Abstract
Exposure of female rats to trichloroethylene (TCE), an environmental toxicant commonly found in ground and surface waters throughout the United States, reduces the fertilizability of oocytes produced by these females compared with oocytes from control females. Localization of cytochrome P450 2E1 and glutathione s-transferase α, TCE-metabolizing enzymes, in the ovary suggests TCE metabolism occurs in the ovary. The production of bioactive TCE metabolites in the ovary may alter female reproductive function by altering ovarian gene transcription and/or protein expression and function. The purpose of the present study was to examine ovarian gene transcription after exposure of female rats to 0.45% TCE (v/v) in 3% Tween. Control rats received 3% Tween. Microarray analysis after 1 and 5 days of exposure indicated ovarian gene transcription was maintained during TCE exposure with the possible exception of a very few genes. Although conclusions for these few genes were ambiguous from the microarray analysis due to the minimal but statistically significant reductions, quantitative real time RT-PCR (qRT-PCR) analysis indicated expression of these genes was unaltered after TCE exposure. Protein analysis confirmed qRT-PCR results. This study suggests TCE-induced reductions in oocyte fertilizability are independent of currently detectable alterations in ovarian gene expression.
Keywords: Trichloroethylene, Ovary, Gene expression, Protein expression, Oocyte plasma membrane
INTRODUCTION
Trichloroethylene (TCE) is a volatile compound commonly used in industrial metal degreasing applications. As a result of manufacturing, usage, and disposal practices, TCE is a common ground and surface water contaminant in the USA (Scott and Cogliano, 2000). The widespread occurrence of TCE, albeit at minimal amounts, in the environment makes the general population vulnerable to exposure. Exposure can occur via inhalation, transdermal absorption, and ingestion. The liver is the primary target organ of toxicity in experimental animals exposed to TCE. Cytochrome P450 dependent oxidation is the major pathway for TCE metabolism given substrate affinity and availability of enzymes (Davidson and Beliles, 1991; Goeptar et al., 1995). Trichloroethylene may also be metabolized by conjugation with glutathione, a pathway that may be relatively more significant in the kidney (Dekant, 1993; Goeptar et al., 1995). Cytochrome P450 2E1 and glutathione s-transferase α (GST α), the primary isoforms of the initial enzymes responsible for TCE metabolism, have been detected in the rodent ovary (Singh and Pandey, 1996; Toft et al., 1997; Cannady et al., 2003). The metabolism of TCE by the ovary into more bioactive and toxic products may adversely effect reproduction. Rat oocyte fertilizability decreases as a result of exposure to TCE (Berger and Horner, 2003; Wu and Berger, 2007); however, the pathway by which this occurs has yet to be fully evaluated.
Earlier studies suggest female reproductive toxicity to TCE may be due to alterations in the composition of the oocyte plasma membrane (Berger and Horner, 2003). Oocytes from female rats exposed to TCE for 4, 5, and 14 days are less capable of binding and being fertilized by normal rat sperm compared with oocytes from control females (Wu and Berger, 2007). Post translational modification of ovarian protein is evident after female rats are exposed to TCE for 4 and 5 days. Modifications of proteins expressed on the oocyte plasma membrane and alterations in protein function may result from change in the expression of the genes encoding those proteins in addition to the post-translational modifications. Alteration of gene expression, or changing gene product stability or turnover mediate some toxic effects (Barker et al., 1994; Aguilar-Mahecha et al., 2001; Mizuyachi et al., 2002; Lock et al., 2006). Microarray analysis is the currently preferred technique to identify candidate genes that might be affected.
The aim of the present study was to evaluate ovarian gene expression after exposure of female rats to TCE as a possible mechanism for the induction of ovarian toxicity to TCE. A toxicogenomic approach was used to examine the effects of TCE-exposure on ovarian gene expression because the ovarian genes that are targeted by TCE-exposure are unknown. The results of the microarray studies were validated with quantitative real-time RT-PCR (qRT-PCR). Proteins, encoded by potentially altered genes, were examined to determine whether mRNA and protein level changes correspond. Four and 5 days of TCE exposure, the shortest intervals evaluated in vivo, are sufficient to reduce oocyte fertilizability by approximately three-fold (Wu and Berger, 2007). Thus, gene expression after 1 and 5 days of TCE exposure were evaluated to consider acute and prolonged effects. Protein levels after 4 to 5 days would most likely be mediated by immediate effects on gene expression given average rates of protein turnover (Pratt et al., 2002). Gene expression analysis after 5 days of exposure allows evaluation of potential prolonged effects of TCE exposure which might contribute to the reduced fertilizability observed 4 or 9 days after a 5 day TCE exposure. The examination of ovarian gene expression after TCE exposure provides new insight on the effects of TCE on the rat ovary.
MATERIALS AND METHODS
Animals
Female Simonson albino rats, a Sprague-Dawley derived strain, from the Department of Animal Science breeding colony at the University of California, Davis were used for this study. Rats were between 28–32 days old at the initiation of TCE treatment. Such animals have not yet ovulated, hence corpora lutea do not contribute to ovarian composition. All rats were housed at 70 ± 2°F and 40–70% humidity under a 14L: 10D light cycle. Rats had ad libitum access to Purina Formulab 5008 rat chow (St. Louis, MO, USA). Between weaning (21 days of age) and treatment with TCE, rats had ad libitum access to deionized water. The University of California, Davis Animal Use and Care Administrative Advisory Committee approved all animal use.
