT2 gene were determined by sequencing of the mouse brain genomic DNA with mVGLUT2-specific primers with the rule of GT-AG24 (data not shown). The transcription start site was determined by primer extension with mouse brain mRNA, which resulted in a single predominant band (Figure 2B). This transcription start site was independently confirmed by sequencing of 5'-rapid amplification of cDNA ends PCR fragment (data not shown). The functional promoter DNA sequence has been submitted to GenBank (Accession number DQ812098).
To identify the minimal promoter and the basal cis-element(s) of the mouse VGLUT2 gene, a series of reporter plasmids containing various lengths of the mVGLUT2 5'-flanking regions (from nt −1884 bp to nt +251 bp or nt +68 bp) upstream of the firefly luciferase (Luc) gene was constructed in pGL-3 vector and transfected into MIN6 cells (Figure 2C). The longer promoter/reporter constructs (−1884+251 to −966/+251) show lower induction despite being a more complete promoter compared to the shorter promoter/reporter constructs (−552/+251 to −96/+251). It is possible that down-regulating factor(s) exist in the intervening region. The construct containing 96 bp of mVGLUT2 promoter sequence and 251 bp of 5'-noncoding region (−96/+251) showed promoter activity similar to that of other longer constructs (−177/+251, −552/+251). However, a further deletion construct (−13/+251) was inactive, suggesting that sequences between −96 to +251 contain the minimal promoter and is critical for the basal transcription of the mVGLUT2 gene. A further deletion on the 3’-end of the construct (−96/+68) did not show any reduction of promoter function. Thus, we conclude that the cis-elements controlling basal transcription of mVGLUT2 is within −96 bp to +68 bp of the 5’-flanking region.
To determine the glucose responsiveness of the cloned mVGLUT2 promoter, two luciferase reporter constructs, −1884/+251 (longer promoter) and −96/+68 (the minimal functional promoter), were transfected into MIN6 cells to measure reporter gene expression under three different glucose concentrations (2.8 mM, 7.5 mM, and 25 mM, respectively). As shown in Figure 2D, glucose significantly stimulates luciferase expression mediated by the minimal functional mVGLUT2 promoter (10 fold increase from 2.8 mM to 25 mM), while no statistical significant stimulation was observed for the cells transfected with pGL3-basic, the promoterless vector. The longer promoter (−1884/+251) also showed a 10-fold stimulation by glucose which is similar to that of the minimal promoter (data not shown). These results indicate that the mVGLUT2 promoter is upregulated by glucose and that the minimal functional mVGLUT2 promoter contains putative glucose-responsive element(s).
Role of Sp1 in Basal Function of the mVGLUT2 Promoter
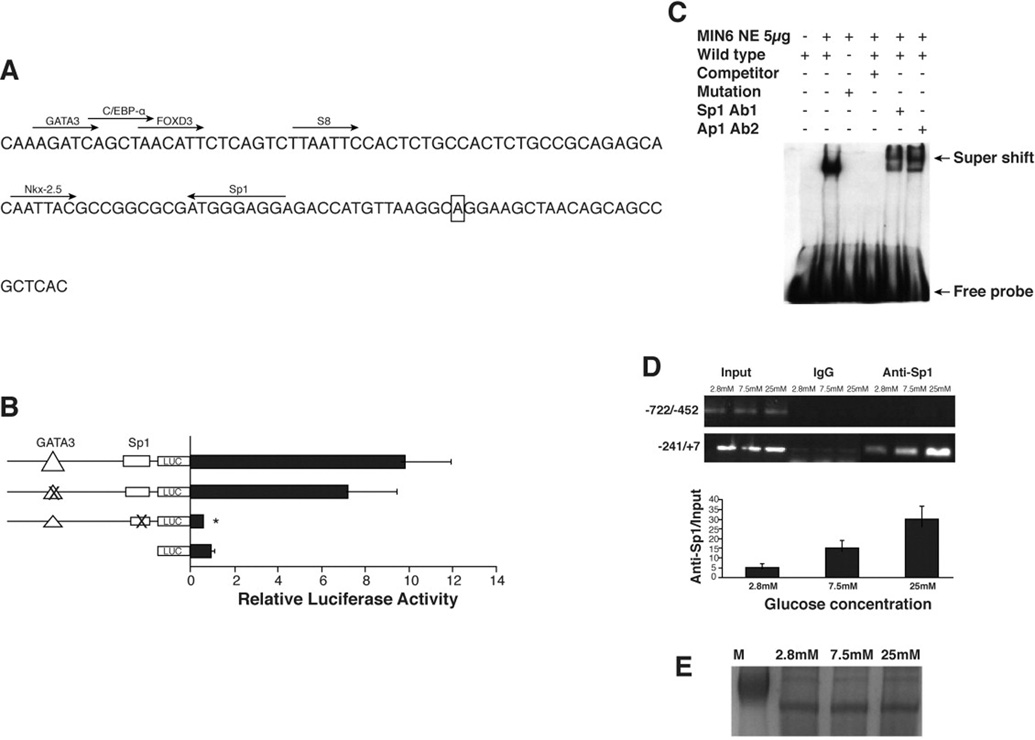
A search for transcriptional factor binding sites within the minimal functional promoter yielded potential consensus binding sites for Sp1, GATA-3, and C/EBPα (Figure 3A). Surprisingly, by searching human and rat genome database, we found that the sequences of the minimal promoter of VGLUT2 are highly conserved among mouse, human, and rat. Two consensus cis-elements, Sp1 and GATA3, are reserved among all three species (data not shown). To determine the role of Sp1 and GATA3 consensus sequences in the transcriptional activation of mVGLUT2, single mutations of these sites were generated by site-directed mutagenesis (Figure 3B). Mutation of GATA-3 site did not result in significant functional change as compared to the wild type promoter. Remarkably, mutation of the single Sp1 site of the minimal promoter completely abolished the promoter activity, suggesting a critical role of Sp1 in the mVGLUT2 transcription.
