Summary
Actin cross-linking domains (ACD) are distinct domains found in several bacterial toxins, including the Vibrio cholerae MARTX toxin. The ACD of V. cholerae (ACDVc) catalyzes the formation of an irreversible iso-peptide bond between lysine 50 and glutatmic acid 270 on two actin molecules in an ATP and Mg/Mn2+-dependent manner. In vivo, cross-linking depletes the cellular pool of G-actin leading to actin cytoskeleton depolymerization. While the actin cross-linking reaction performed by these effector domains has been significantly characterized, the ACDVc catalytic site has remained elusive due to lack of significant homology to known proteins. Using multiple genetic approaches, we have identified regions and amino acids of ACDVc required for full actin cross-linking activity. Then, using these functional data and structural homology predictions, it was determined that several residues demonstrated to be important for ACDVc activity are conserved with active site residues of the glutamine synthetase family of enzymes. Thus, the ACDs are a family of bacterial toxin effectors that may be evolutionarily related to ligases involved in amino acid biosynthesis.
Keywords: Vibrio cholerae, ACD, actin, glutamine synthetase
Introduction
MARTX (Multifunctional-Autoprocessing Repeats-in-ToXin) toxins are a newly recognized distinct class of secreted bacterial protein toxins identified in the genomes of at least 11 Gram-negative bacterial species (Satchell, 2007, Satchell & Geissler, 2009). The best-characterized MARTX toxin is produced by both clinical and environmental isolates of Vibrio cholerae and is associated with the ability of V. cholerae to depolymerize the eukaryotic cell actin cytoskeleton (Lin et al., 1999, Fullner & Mekalanos, 2000). The depolymerization of actin stress fibers is due to covalently cross-linking of actin monomers into oligomers by incorporation of an iso-peptide bond between the side chains of glutamic acid 270 and lysine 50 (Fullner & Mekalanos, 2000, Kudryashov et al., 2008b).
The ability of MARTX to induce cross-links in actin was isolated to a single domain at amino acids (aa) 1963–2341, a domain now termed the actin cross-linking domain (ACDVc). This domain introduces cross-links in actin in vivo when transiently expressed in cells as a fusion to EGFP (Sheahan et al., 2004) or when purified ACDVc protein is delivered to cells in the absence of the remainder of MARTX (Cordero et al., 2006). Purified ACDVc has also been demonstrated to cross-link monomeric G-actin in vitro in the absence of any other eukaryotic proteins, demonstrating that ACDVc is the ligase enzyme that introduces the iso-peptide bond between actin molecules. The in vitro reaction requires ATP and Mg2+ or Mn2+ (Cordero et al., 2006). Coincident with actin cross-linking, ATP is hydrolyzed resulting in the release of inorganic phosphate (Pi). The resulting multimeric actin molecules lack the linear structure normally associated with polymerized actin and are unable to be incorporated into filaments. The resulting depletion of the G-actin pool causes a net disassembly of the actin cytoskeleton, thereby providing a mechanism for the rounding of tissue culture cells exposed to ACD (Kudryashov et al., 2008b).
Other bacterial toxins also contain ACDs including MARTX toxins from Aeromonas hydrophila (Satchell, 2007), V. campbellii (GenBank: ABGR00000000), and V. vulnificus Biotype 2 (Lee et al., 2008). In addition, V. cholerae expresses a Type VI secretion system effector with an ACD that has been demonstrated to cross-link actin both in vivo (Sheahan et al., 2004) and in vitro (Pukatzki et al., 2007).
Three of the ACD containing toxins have been linked with evasion of innate immunity. The V. cholerae MARTX is one of three toxins required for V. cholerae to persistently colonize the small intestine (Cordero et al., 2007, Rahman et al., 2008, Olivier et al., 2007), VgrG-1 has been shown to target only phagocytic cells (Ma et al., 2009), and the V. vulnificus Biotype 2 MARTX toxin is essential for bacterial survival in eel blood (Lee et al., 2008). Therefore, the ACD and cross-linking of actin could play a conserved role in immune evasion by pathogenic Aeromonads and Vibrios (Satchell & Geissler, 2009).
Despite this in-depth knowledge of the actin cross-linking reaction and its role in pathogenesis, little is known about the ACD enzymes. Extensive bioinformatics analyses have not revealed potential mechanisms or conservation with known peptide ligase enzymes. The ability to cross-link actin is itself a novel biochemical reaction so there are no biochemical, database, or structural models that can be used to identify essential regions of the protein. Thus, in this study we undertook a multifaceted genetic approach to identify regions of ACDVc that are required for functional activity in vivo, and in doing so, have identified residues essential for activity that are candidates for forming the catalytic site. A database search and structural-based alignment method revealed conservation of ACDVc essential residues with other ACDs and with the active sites of glutamine synthetases (GS) and γ-glutamylcysteine synthetases (GCS). Thus, we propose that ACDVc cross-links the γ-hydroxyl group of actin E270 to the γ-NH2 group of K50 in a mechanism related to that used to add ammonia to glutamic acid to form glutamine or to link glutamic acid to cysteine to form γ-glutamylcysteine.