Chemicals
Trichloroethylene (ACS reagent ≥ 99.5%), and polyoxyethylenesorbitan monolaurate (Tween-20) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Water was obtained from a Milli-Q Synthesis system (Millipore, Bedford, MA, USA). Media components for immunocytochemistry were cell-culture tested or molecular biology grade and purchased from Fisher Scientific (Pittsburgh, PA, USA) or Sigma-Aldrich. Pregnant mare serum gonadotropin (PMSG) was acquired from Sigma-Aldrich. Human chorionic gonadotropin (hCG) was obtained from Intervet, Inc. (Millsboro, DE, USA). Goat anti-ALCAM was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit anti-Itmap1 was a generous gift from Dr. Louis Muglia (Washington University School of Medicine and St. Louis Children’s Hospital, St. Louis, MO, USA). Mouse anti-human CD9 was acquired from BioLegend (San Diego, CA, USA). Secondary antibodies, peroxidase-conjugated AffiniPure donkey anti-goat IgG, peroxidase-conjugated AffiniPure donkey anti-rabbit IgG, and FITC-conjugated donkey anti-mouse IgG were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Chemiluminescence reagent was purchased from Perkin Elmer (Waltham, MA, USA). A RNAqueous kit was acquired from Ambion, Inc. (Austin, TX, USA). A GeneChip One-Cycle cDNA synthesis kit was purchased from Affymetrix, Inc. (Santa Clara, CA, USA). Protoscript First Strand cDNA Synthesis kit was obtained from New England Biolabs, Inc. (Ipswich, MA, USA). The 2X RT2 Real-Time SYBR Green PCR master mix and RT2 primer sets for activated leukocyte cell adhesion molecule (ALCAM), CEA-related cell adhesion molecule 1 (Ceacam1), CUB and zona pellucida-like domains 1 (Cuzd1), and transmembrane 4 superfamily member 3 (Tm4sf3) were purchased from SuperArray Bioscience Corporation (Frederick, MD, USA). Custom designed primers were synthesized and purchased from Sigma-Proligo (The Woodlands, TX, USA).
Treatment
Females receiving TCE-treatment drank deionized water containing 0.45% TCE (v/v) and 3% Tween from glass bottles. Control females received 3% Tween (vehicle) from glass bottles (Berger and Horner, 2003). Four replicates of 1 day TCE-exposure and 3 replicates of 5 day TCE-exposure were assessed by microarray. Each female in the TCE-exposure group received an estimated 0.66 g TCE/kg body weight/day based on the average water consumption of 10 mL/100 g body weight (van Zutphen et al., 2001; Berger and Horner, 2003). The 0.45% TCE (v/v) in this acute exposure is in the range of typical experimental exposures (National Toxicology Program (NTP), 1988; Barton and Das, 1996), but much higher than the background levels of TCE in large bodies of water which range from 0.001 to 0.007 ppb (μg/L) (Gist and Burg, 1995). The similar liver weights of TCE-treated females and vehicle-control females after 3 days of TCE exposure indicate systemic toxicity is unlikely (5.68 g vs. 5.18 g., respectively, P>0.10) (Lamb and Hentz, 2006; Wu and Berger, 2007). Water removal by the rats from drinking water bottles was consistent with the reported average water consumption. Water was refilled every 24–48 hours to minimize headspace and volatilization of TCE.
Microarrays
Females were sacrificed by CO2 asphyxiation followed by cervical dislocation immediately prior to the collection of ovaries. Ovaries were collected using sterile surgical instruments and immediately snap frozen in liquid nitrogen. Six littermate females were used per replicate; ovaries from three TCE-treated females were pooled prior to RNA isolation and ovaries from three vehicle-control females were similarly pooled. Subsequently, RNA was isolated from the ovaries using RNAqueous according to the manufacturer’s protocol. Each RNA pellet was resuspended in nuclease-free water. RNA concentration was assessed by measuring UV absorbance at 260 nm with a Shimadzu UV-1700 Pharma Spec spectrophotometer (Columbia, MD, USA). The purity of RNA was assessed by the ratio of A260 and A280 values; all values were between 1.8 to 2.1. The RNA quality was assessed by electrophoresis on a 1% agarose gel with formaldehyde in 1X MOPS buffer (400 mM MOPS, pH 7.0, 100 mM sodium acetate, 10 mM EDTA). The gel was visualized with a UV transilluminator (ChemiImager 4400, Alpha Innotech Corporation, San Leandro, CA, USA).
Ovarian RNA was stored at −80°C in aliquots of 2.5 μg/μl nuclease-free water. The cDNA was synthesized from ovarian RNA samples using a GeneChip One-Cycle cDNA Synthesis Kit. Rat ovarian gene expression was evaluated using Affymetrix GeneChip Rat Genome 230 2.0 arrays. The Microarray Core Facility at the University of California, Davis performed the hybridization of samples to the microarrays. Image data were analyzed using Affymetrix GeneChip Operating Software (GCOS) version 1.3; quality assessment of each array was conducted and arrays being compared had comparable background and noise values; hence, one sample from the 5-day experiment was not included in the analysis. Analysis was performed with DNA-Chip Analyzer (dChip 1.4). Intensity values were normalized by the model-based expression analysis method (Li and Wong, 2001).