Figure 3. Sp1 mediates basal regulation of mVGLUT2 promoter.
(A) Putative consensus binding sites for transcriptional factors are indicated in the minimal functional promoter region (−96 to +68). The transcription start site is marked by box. (B) Relative luciferase activities of single mutations for Sp1 and GATA3 of mVGLUT2 promoter were measured in MIN6 cells. Values are means ± SD of duplicate data from 3 experiments. *P < 0.001 vs. wild-type promoter constructs. (C) Electrophoretic mobility shift assay with purified MIN6 nuclear extract. The DNA-protein binding was performed with DIG-labeled wild-type or mutant oligonucleotides corresponding to the nucleotide sequence −41 to −9 of the mVGLUT2 promoter. Competition for binding was performed by including a 125-fold molar excess of unlabeled probe. Identification of the DNA-protein complex was characterized by supershift analysis with 2 specific antibodies (Ab1 and Ab2). (D) The in vivo interaction of Sp1 and mVGLUT2 promoter was evaluated by Chromatin immunoprecipitation (CHIP) assays with the basal promoter DNA (−241/+7) and the control DNA (−722/−452) in MIN6 cells treated with different glucose concentrations. The ChIP assays were repeated three times on independent MIN-6 preparations. (E) SDS-PAGE indicating equal protein loading used for ChIP assay.
Electrophoretic mobility shift assay was then used to identify and characterize potential protein binding activity associated with the −96/+68 bp region. An oligonucleotide encompassing the consensus Sp1 binding sequence of the minimal promoter was used as a probe for the mobility shift assays. As shown in Figure 3C, one predominant shifted band was observed when the probe was incubated with nuclear extracts from MIN6 cells. The presence of a 125-fold molar excess of unlabeled probe abolished the interaction between the probe and nuclear proteins. Additionally, a probe containing a mutation of the consensus Sp1 binding sites lost the binding ability to the nuclear protein. Furthermore, the band was supershifted by two independent Sp1 specific antibodies, indicating the specific binding of Sp1 protein to the consensus Sp1 site on the mVGLUT2 promoter. Antibodies of Sp2, Sp3, and Sp4, other members of Sp1 family, did not recognize the DNA-protein complex (data not shown). The interaction between Sp1 and mVGLUT2 promoter was further confirmed by chromatin immunoprecipitation (ChIP) assays. As shown in Figure 3D, the DNA-protein complexes were immunoprecipitated using either rabbit IgG as a control or antibodies to Sp1. Using primers to the −241/+7 region of the VGLUT2 gene, we observed a strong PCR product with DNA immunoprecipitated with Sp1 antibody. This binding is enhanced by increase of glucose concentration. To verify the specificity of the Sp1 binding to the basal promoter region of VGLUT2 promoter, a control with more upstream DNA region (−772/−452) of VGLUT2 promoter was used for the ChIP assays. The control DNA region did not generate immunoprecipitation with Sp1 antibody nor glucose regulation. A SDS-PAGE was done to confirm the same amount of cell lysate used in the experiment (Figure 3E). Results from above demonstrate an important role of Sp1 in glucose regulation of the mVGLUT2 gene.