Results
Linker Scanning Mutagenesis Identifies Functional Regions of ACD
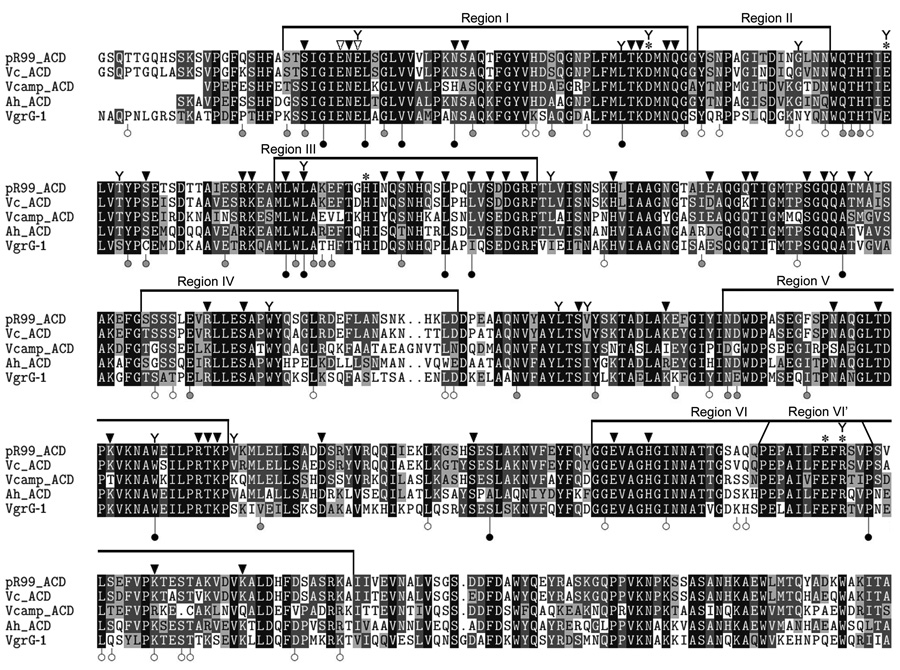
PSI-BLAST (Altschul et al., 1997) searches have identified 5 protein toxins with an ACD. Analysis of an alignment of the five ACD proteins indicated that 200 out of 447 residues are identical between the five orthologs, highlighting amino acids of the proteins that could be essential for cross-linking activity (Fig. 1). To further clarify regions essential for cross-linking activity, a linker scanning mutagenesis (LSM) screen was conducted using the ACD from V. cholerae MARTX (ACDVc). LSM is a commonly used genetic technique to investigate proteins that involves introduction of 5 aa linkers into a given protein sequence followed by functional analyses to reveal important regions of the protein. Initially, the ACDVc coding region (aa 1963–2374) was cloned into pENTR/D-TOPO and the resulting plasmid was subjected to LSM mutagenesis (Experimental Procedures). A total of 70 unique mutants with in-frame 5 codon insertions within ACDVc were generated and then recombined into pDEST47 to fuse GFP to the C-terminus of ACDVc in a eukaryotic expression vector, resulting in pDEST47-ACD-GFP (pDEST-ACD). The mutant forms of pDEST-ACD were individually transiently transfected into COS-7 cells, then cell rounding and actin cross-linking were monitored by western blotting using anti-actin antibodies. COS-7 cells transfected with wild-type pDEST-ACD or 30 of the mutant plasmids contained cross-linked forms of actin, indicating that disruption of these regions of ACDVc did not block activity (data not shown, Fig. 2, lower unfilled markers). This group included 7 insertions in the 3’ end of acd. Subsequently, it was shown that 73 codons could be deleted from the originally characterized C-terminus of ACDVc without affecting actin cross-linking activity (data not shown).
Fig. 1.