Quantitative Real Time RT-PCR
Two micrograms of total RNA from the ovaries of rats treated with TCE or vehicle for 1 day was reverse transcribed using Moloney murine leukemia virus-reverse transcriptase in a Protoscript First Strand cDNA Synthesis Kit. The final cDNA product was diluted to a final volume of 50 μL with nuclease-free water. A 1 μL aliquot of diluted cDNA was analyzed using 2X RT2 Real-Time SYBR Green PCR master mix by an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol. The standard PCR mode was used on the ABI 7500 with the following thermal cycler program: Stage 1 Rep 1, 50.0°C for 2 minutes; Stage 2 Rep 1, 95.0°C for 15 minutes; Stage 3 Reps 40, 95.0°C for 30 seconds, 60.0°C for 30 seconds, and 72.0° for 30 seconds; Stage 4 Rep 1, 95.0°C for 15 seconds, 60.0°C for 1 minute, and 95.0°C for 15 seconds. Seven genes were evaluated by qRT-PCR using SuperArray RT2 PCR Primer sets for ALCAM, Ceacam1, Cuzd1, and Tm4sf3 and PCR primers designed using the following sequences: rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward: AGGGCTGCCTTCTCTTGTGAC, reverse: TGGGTAGAATCATACTGGAACATGTAG, an expressed sequence tag (Accession no. AI236482), forward: GCTCCCAGGAGTGTAGCTTG, reverse: CCACACCACACACAGAGTCC, and a second expressed sequence tag (Accession no. AI599133), forward: GCCAATCTGAACTTGCGTTT, reverse: GGTTTGGCCAGAAATCTGAA. The PCR was conducted in 96-well optical reaction plates (Applied Biosystems), and each well contained a 25 μl reaction mixture consisting of 12.5 μl of SYBR green PCR master mix, 1 μl of first strand cDNA template, 1 μl RT2 PCR Primer set, and 10.5 μl nuclease-free water. For the GAPDH housekeeping gene and expressed sequence tags (EST’s), 0.5 μl of the forward and 0.5 μl of the reverse primer were used. The SYBR green dye was measured at 530 nm during the extension phase. Quantitative real time RT-PCR results were assessed using 7500 Fast System SDS Software version 1.3 (Applied Biosystems). The relative amount of mRNA in each sample was calculated based on its threshold cycle (Ct) in comparison with the Ct of the housekeeping gene (HKG). Relative expression in samples from treated animals and vehicle-control animals were compared by the ratio method (Pfaffl, 2001) and by the standard curve method (SuperArray Bioscience Corporation). Results were similar and values from the ratio method are reported. The purity of the amplified product was determined by a single peak in the dissociation curve. Quantitative real time RT-PCR was conducted in duplicate for each sample.
Quantitative Assessment of ALCAM and Cuzd1 Protein
The ALCAM and Cuzd1 ovarian protein were quantitatively assessed using Western blots. Three replicates were evaluated. A replicate consisted of ovaries from 1 rat treated with TCE for 5 days, and ovaries from a littermate treated with vehicle for 5 days. The ALCAM and Cuzd1 protein were also quantitatively assessed in ovaries from female rats treated with TCE for 2 days and vehicle for 2 days. After collection with sterile surgical instruments, ovaries were immediately immersed in 1 ml of 0.1 M phosphate buffer (pH 7.0) on ice. Tissue was homogenized using a glass tube and Teflon pestle for approximately 2 minutes until the tissue was completely homogenized. The suspension was centrifuged at 10,000 × g in a Fisher Scientific Micro-Centrifuge Model 59A (Pittsburgh, PA, USA) for 20 minutes. The supernate was collected and stored at −20°C until use. Protein concentration was assessed using the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA). Ovarian protein samples were solubilized with 0.9% SDS (w/v), and separated on an 8% SDS-PAGE gel (ALCAM), or a 10% SDS-PAGE gel (Cuzd1). Proteins were electroblotted to 0.45μm polyvinylidene fluoride (PVDF) membrane with a wet transfer tank (Hoefer, Inc, San Francisco, CA, USA). Duplicate blots stained with Coomassie blue were assessed for equal loading of proteins.
Membranes probed with goat anti-ALCAM antibody were blocked with 10% normal donkey serum (NDS) in TTBS (100 mM Tris, 0.9% NaCl, 0.1% Tween-20, pH 7.5) for 1 hour. Activated leukocyte cell adhesion molecule was detected with goat anti-ALCAM (1 μg/ml in TTBS with 10% NDS for 1 hour at room temperature). Normal goat serum was used as a negative control (1:4000 in TTBS with 10% NDS). After rinsing with TTBS, blots were incubated in peroxidase-conjugated donkey anti-goat IgG (1:25,000 in TTBS with 10% NDS) for 1 hour. For the quantitative assessment of Cuzd1, membranes were blocked with 1% bovine serum albumin (BSA), 1% normal donkey serum in PBST (PBS containing 0.5% Tween-20, pH 7.4) for 1 hour. The Cuzd1 (also known as integral membrane-associated protein1, Itmap1) was detected with rabbit anti-Itmap1 (1:1000 in 1% BSA-PBST incubated overnight at 4°C) (Imamura et al., 2002). Normal rabbit serum was used as a negative control. After rinsing with PBST, membranes were incubated with peroxidase-conjugated donkey anti-rabbit IgG (1:10,000 in 1% BSA-PBST) for 1 hour at room temperature. All immunoreactive bands were visualized using chemiluminescence reagent and x-ray film. Densitometric measurements of ALCAM and Cuzd1 were analyzed using Image Quant software (Molecular Dynamics, Sunnyvale, CA, USA).