Role of Sp1 in Glucose-induced Regulation of mVGLUT2
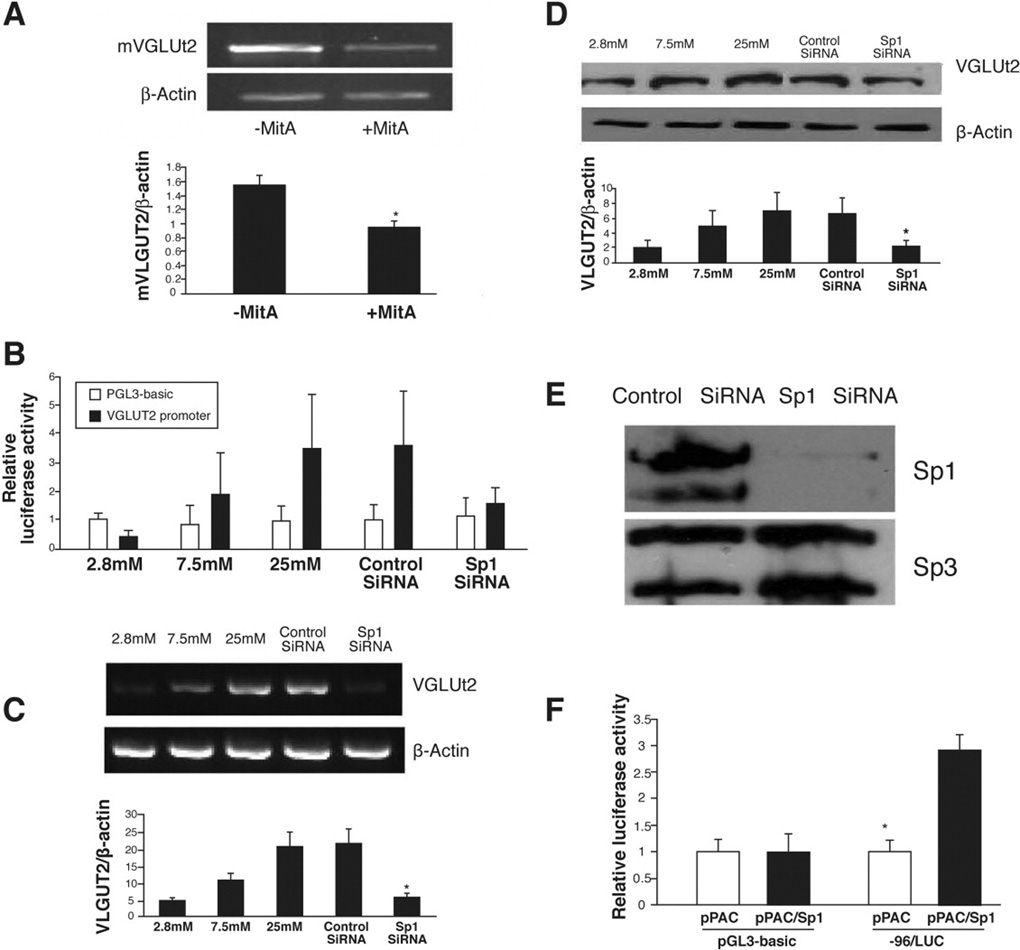
Several reports have suggested that the ubiquitously expressed transcription factor Sp1 may also provide a mechanism for glucose responsiveness25–27. Members of the Sp1 family were reported to be involved in regulation of glucose/insulin-dependent genes encoding leptin, fatty acid synthase, and ATP citrate-lyase28–29. To determine the role of Sp1 in glucose-induced transcriptional activation of mVGLUT2, we investigated the regulation of mVGLUT2 gene by Sp1 under condition of glucose stimulation. Inhibition of the Sp1 binding to the GC boxes of promoter by mithramycin A, a Sp1 specific inhibitor, significantly reduced glucose-induced expression of the endogenous VGLUT2 gene in MIN6 cells (Figure 4A). Furthermore, the magnitude of VGLUT2 induction by glucose and the effects of the Sp1-specific siRNA were determined at promoter, mRNA and protein levels. Transfection of Sp1-specific siRNA significantly reduced glucose-induced mVULUT2 promoter activity (Figure 4B), glucose-induced endogenous VGLUT2 mRNA (Figure 4C) and VLGUT2 protein expression (Figure 4D) in MIN6 cells. Meanwhile, Sp1 protein, but not Sp3 protein, was also significantly reduced (by 90%) by Sp1-specific siRNA (Fig. 4E), suggesting the specific knockdown of Sp1. These results indicated an important role of Sp1 in glucose-mediated mVGLUT2 gene transcription.
Figure 4. Role of Sp1 in glucosed-induced transcription of mVGLUT2.