Summary of alignment and mutagenesis results to identify essential residues. CLUSTAL W alignment of all confirmed and predicted ACD protein sequences. pR99_ACD, V. vulnificus pR99 rtxA (NC_009701); Vc_ACD, V. cholerae rtxA (NC_002505.1); Vcamp_ACD, V. campbellii rtxA (NZ ABGR01000041.1); Ah_ACD, A. hydrophila rtxA (NC_008570.1); VgrG-1, V. cholerae vgrG-1 (NC_002505.1). The degree of shading indicates aa identity (black = 100% identical, white = no identity). The positions of the LSM mutants are indicated below the sequence, according to the more detailed description in Fig. 2. The location of each individual mutant acquired by epPCR as detailed in Fig. 3 is indicated with a Y above the sequence. Regions represent susceptibility to insertion mutations (see text). Inverted black arrows indicate residues not essential for actin cross-linking in HeLa cells or prevention of colony formation in yeast after mutation to Ala; white arrows represent residues essential for both cross-linking in HeLa cells and prevention of colony formation in yeast; asterisks indicate residues partially defective for actin cross-linking in HeLa cells and that prevent colony formation in yeast.
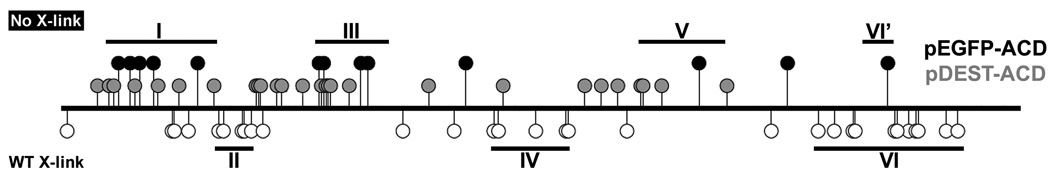
Fig. 2.
LSM reveals regions of ACDVc important for actin cross-linking. The location of each insertion is plotted along the length of ACDVc. Circles above the bar indicate insertions that blocked cross-linking and the circles below indicate insertions that displayed wild-type cross-linking. ACDVc was transfected and expressed as a GFP fusion in two separate plasmids: filled gray circles are insertions that did not cross-link when expressed from pDEST-ACD, but cross-linked when expressed from pEGFP-ACD; filled black circles are insertions that blocked cross-linking when expressed from both plasmids.
Western blotting showed that actin was not cross-linked in cells transfected with 40 of the pDEST-ACD mutant constructs (Fig. 2, upper grey markers, Table S2). Over the course of the screening, the transfection efficiency in COS-7 cells with the pDEST-ACD-based plasmid was poor, therefore expressed protein was not reproducibly detected by either GFP or ACD immunoblotting. Attempts to increase efficiency by changing transfection conditions and cell lines did not sufficiently improve reproducibility of detection. Thus, to confirm that a stable fusion protein was expressed in transfected cells, the 40 ACDVc coding regions containing linker that disrupted actin cross-linking when expressed from pDEST-ACD were transferred into pEGFP-ACD. pEGFP-ACD expresses ACD-EGFP in COS-7 cells at levels that are reproducibly detectable after transient transfection (Sheahan et al., 2004). When expressed from pEGFP-ACD, only 13 of the 40 ACDVc LSM insertion mutants were defective for actin cross-linking (Fig. 2, solid black markers, Table S2) and all produced detectable ACD-EGFP fusion protein (data not shown).
In Fig. 2, three large regions (I, III, and V) within ACDVc can be identified that contain multiple disruptive insertions. The aa conservation within these regions between five ACD family members was found to be very high (65% identity), again highlighting the importance of these regions (Fig. 1). In support of this, 10 of the 13 disruptive insertions occurred at 100% conserved residues. Three additional regions of ACDVc (II, IV, and VI) can be identified that are permissive to insertional mutagenesis (Fig. 2), suggesting that these regions of ACDVc are not involved in catalytic activity. Consistent with this conclusion, the aa conservation in these regions is low (37% identity) (Fig. 1). Despite 11 non-disruptive insertions, one disruptive insertion occurred in a highly conserved portion of Region VI, prompting us to consider that a sub-region (Region VI’) could be involved in ACDVc activity (Fig. 1 and 2).
The LSM approach was thus successful in identifying regions of ACDVc likely involved in its overall activity and reduced the number of potential essential aa to less than 100, or below 25% of the ACDVc. However since these regions were still very broad, a parallel non-biased genetic approach that would identify individual residues responsible for ACDVc function was employed.