Immunocytochemical Detection of a Transmembrane 4 Superfamily 3 Protein (CD9)
Two female rats received drinking water containing TCE for the 3 days prior to oocyte recovery, and two littermate females received vehicle-control water for the corresponding 3 days in each replicate. A total of 12 females were used in 3 replicates. Ovulation was induced while females received TCE and vehicle-control water with 15 IU of PMSG followed by 15 IU of hCG 48 hours later. Oviducts were removed and cumulus masses were released into warm saline-BSA (0.9% NaCl, 1 mg BSA/ml). Cumulus masses and zonae pellucidae were removed with hyaluronidase (1 mg hyaluronidase/ml saline-BSA) and acid Tyrode’s (Nicolson et al., 1975), respectively as previously described (Berger and Horner, 2003), then oocytes were rinsed with saline-BSA.
Each female ovulated a mean of 15 oocytes, which provided approximately 30 oocytes from each treatment in each replicate. Non-specific binding was blocked by incubating with 15% heat-treated normal donkey serum (HT-NDS) in HEPES-saline (10 mM HEPES in 0.9% NaCl) for 30 minutes in a warm room (approximately 30°C). Oocytes were incubated with mouse anti-human CD9 (0.95 μg anti-CD9 per 66 μl saline-BSA with 2% HT- NDS) for 2 hours in a warm room. Other oocytes from the same females were incubated in normal mouse IgG (Sigma-Aldrich) (0.95 μg per 68.5 μl saline-BSA with 2% HT- NDS). Following three rinses in saline-BSA, oocytes were incubated with FITC-conjugated donkey anti-mouse IgG (1:150 in 2% HT-NDS in saline-BSA) for 1 hour. After oocytes were rinsed 3 times in saline-BSA, oocytes were fixed in 2% paraformaldehyde for 5 minutes. Subsequently, oocytes were rinsed in saline-BSA, transferred to mounting medium (5 mg p-phenylenediamine, 5 mg sodium azide in 0.5 ml phosphate buffered saline with 4.5 ml glycerol, pH 8.0), and loaded into 0.1 mm square microcells (Vitro Dynamics Inc, Rockway, NJ, USA). Immunolabelled oocytes were visualized using a BioRad Radiance 2100 laser scanning confocal microscope. Images were captured with Laser Sharp 2000 software (BioRad Cell Science Division, now part of Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA). Densitometric measurements of immunofluorescent CD9-labeled oocytes were collected using Image Quant software.
Statistical Analysis
Gene expression data from qRT-PCR experiments and densitometry values from Western blots and confocal microscopy were subjected to statistical analysis with SAS version 9 (SAS Statistical Programs, Cary, NC, USA). Treatment was a fixed factor and replicates within a treatment were random for the analysis of variance (ANOVA). P-values are reported from dChip for the microarray data, and were computed based on the t-distribution, and the degrees of freedom are set according to the Welch modified two-sample t-test. P-values less than 0.08 were considered significant.
RESULTS
Ovarian Gene Expression after Exposure of Female Rats to TCE
Microarray analysis of over 31 thousand known rat genes and EST’s indicated TCE exposure had little if any effect on ovarian gene expression, after either 1 or 5 days of exposure. Although 13 genes demonstrated statistically significant down-regulation after 1 day of TCE exposure, alterations were less than 2-fold (Table 1). The ALCAM (P=0.036), Ceacam1 (P=0.035), Cuzd1 (P=0.071), and Tm4sf3 (P=0.007) were further evaluated because a connection to reproduction was previously identified and these genes belong to the integral to membrane gene ontology. Seven EST’s had microarray-detected changes in gene expression in the −1.35 to −2-fold range, the largest fold change observed for any genes by microarray analysis. Analysis of gene expression data from the 5-day microarrays revealed no statistically significant alteration in expression of any known genes or EST’s nor were there any significant changes greater than two-fold.
Table 1.