(A) Inhibition of mVGLUT2 mRNA expression by Sp1 specific inhibitor mithramycin A. MIN6 cells cultured in a high-glucose-containing medium (25mM) were treated with or without 100nM mithramycin A for 24 hours before harvest. (B) Effects of Sp1 siRNA on mVGLUT2 promoter activity. (C) Inhibition of mVGLUT2 mRNA expression by Sp1 siRNA. Scramble (control) or Sp1 siRNA was transfected for 48 hours before next transfection with mVGLUT2 promoter. Results are means ±SE from 3 separate experiments. * p< 0.05. (D) Inhibition of mVGLUT2 protein expression by Sp1 siRNA. (E) Down-regulation of Sp1 protein by Sp1-specific siRNA in MIN6 cells. Western-blots were performed with Sp1 or Sp3 antibody and MIN6 cells transfected with control or Sp1 specific siRNA. Sp1 protein in MIN6 cells was inhibited by 90% by Sp1-specific siRNA while Sp3 protein was not affected. (F) Effects of Sp1 on mVGLUT2 transcription in Drosophila SL2 cells. One microgram of mVGLUT2 promoter construct (−96/+68) or promoterless construct (pGL3-basic) was cotransfected with 0.5 µg of Drosophila Sp1 expression vectors, pPAC/Sp1 or the blank pPAC vector.
The ubiquitous expression of Sp1 could affect the interpretation of above experimental results. We therefore studied gene regulation by Sp transcription factors in Drosophila SL2 cell, known as lacking of endogenous Sp130. Under high glucose concentration (25 mM), we cotransfected the minimal promoter construct 96/+68 along with Drosophila expression vectors pPacSp1 into Drosophila SL2 cells. As shown in Figure 4F, the minimal promoter alone was unable to drive glucose-induced transcription in the SL2 cells. Addition of the Sp1 expression vector drastically increased transcription, indicating that the transcription factor Sp1 is essential for the formation of the transcriptional initiation complex of the mVGLUT2 gene. Expression of Sp2, Sp3, or Sp4 in SL2 cells did not significantly change transcription of the minimal promoter (data not shown).
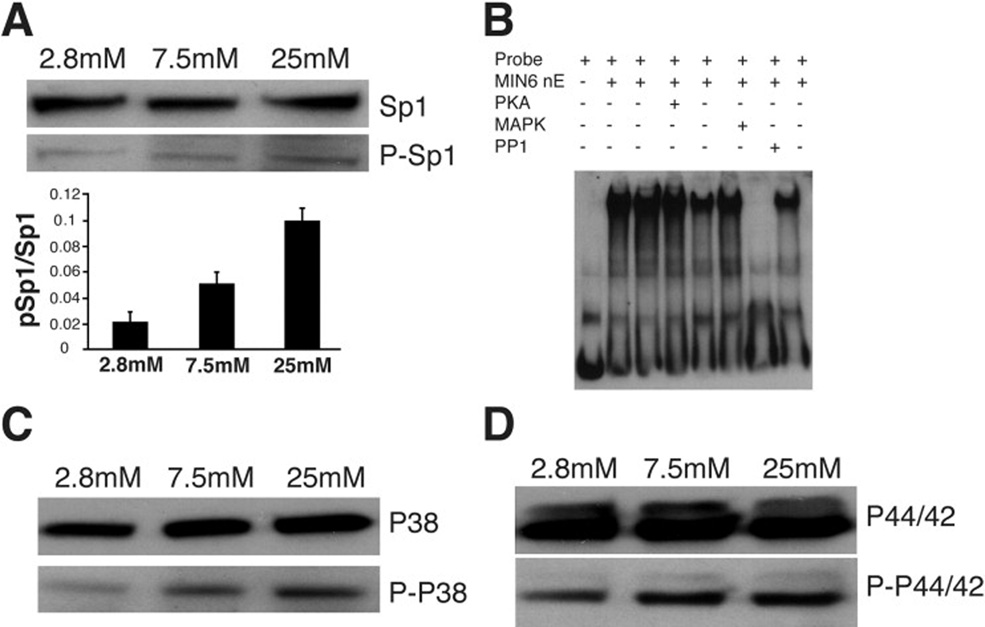
The mechanism of Sp1-mediated glucose activation of mVGLUT2 was further explored. As shown in Figure 5A, glucose treatment does not change the amount of total Sp1 but increases the phosphorylation of Sp1 in the nucleus of MIN6 cells. Phosphorylation of Sp1 by mitogen activated kinase (MAPK) but not protein kinase A in MIN6 cells significantly increases Sp1 binding activity, while dephosphorylation of Sp1 by protein phosphatase 1 (PP1) dramatically decreases its binding activity to the mVGLUT2 promoter (Figure 5B). Further studies regarding MAPK pathway in the activation of Sp1 suggested that the activation of MAPK by glucose is through phosphorylation of p38 and p44/42 kinases (Figure 5C and 5D, respectively). These results suggest that glucose may induce phosphorylation of Sp1 through MAPK P38 and P44/42, which increases Sp1 binding capacity to the mVGLUT2 promoter and subsequently leads activation of the mVGLUT2 gene.
Figure 5. Glucose stimulates phosphorylation of Sp1 and results in increase of Sp1 binding to mVGLUT2 promoter.