Microarray Analysis of Ovarian Gene Expression Changes After 1 Day Exposure to TCE
| Gene | Accession Number | Mean Fold Change ■ |
P-value |
|---|---|---|---|
| Period homolog 2 (Drosophila) | NM_031678 | −1.90 | 0.008 |
| gb:AI599133/DB_XREF=gi:4608181/DB_XREF=EST250836/CLONE=REMEB39/FEA=EST/CNT=11/TID=Rn.17221.1/TIER=Stack/STK=6/UG=Rn.17221/UG_TITLE=ESTs | AI599133 | −1.90 | 0.099 |
| gb:AW524669/DB_XREF=gi:7167054/DB_XREF=UI-R-BO0-aib-a-09 0-UI.s1/CLONE=UI-R-BO0-aib-a-09-0-UI/FEA=EST/CNT=1/TID=Rn.45901.1/TIER=ConsEnd/STK=1/UG=Rn.45901/UG_TITLE=EST | AW524669 | − 1.85 | 0.001 |
| CUB and zona pellucida-like domains 1 (Cuzd1)* | NM_054005 | − 1.60 | 0.071 |
| gb:AI236482/DB_XREF=gi:3829988/DB_XREF=EST233044/CLONE=ROVDG72/FEA=EST/CNT=4/TID=Rn.25530.1/TIER=ConsEnd/STK=1/UG=Rn.25530/UG_TITLE=ESTs | AI236482 | −1.58 | 0.208 |
| Ubiquitin specific protease 42 (predicted) | BM388096 | −1.49 | 0.039 |
| Glutamic pyruvic transaminase 1, soluble | NM_031039 | − 1.48 | 0.039 |
| CEA-related cell adhesion molecule 1 (Ceacam1)*///CEA-related cell adhesion molecule 10 | U23056 | −1.46 | 0.035 |
| Thyroid hormone responsive protein | AI169092 | −1.46 | 0.031 |
| Activated leukocyte cell adhesion molecule (ALCAM)* | NM_031753 | −1.45 | 0.036 |
| FXYD domain-containing ion transport regulator 3 | AI009597 | −1.45 | 0.026 |
| Ng22 protein | BI289103 | −1.43 | 0.017 |
| Tektin 1 | NM_053508 | −1.42 | 0.037 |
| Guanine nucleotide binding protein (G protein), gamma 10 | AI407487 | −1.39 | 0.011 |
| Potassium voltage-gated channel, subfamily H (eag-related), member 1 | BF394600 | −1.39 | 0.037 |
| gb:BF393275/DB_XREF=gi:11378135/DB_XREF=UI-R-CA0-bfs-h-08-0-UI.s1/CLONE=UI-R-CA0-bfs-h-08-0-UI/FEA=EST/CNT=2/TID=Rn.45387.1/TIER=ConsEnd/STK=0/UG=Rn.45387/UG_TITLE=ESTs | BF393275 | − 1.39 | 0.051 |
| Phosphatidylinositol transfer protein, membrane-associated | BF419876 | −1.38 | 0.054 |
| gb:BF406350/DB_XREF=gi:11394325/DB_XREF=UI-R-CA1-bjf-p-110-UI.s1/CLONE=UI-R-CA1-bjf-p-11-0-UI/FEA=EST/CNT=2/TID=Rn.60836.1/TIER=ConsEnd/STK=2/UG=Rn.60836/UG_TITLE=ESTs | BF406350 | −1.38 | 0.021 |
| gb:AI236895/DB_XREF=gi:3830401/DB_XREF=EST233457/CLONE=ROVDL91/FEA=EST/CNT=3/TID=Rn.25401.1/TIER=ConsEnd/STK=0/UG=Rn.25401/UG_TITLE=ESTs | AI236895 | −1.37 | 0.017 |
| gb:BE118251/DB_XREF=gi:8510356/DB_XREF=UI-R-BJ1-azo-e-03- | |||
| 0-UI.s1/CLONE=UI-R-BJ1-azo-e-03-0-UI/FEA=EST/CNT=12/TID=Rn.7363.1/TIER=Stack/STK=6/UG=Rn.7363/UG_TITLE=ESTs | BE118251 | −1.36 | 0.011 |
| Transmembrane 4 superfamily member 3 (Tm4sf3)* | NM_133526 | − 1.35 | 0.007 |
4 replicates.
Reputed reproductive function.
An Absence of Altered Gene Expression by Quantitative Real Time RT-PCR
Although qRT-PCR analysis indicated arithmetically higher values of ALCAM in ovaries from TCE-exposed females (30% higher), this was not different from the level of ALCAM in vehicle controls (P=0.65). The values for Ceacam1 mRNA and Cuzd1 mRNA were not statistically different (75% and 15% higher in treated animals, P=0.31 and P=0.57, respectively). The levels of mRNA for Tm4sf3 was 8% higher in ovaries from treated animals compared with the vehicle controls; these values were also not different (P=0.91). Similarly, qRT-PCR analysis indicated two randomly chosen EST’s of the 7 identified by microarray analysis were not altered (P=0.36 and P=0.35) (Table 2).
Table 2.
Quantitative Real Time RT-PCR evaluation of microarray-detected changes in ovarian gene expression after 1 day of TCE exposure
| Gene | Accession Number | Mean Fold Change ■ |
P-value |
|---|---|---|---|
| gb:AI599133/DB_XREF=gi:4608181/DB_XREF=EST250836/CLONE=REMEB39/FEA=EST/CNT=11/TID=Rn.17221.1/TIER=Stack/STK=6/UG=Rn.17221/UG_TITLE=ESTs | AI599133 | 1.89 | 0.36 |
| CUB and zona pellucida-like domains 1 (Cuzd1)* | NM_054005 | 1.15 | 0.57 |
| gb:AI236482/DB_XREF=gi:3829988/DB_XREF=EST233044/CLONE=ROVDG72/FEA=EST/CNT=4/TID=Rn.25530.1/TIER=ConsEnd/STK=1/UG=Rn.25530/UG_TITLE=ESTs | AI236482 | 2.15 | 0.35 |
| CEA-related cell adhesion molecule 1 (Ceacam1) *///CEA-related cell adhesion molecule 10 | U23056 | 1.75 | 0.31 |
| Activated leukocyte cell adhesion molecule (ALCAM)* | NM_031753 | 1.30 | 0.65 |
| Transmembrane 4 superfamily member 3 (Tm4sf3)* | NM_133526 | 1.08 | 0.91 |
Values represent the mean of 4 replicates with each sample run in duplicate.
Reputed reproductive function.
Evaluation of Corresponding ALCAM, Cuzd1, and CD9 Proteins
Treatment of females with TCE for 5 days did not affect relative amounts of ALCAM protein compared with vehicle controls. The mean densitometry measurement from three replicates was 1688 arbitrary density units (ADU) in ovarian protein from rats treated with TCE for 5 days, and 1621 ADU in ovarian protein from vehicle-control females (P=0.84, SEM=322). Five days was chosen for protein analysis due to estimated protein turnover, but similar results were observed after 2 days of exposure.
The immunoreactive bands representing Cuzd1 in ovarian protein from females treated with TCE for 5 days and vehicle-control females were similar demonstrating TCE had no effect on Cuzd1 protein expression (mean 361 ADU for ovarian protein from TCE-treated rats versus 190 ADU in ovarian protein from vehicle-control rats, P=0.32, SEM=152, n=3). The Cuzd1 protein levels were also similar after 2 days of TCE exposure.
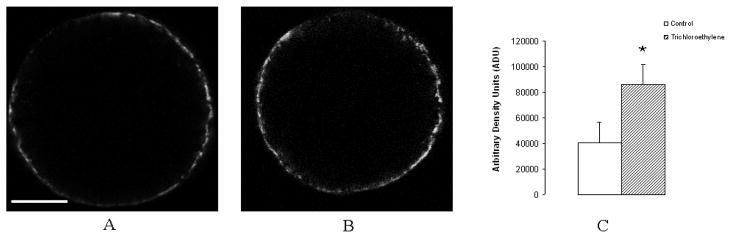
CD9 was evaluated as a representative transmembrane 4 superfamily member 3 protein. Oocytes from females exposed to TCE for 3 days had more CD9 protein on the oocyte surface compared with oocytes from vehicle controls (86085 ADU versus 40707 ADU, P=0.0074, SEM =15754, Figure 1).
Figure 1.
Immunolocalization of CD9 on the plasma membrane of oocytes from vehicle-control and TCE-treated rats. Females were exposed to TCE for 3 days via drinking water, and received an estimated 0.66 g TCE/kg body weight/day based on the average water consumption of 10 mL/100 g body weight. (A) Representative oocyte from vehicle-control female showing immunofluoresent labeling of CD9. Scale bar represents 20 microns. (B) Representative oocyte from TCE-treated female showing immunofluoresent labeling of CD9. (C) Average densitometry values from oocytes of vehicle-control and TCE-treated rats. Three replicates were assessed. After exposure of females to TCE, oocytes expressed significantly more CD9 as determined by immunofluorescence measurements. ★ indicates P<0.05 compared with the vehicle control.
DISCUSSION
Altered gene expression might be a consequence of direct effects on proteins (Song et al., 2002; Girardot et al., 2004) and contribute to prolonged effects on oocyte fertilizability (Wu and Berger, 2007). The previously observed initial reductions in oocyte fertilizability may have resulted from alteration in gene expression in addition to the known increase in protein oxidation. In our previous work, a 4 to 5 day in vivo exposure to TCE dramatically reduced oocyte fertilizability regardless of whether the exposure occurred in the early, mid, or late phase of oocyte growth during meiotic nuclear arrest (Wu and Berger, 2007). A reduced ability to bind sperm and sperm plasma membrane, and increased oxidation of granulosa cell proteins were observed. The present study examined whether TCE exposure alters ovarian mRNA transcription after acute and more prolonged exposures. Ovarian RNA was examined as a whole since alterations in granulosa cell gene expression might mediate the effects of TCE-induced reduction in oocyte fertilizability.
Microarray analysis allows one to screen the expression of a large number of genes. Previous studies examining changes in gene expression after exposure to TCE or a TCE metabolite have been conducted on cardiac, liver, kidney, and skin tissue (Bartosiewicz et al., 2001a; Collier et al., 2003; Chen et al., 2006; Lock et al., 2006). Studies in these organs showed minimal to no changes in gene expression, and toxic organ effects from TCE exposure. Only the highest dose of TCE (1000 mg/kg) altered hepatic gene expression, and only three genes were mildly affected (Bartosiewicz et al., 2001a). Kidney cells exposed to S-(1,2-dichlorovinyl)-L-cysteine, a metabolite of TCE, had minor changes in gene expression (Lock et al., 2006). Exposure to TCE produced inconsistent and negligible changes in rat skin gene expression (Chen et al., 2006). Ovarian gene expression changes were minimal following exposure to TCE for 1 or 5 days. Since small alterations in expression might be biologically significant, four known genes that had a statistically significant reduced expression by microarray analysis were further analyzed by qRT-PCR. The qRT-PCR analysis indicated expression of these four genes increased by 8% or more, albeit not significantly. This suggests the results from the microarray analysis were false “positives” rather than small, biologically significant effects. The qRT-PCR analysis of the two EST’s with the largest (albeit statistically insignificant) reduction via microarray analysis also indicated no change in gene expression induced by TCE exposure. Protein expression encoded by three of these genes was also evaluated and down-regulation was not observed. We cannot exclude the possibility that gene expression is altered in a minor cell type within the ovary (Fielden and Zacharewski, 2001). However, granulosa cells are the common cell type within the ovary, the cell type which contains the enzymes necessary for TCE metabolism (Tiltman and Haffajee, 1999; Cannady et al., 2003), and the cell type most likely to affect oocyte quality.