(A). Glucose stimulates Sp1 phosphorylation. Nuclear protein was purified from MIN6 cells cultured in a medium containing indicated glucose concentrations for 48 hours. Western blot was performed with antibodies for Sp1 (Sp1) or phosphorylated Sp1 (p-Sp1). (B). Phosphorylation of Sp1 increases its binding activity to mVGLUT2 promoter. Electrophoretic mobility shift assay was performed with nuclear extract of MIN6 cells treated with protein kinase A (PKA), MAP kinase (MAPK) or protein phosphotase 1(PP1) and DIG-labeled oligonucleotides corresponding to the nucleotide sequence −41 to −9 of the mVGLUT2 promoter. (C). Glucose activation of MAP kinase P-38. Western blot was performed with nuclear protein from MIN6 treated with different glucose concentrations and antibodies against P-38 (P-38) or phosphorylated P-38 (pP-38). (D). Glucose activation of MAP kinase P-44/42. Western blot was performed with nuclear protein from MIN6 treated with different glucose concentrations and antibodies against P-44/42 (P-44/42) or phosphorylated P-44/42 (pP-44/42).
Role of MAPK P38 and P44/42 in Glucose-induced Activation of mVGLUT2
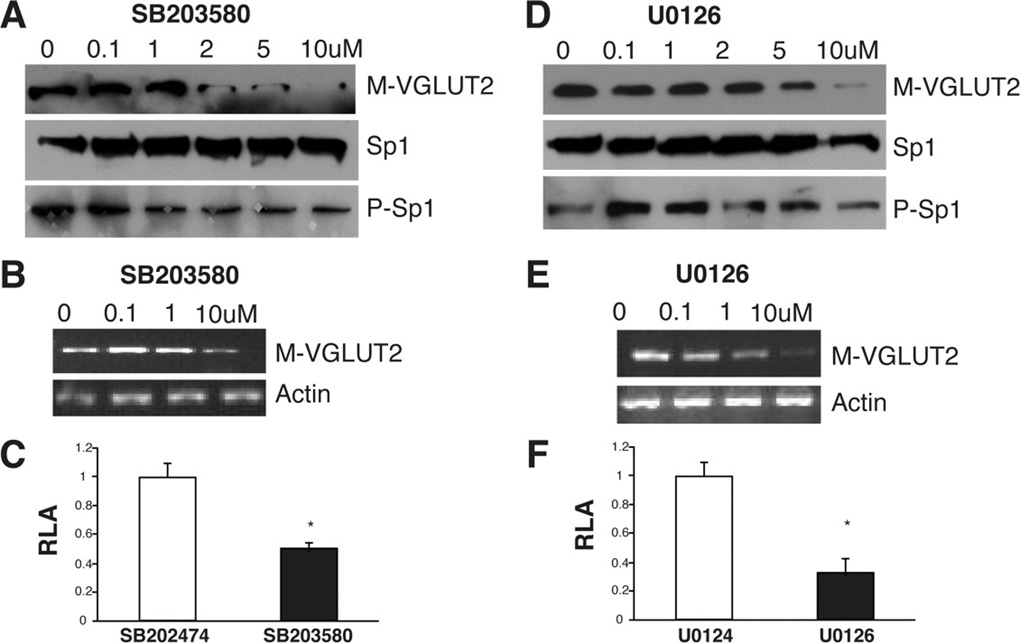
Sp1 as a target of signal transduction cascades in gene activation has been confirmed in many genes. Changes in Sp1 phosphorylation provide one potential mechanism for manipulating activity of this protein31. To confirm the observation that MAPK P38 and P44/42 are involved in Sp1-mediated glucose activation of mVGLUT2, we examine the effects MAPK P38 and P44/42 inhibitors on glucose-induced expression of Sp1 and mVGLUT2. SB203580, a P38 inhibitor, decreases glucose-induced phosphorylation of Sp1 (Figure 6A) without change the amount of total Sp1. As expected, the P38 inhibitor significantly reduces glucose-induced mVGLUT2 protein (Figure 6A), mRNA (Flgure6B), and promoter activity (Figure 6C) in MIN6 cells. Similar patterns of reduction of glucose-induced Sp1 phosphorylation and VGLUT2 expression were observed with MAPK P44/42 inhibitor, U0126 (Figure 6D, 6E and 6F).
Figure 6. Inhibition of Sp1 phosphorylation reduces glucose-induced mVGLUT2 expression.
(A). MAPK P38 inhibitor SB203580 decreases Sp1 phosphorylation and mVGLUT2 expression in MIN6 cells with dose dependence. Expression of mVGLUT2, Sp1 and phosphorylated Sp1 were detected by Western blot using specific antibodies. (B). mRNA of mVGLUT2 in MIN6 cells treated with P38 inhibitor SB203580 was detected by RT-PCR with mVGLUT2 specific primers. (C). Dual luciferase assay was measured in MIN6 cells expressing mVGLUT2 promoter (−96/+68) and treated with P38 inhibitor, SB203580, or the control agent (SB202474) for 24 hours. *P < 0.01. (D). MAPK P44/42 inhibitor U0126 decreases Sp1 phosphorylation and mVGLUT2 expression in MIN6 cells with dose dependence. Expression of mVGLUT2, Sp1 and phosphorylated Sp1 were detected by Western blot using specific antibodies. (E). mRNA of mVGLUT2 in MIN6 cells treated with P44/42 inhibitor U0126 was detected by RT-PCR with mVGLUT2 specific primers. (F). Dual luciferase assay was measured in MIN6 cells expressing mVGLUT2 promoter (−96/+68) and treated with P344/42 inhibitor, U0126, or the control agent (U0124) for 24 hours. *P < 0.01.
Discussion
The present study was designed to determine the molecular mechanism of glucose-induced transcriptional regulation of mVGLUT2 and the potential role of VGLUT2 in glucose-induced insulin secretion. We first cloned the mouse VGLUT2 promoter, and demonstrated a single transcriptional start site and the minimal functional promoter region. The transcription factor Sp1 was shown to mediate the basal regulation of transcriptional expression of the mVGLUT2 gene. Phosphorylation of Sp1 by MAPK P38 and P44/42 increases the binding of Sp1 to the mVGLUT2 promoter and is necessary for glucose-induced transcriptional activation of mVGLUT2.
Sp1 consists of three contiguous zinc-finger domains that bind to the consensus sequence KRGGMGKRRY, which is referred to as a GC box32. Additional transcription factors (Sp2, Sp3, and Sp4), with structural and transcriptional properties similar to those of Sp1, have been cloned, and together they form a Sp1 multigene family33. Functional studies have shown that Sp1 and Sp4 generally act as transcriptional activators30,34, while Sp3 acts as both a repressor and an activator35,36. Deletion and mutation analysis of putative Sp1 cis-elements of mVGLUT2 suggested a potential role of Sp1 involved in the basal promoter activity. Gel mobility shift assays were used to determine whether protein-DNA binding to the GC element correlated with the function of the promoter. Since a number of proteins bind the GC element, including other members of the Sp1 family, the specific antibodies against Sp1, Sp2, Sp3, and Sp4 were used to identify individual components in the complexes. Super shift assay with the specific antibodies further confirmed the direct binding of Sp1, but not other members of the Sp1 family, to the Sp1 cis-element of the mVGLUT2 promoter. In Vivo binding of Sp1 to the mVGLUT2 promoter was further confirmed by ChIP analyses. From these studies, we established an essential role of Sp1 in the basal transcription of the mVGLUT2 gene.
Consistent with our previous findings in other pancreatic cells, mVGLUT2 promoter exhibits glucose responsiveness (Fig. 2D). The region of mVGLUT2 promoter responsible for conferring glucose-stimulated transcription contains several consensus binding sites including Sp1 and GATA3. Site-directed mutagenesis of these sites indicates that Sp1 consensus sequence mediates glucose-induced mVGLUT2 activation, while the GATA3 site does not. Utilizing Drosophila SL2 cell, we confirmed that Sp1, but not Sp2, Sp3 or Sp4, is required for glucose-induced activation of the mVGLUT2 promoter. The critical role of Sp1 in mVGLUT2 activation was further supported by the evidence that mVGLUT2 mRNA of MIN6 cells was down-regulated by blocking of Sp1 binding with mithramycin A or reducing of Sp1 expression by Sp1-specific siRNA. Taken together, it appears that Sp1 site may provide a mechanism for the glucose responsiveness of the mVGLUT2 gene.
It has been suggested that changes in Sp1 phosphorylation provide one potential mechanism for manipulating activity of this protein. The Sp1 protein contains consensus phosphorylation sites for many kinases including PKA, protein kinase C, MAPK, casein kinases (CK1 and CK2), and calmodulin kinases (CamKs). It is likely that different kinases or phosphatases may target selected motif in response to extracelular stimulus and thus alter Sp1 phosphorylation in a cell- or gene-specific manner. It has been shown that the DNA binding activity of Sp1 can be modulated either positively or negatively by phosphorylation37–39. For glucose-responsive genes, a glucose-dependent Sp1 dephosphorylation has been suggested to be responsible for glucose-mediated activation of the aldolase A gene40. We have found that glucose treatment increases phosphorylation of MAP kinases P38 and P44/42, and thus induces phosphorylation of the existing Sp1 protein without changing the amount of total Sp1 protein. Phosphorylation of Sp1 by MAPK significantly increases Sp1 binding activity to the mVGLUT2 promoter, while dephosphorylation of Sp1 by protein phosphatase 1 (PP1) dramatically decreases its binding activity to the mVGLUT2 promoter. Inhibition of phosphorylation of Sp1 by inhibitors of MAPK (P38 and P44/42) dramatically reduces glucose-induced activation of the mVGLUT2 gene. Taken together, in MIN6 cells, glucose increases phosphorylation of Sp1 via MAPK P38 and P44/42, resulting in an increase of binding of Sp1 to the mVGLUT2 promoter and allowing activation of the mVGLUT2 gene. This mechanism is in contrast to the previous finding in which dephosphorylation of Sp1 increases its binding activity to aldose A gene in a rat hepatoma cell line. The discrepancy of Sp1 phosphorylation status on its binding activity and the effect on the target genes suggests that the function of Sp1 on gene transcription is cell-type- and gene-dependent31. In conclusion, our results demonstrated a novel transcriptional mechanism of glucose-induced activation of mVGLUT2.