Quantitative real time RT-PCR validation is the technique most often used to confirm microarray results because the method requires the least amount of RNA compared with Northern blots, ribonuclease protection assays, and conventional RT-PCR (Rajeevan et al., 2001; Dallas et al., 2005). A poor correlation for fold-changes between microarrays and qRT-PCR has been noted for genes that exhibited a fold-change of less than 1.5 on microarrays (Hughes et al., 2000; Dallas et al., 2005). The large number of genes analyzed and the short sequences used for analysis contribute to false positives. In addition, computational tools and algorithms used to analyze raw data from each technique employ distinct criteria which may add variation (Rockett and Hellmann, 2004). Genes with strong hybridization signals and a minimum 2-fold to 4-fold difference are likely to be validated by qRT-PCR (Rajeevan et al., 2001; Chuaqui et al., 2002).
Changes in protein expression correlate with differences in mRNA levels less than 50% of the time (Chuaqui et al., 2002). Discrepancies may be due to inaccurate mRNA results, differences in protein levels compared with mRNA levels, and sensitivity and dynamic range of the methodology used to evaluate protein expression (Chuaqui et al., 2002). In these experiments, microarray analysis suggested a decrease in CD9, the qRT-PCR hinted at an increase but was inconclusive, and analysis of protein expression indicated a small but significant increase in oocytes from treated animals.
Although gene expression is frequently affected by exposure to toxicants (Aguilar-Mahecha et al., 2001; Bartosiewicz et al., 2001a; Bartosiewicz et al., 2001b; Mizuyachi et al., 2002), transcription and post-translational modification of proteins may mediate toxic effects instead of or in addition to effects on gene expression. Other data suggests post-translational modification of proteins within the ovary including oxidation of granulosa cell proteins may partially explain the reduced oocyte fertilizability following TCE exposure (Wu and Berger, 2007). This study suggests toxic effects of TCE exposure on ovarian function are mediated by a pathway that is independent of alterations in gene expression.
Acknowledgments
Portions of this paper were presented at the 39th Annual Meeting of the Society for the Study of Reproduction, Omaha, Nebraska, July 29-August 1, 2006. Funding for KLW was provided in part by the National Institutes of Health, National Institute of Child Health and Human Development (NICHD) training grant in Fertilization and Early Development (T32 HD071131), and the University of California Toxic Substances Research and Teaching Program (UC TSR&TP) student fellowship.
The authors thank Barbara Jean Nitta-Oda for her assistance with Western blots.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no financial, personal, or other relationships that may be considered conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar-Mahecha A, Hales BF, Robaire B. Acute cyclophosphamide exposure has germ cell specific effects on the expression of stress response genes during rat spermatogenesis. Mol Reprod Dev. 2001;60:302–311. doi: 10.1002/mrd.1092. [DOI] [PubMed] [Google Scholar]
- Barker CW, Fagan JB, Pasco DS. Down-regulation of P4501A1 and P4501A2 mRNA expression in isolated hepatocytes by oxidative stress. J Biol Chem. 1994;269:3985–3990. [PubMed] [Google Scholar]
- Barton HA, Das S. Alternatives for a risk assessment on chronic noncancer effects from oral exposure to trichloroethylene. Regul Toxicol Pharmacol. 1996;24:269–285. doi: 10.1006/rtph.1996.0140. [DOI] [PubMed] [Google Scholar]
- Bartosiewicz M, Penn S, Buckpitt A. Applications of gene arrays in environmental toxicology: fingerprints of gene regulation associated with cadmium chloride, benzo(a)pyrene, and trichloroethylene. Environ Health Perspect. 2001a;109:71–74. doi: 10.1289/ehp.0110971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosiewicz MJ, Jenkins D, Penn S, Emery J, Buckpitt A. Unique gene expression patterns in liver and kidney associated with exposure to chemical toxicants. J Pharmacol Exp Ther. 2001b;297:895–905. [PubMed] [Google Scholar]
- Berger T, Horner CM. In vivo exposure of female rats to toxicants may affect oocyte quality. Reprod Toxicol. 2003;17:273–281. doi: 10.1016/s0890-6238(03)00009-1. [DOI] [PubMed] [Google Scholar]
- Cannady E, Dyer CA, Christian PJ, Sipes IG, Hoyer PB. Expression and activity of cytochromes P450 2E1, 2A, and 2B in the mouse ovary: the effect of 4-vinylcyclohexene and its diepoxide metabolite. Toxicol Sci. 2003;73:423–430. doi: 10.1093/toxsci/kfg077. [DOI] [PubMed] [Google Scholar]
- Chen XY, Zhuang ZX, Wang XH, Zhang JZ. Immune responses to trichloroethylene and skin gene expression profiles in Sprague Dawley rats. Biomed Environ Sci. 2006;19:346–352. [PubMed] [Google Scholar]
- Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM, Phillips JL, Krizman DB, Tangrea MA, Ahram M, Linehan WM, Knezevic V, Emmert-Buck MR. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32(Suppl):509–514. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- Collier JM, Selmin O, Johnson PD, Runyan RB. Trichloroethylene effects on gene expression during cardiac development. Birth Defects Res A Clin Mol Teratol. 2003;67:488–495. doi: 10.1002/bdra.10073. [DOI] [PubMed] [Google Scholar]
- Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ, Kees UR. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR -- how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IW, Beliles RP. Consideration of the target organ toxicity of trichloroethylene in terms of metabolite toxicity and pharmacokinetics. Drug Metab Rev. 1991;23:493–599. doi: 10.3109/03602539109029772. [DOI] [PubMed] [Google Scholar]
- Dekant W. Bioactivation of nephrotoxins and renal carcinogens by glutathione S-conjugate formation. Toxicol Lett. 1993;67:151–160. doi: 10.1016/0378-4274(93)90052-y. [DOI] [PubMed] [Google Scholar]
- Fielden MR, Zacharewski TR. Challenges and limitations of gene expression profiling in mechanistic and predictive toxicology. Toxicol Sci. 2001;60:6–10. doi: 10.1093/toxsci/60.1.6. [DOI] [PubMed] [Google Scholar]
- Girardot F, Monnier V, Tricoire H. Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics. 2004;5:74. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gist GL, Burg JR. Trichloroethylene--a review of the literature from a health effects perspective. Toxicol Ind Health. 1995;11:253–307. doi: 10.1177/074823379501100301. [DOI] [PubMed] [Google Scholar]
- Goeptar AR, Commandeur JN, van Ommen B, van Bladeren PJ, Vermeulen NP. Metabolism and kinetics of trichloroethylene in relation to toxicity and carcinogenicity. Relevance of the mercapturic acid pathway. Chem Res Toxicol. 1995;8:3–21. doi: 10.1021/tx00043a001. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Imamura T, Asada M, Vogt SK, Rudnick DA, Lowe ME, Muglia LJ. Protection from pancreatitis by the zymogen granule membrane protein integral membrane-associated protein-1. J Biol Chem. 2002;277:50725–50733. doi: 10.1074/jbc.M204159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JC, Hentz KL. Toxicological review of male reproductive effects and trichloroethylene exposure: Assessing the relevance to human male reproductive health. Reproductive Toxicology. 2006;22:557–563. doi: 10.1016/j.reprotox.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock EA, Barth JL, Argraves SW, Schnellmann RG. Changes in gene expression in human renal proximal tubule cells exposed to low concentrations of S-(1,2-dichlorovinyl)-l-cysteine, a metabolite of trichloroethylene. Toxicol Appl Pharmacol. 2006;216:319–330. doi: 10.1016/j.taap.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Mizuyachi K, Son DS, Rozman KK, Terranova PF. Alteration in ovarian gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin: reduction of cyclooxygenase-2 in the blockage of ovulation. Reprod Toxicol. 2002;16:299–307. doi: 10.1016/s0890-6238(02)00024-2. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (NTP) NTP Toxicology and Carcinogenesis Studies of Trichloroethylene (CAS No. 79-01-6) in Four Strains of Rats (ACI, August, Marshall, Osborne-Mendel) (Gavage Studies) Natl Toxicol Program Tech Rep Ser. 1988;273:1–299. [PubMed] [Google Scholar]
- Nicolson GL, Yanagimachi R, Yanagimachi H. Ultrastructural localization of lectin-binding sites on the zonae pellucidae and plasma membranes of mammalian eggs. J Cell Biol. 1975;66:263–274. doi: 10.1083/jcb.66.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JM, Petty J, Riba-Garcia I, Robertson DH, Gaskell SJ, Oliver SG, Beynon RJ. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett JC, Hellmann GM. Confirming microarray data--is it really necessary? Genomics. 2004;83:541–549. doi: 10.1016/j.ygeno.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CS, Cogliano VJ. Trichloroethylene health risks--state of the science. Environ Health Perspect. 2000;108(Suppl 2):159–160. [PMC free article] [PubMed] [Google Scholar]
- Singh D, Pandey RS. Glutathione-S-transferase in rat ovary: its changes during estrous cycle and increase in its activity by estradiol-17 beta. Indian J Exp Biol. 1996;34:1158–1160. [PubMed] [Google Scholar]
- Song KH, Lee K, Choi HS. Endocrine disrupter bisphenol a induces orphan nuclear receptor Nur77 gene expression and steroidogenesis in mouse testicular Leydig cells. Endocrinology. 2002;143:2208–2215. doi: 10.1210/endo.143.6.8847. [DOI] [PubMed] [Google Scholar]
- Tiltman AJ, Haffajee Z. Distribution of glutathione S-transferases in the human ovary: an immunohistochemical study. Gynecol Obstet Invest. 1999;47:247–250. doi: 10.1159/000010115. [DOI] [PubMed] [Google Scholar]
- Toft E, Becedas L, Soderstrom M, Lundqvist A, Depierre JW. Glutathione transferase isoenzyme patterns in the rat ovary. Chem Biol Interact. 1997;108:79–93. doi: 10.1016/s0009-2797(97)00095-1. [DOI] [PubMed] [Google Scholar]
- van Zutphen LFM, Baumans V, Beynen AC. Principles of Laboratory Animal Science: a Contribution to the Humane Use and Care of Animals and to the Quality of Experimental Results. Elsevier; New York: 2001. [Google Scholar]
- Wu KL, Berger T. Trichloroethylene Metabolism in the Rat Ovary Reduces Oocyte Fertilizability. Chem Biol Interact. 2007;170:20–30. doi: 10.1016/j.cbi.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]