Acknowledgments
Grant Support: This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK063142 to L. Bai and R01-DK033209 to F. K. Ghishan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There is no conflict of interest to disclose.
References
- 1.Henquin JC. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes. 2000;49:1751–1760. doi: 10.2337/diabetes.49.11.1751. [DOI] [PubMed] [Google Scholar]
- 2.Rorsman P. The pancreatic beta-cell as a fuel sensor: an electrophysiologist’s viewpoint. Diabetologia. 1997;40:487–495. doi: 10.1007/s001250050706. [DOI] [PubMed] [Google Scholar]
- 3.Lang J. Molecular mechanisms and regulation of insulin exocytosis as a paradigm of endocrine secretion. Eur J Endocrinol. 1999;259:3–17. doi: 10.1046/j.1432-1327.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- 4.Wollheim CB. Beta-cell mitochondria in the regulation of insulin secretion a new culprit in Type II diabetes. Diabetologia. 2000;43:265–277. doi: 10.1007/s001250050044. [DOI] [PubMed] [Google Scholar]
- 5.Maechler P, Wollheim CB. Mitochondrial glutamate acts as a messenger in glucose-induced insulin exocytosis. Nature (Lond) 1999;402:685–689. doi: 10.1038/45280. [DOI] [PubMed] [Google Scholar]
- 6.Maechler P, Gjinovci A, Wollheim CB. Implication of glutamate in the kinetics of insulin secretion in rat and mouse perfused pancreas. Diabetes. 2002;51:S99–S102. doi: 10.2337/diabetes.51.2007.s99. [DOI] [PubMed] [Google Scholar]
- 7.Rubi B, Ishihara H, Hegardt FG, Wollheim CB, Maechler P. GAD65-mediated glutamate decarboxylation reduces glucose-stimulated insulin secretion in pancreatic β cells. J. Biol. Chem. 2001;276:36391–36396. doi: 10.1074/jbc.M104999200. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand G, Gross R, Puech R, Loubatieres-Mariani MM, Bockaert J. Evidence for a glutamate receptor of the AMPA subtype which mediates insulin release from rat perfused pancreas. Br J Pharmacol. 1992;106:354–359. doi: 10.1111/j.1476-5381.1992.tb14340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrand G, et al. Glutamate stimulates glucagon secretion via an excitatory amino acid receptor of the AMPA subtype in rat pancreas. Eur J Pharmacol. 1993;237:45–50. doi: 10.1016/0014-2999(93)90091-u. [DOI] [PubMed] [Google Scholar]
- 10.Weaver CD, Gundersen V, Verdoorn TA. A high affinity glutamate/aspartate transport system in pancreatic islets of Langerhans modulates glucose-stimulated insulin secretion. J Biol Chem. 1998;273:1647–1653. doi: 10.1074/jbc.273.3.1647. [DOI] [PubMed] [Google Scholar]
- 11.Bai L, Zhang X, Ghishan FK. Characterization of vesicular glutamate transporter in pancreatic α- and β -cells and its regulation by glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G808–G814. doi: 10.1152/ajpgi.00333.2002. [DOI] [PubMed] [Google Scholar]
- 12.Skerry TM, Genever PG. Glutamate signalling in non-neuronal tissues. Trends Pharmacol. Sci. 2001;22:174–181. doi: 10.1016/s0165-6147(00)01642-4. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi M, Otsuka M, Morimoto R, Hirota S, Yatsushiro S, Takeda J, Yamamoto A, Moriyama Y. Differentiation-associated Na+-dependent inorganic phosphate cotransporter (DNPI) is a vesicular glutamate transporter in endocrine glutamatergic systems. J Biol Chem. 2001;276:43400–43406. doi: 10.1074/jbc.M106244200. [DOI] [PubMed] [Google Scholar]
- 14.Inagaki N, et al. Expression and role of ionotropic glutamate receptors in pancreatic islet cells. FASEB J. 1995;9:686–691. [PubMed] [Google Scholar]
- 15.Liu HP, Tay SS, Leong SK. Localization of glutamate receptor subunits of the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) type in the pancreas of newborn guinea pigs. Pancreas. 1997;14:360–368. doi: 10.1097/00006676-199705000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Weaver CD, Yao TL, Powers AC, Verdoorn TA. Differential expression of glutamate receptor subtypes in rat pancreatic islets. J Biol Chem. 1996;271:12977–12984. doi: 10.1074/jbc.271.22.12977. [DOI] [PubMed] [Google Scholar]
- 17.Otis TS. Vesicular glutamate transporters in cognito. Neuron. 2001;29:11–14. doi: 10.1016/s0896-6273(01)00176-3. [DOI] [PubMed] [Google Scholar]
- 18.Bai L, Xu H, Collins JF, Ghishan FK. Molecular and functional analysis of a novel neuronal vesicular glutamate transporter. J. Biol. Chem. 2001;276:36764–36769. doi: 10.1074/jbc.M104578200. [DOI] [PubMed] [Google Scholar]
- 19.Israel M, Tomasi M, Bostel S, Meunier FM. Cellular resistance to Evans blue toxicity involves an up-regulation of a phosphate transporter implicated in vesicular glutamate storage. J. Neurochem. 2001;78:658–663. doi: 10.1046/j.1471-4159.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki J, et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 21.Bai L, Collins JF, Xu H, Ghishan FK. Transcriptional regulation of rat Na(+)/H(+) exchanger isoform-2 (NHE-2) gene by Sp1 transcription factor. Am. J. Physiol. 2001;280:C1168–C1175. doi: 10.1152/ajpcell.2001.280.5.C1168. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Walsh JR, Ghishan FK, Bai L. Molecular cloning and characterization of a human urate transporter (hURAT1) gene promoter. Bioch. Biophy. Acta. 2004;1681:53–58. doi: 10.1016/j.bbaexp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Jurado LA, Song S, Roesler WJ, Park EA. Conserved amino acids within CCAAT enhancer-binding proteins (C/EBP(alpha) and beta) regulate phosphoenolpyruvate carboxykinase (PEPCK) gene expression. J. Biol. Chem. 2002;277:27606–27612. doi: 10.1074/jbc.M201429200. [DOI] [PubMed] [Google Scholar]
- 24.Breathnach R, et al. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. U. S. A. 1978;75:4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniel S, Zhang S, DePaoli-Roach AA, Kim KH. Dephosphorylation of Sp1 by protein phosphatase 1 is involved in the glucose-mediated activation of the acetyl-CoA carboxylase gene. J. Biol. Chem. 1996;271:14692–14697. doi: 10.1074/jbc.271.25.14692. [DOI] [PubMed] [Google Scholar]
- 26.Daniel S, Kim KH. Sp1 mediates glucose activation of the acetyl-CoA carboxylase promoter. J. Biol. Chem. 1996;271:1385–1392. doi: 10.1074/jbc.271.3.1385. [DOI] [PubMed] [Google Scholar]
- 27.Chen YQ, et al. Sp1 sites mediate activation of the plasminogen activator inhibitor-1 promoter by glucose in vascular smooth muscle cells. J. Biol. Chem. 1998;273:8225–8231. doi: 10.1074/jbc.273.14.8225. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda H, Iritani N. Transcriptional regulation of leptin gene promoter in rat. FEBS Letters. 1999;455:165–169. doi: 10.1016/s0014-5793(99)00877-7. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda H, Noguchi T, Iritani N. Transcriptional regulation of fatty acid synthase gene and ATP citrate-lyase gene by Sp1 and Sp3 in rat hepatocytes. FEBS Letters. 1999;464:113–117. doi: 10.1016/s0014-5793(99)01700-7. [DOI] [PubMed] [Google Scholar]
- 30.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–888. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 31.Samson SL, Wong NC. Role of Sp1 in insulin regulation of gene expression. J. Mol. Endocrin. 2002;29:265–279. doi: 10.1677/jme.0.0290265. [DOI] [PubMed] [Google Scholar]
- 32.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 33.Cook T, Gebelein B, Urrutia R. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Annals of the New York Academy of Sciences. 1999;880:94–102. doi: 10.1111/j.1749-6632.1999.tb09513.x. [DOI] [PubMed] [Google Scholar]
- 34.Hagen G, et al. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J. Biol. Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- 35.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majello B, Luca PD, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 37.Armstrong SA, et al. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- 38.Rohlff C, et al. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- 39.Merchant JL, Du M, Todisco A. Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem. & Biophy. Res. Comm. 1999;254:454–461. doi: 10.1006/bbrc.1998.9964. [DOI] [PubMed] [Google Scholar]
- 40.Schafer D, Hamm-Kunzelmann B, Brand K. Glucose regulates the promoter activity of aldolase A and pyruvate kinase M2 via dephosphorylation of Sp1. FEBS Letters. 1997;417:325–328. doi: 10.1016/s0014-5793(97)01314-8. [DOI] [PubMed] [Google Scholar]