S. cerevisiae can be Used as a Model System to Study ACD Activity
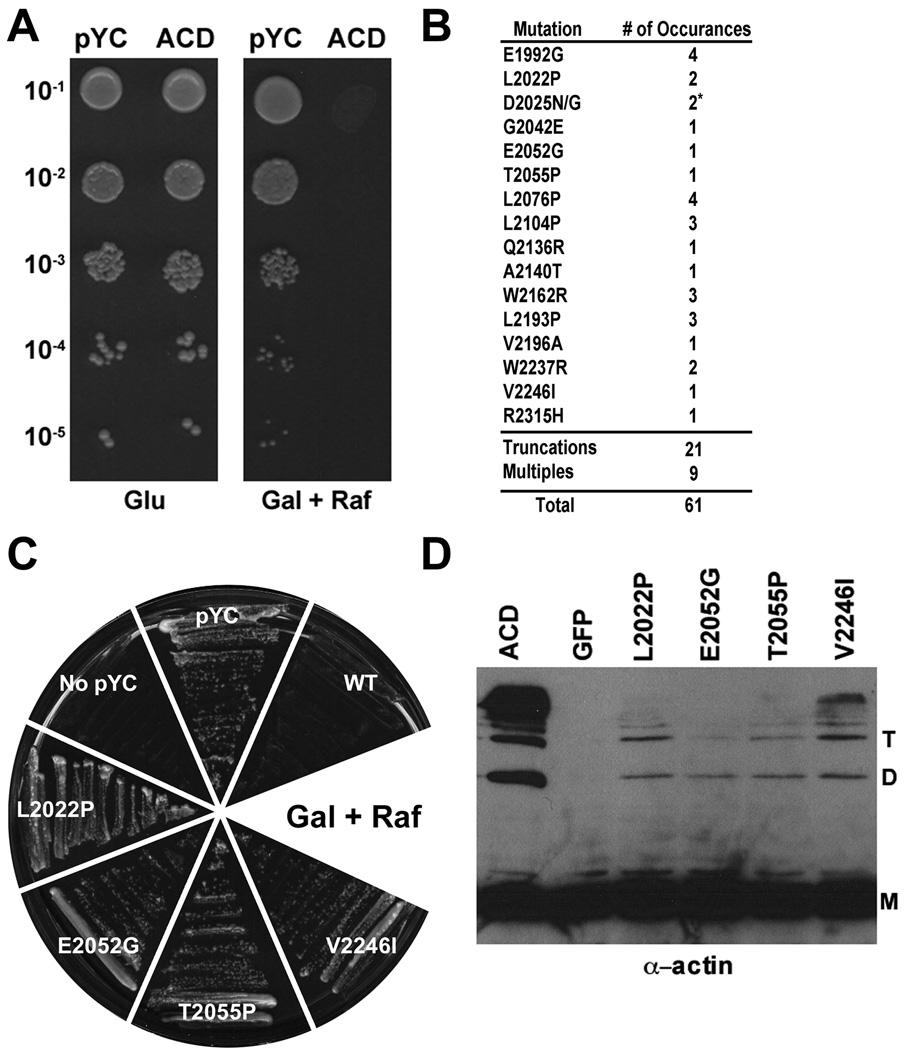
Following the identification of regions involved in actin cross-linking activity using LSM, a Saccharomyces cerevisiae (Sc) based strategy utilizing error-prone PCR (epPCR) coupled with yeast gap repair cloning was used to identify functional aa within ACDVc (Hua et al., 1997). It has been previously demonstrated that ACDVc can cross-link yeast actin in vitro (Kudryashov et al., 2008b), suggesting that expression of ACDVcwould compromise actin polymerization in growing yeast cells, thereby preventing cell division. In order to determine if ACDVc affects growth of Sc, ACDVc was cloned into the yeast expression plasmid, pYC2/NT A (pYC) to yield pYC-ACD. pYC contains the GAL1 promoter that is repressed when grown in media containing glucose (glu), but its expression is fully induced when grown in media containing galactose (gal). Sc+pYC-ACD grew normally on glu agar, but was unable to form colonies on gal agar, whereas Sc+pYC grew normally on either media (Fig. 3A). This result indicated that expression of ACDVc within Sc from pYC prevented colony formation under induction conditions.
Fig. 3.
Mutations in ACDVc isolated by epPCR that allow yeast colony formation, reduce actin cross-linking activity. (A) Dilutions of Sc containing either pYC or pYC-ACD (ACD) were spotted onto plates containing either Glu or gal and raf (Gal + Raf) and grown for 48 hrs at 30°C. (B) The location and number of isolates of each single mutation identified from pYC-ACD by epPCR (see Experimental Procedures for details). The number of individual colonies containing plasmids encoding truncations and multiple mutations identified in the screen are also listed. * two separate mutations in D2025 were isolated, one time each. (C) Sc containing plasmids expressing the indicated mutation in ACD were plated onto Gal + Raf; Sc without plasmid (No pYC), empty vector (pYC) and wild-type ACD (WT) are included as controls. (D) HeLa cells were transfected with pEGFP (GFP), pEGFP-ACD (WT) or pEGFP-ACD with the indicated mutation and processed for immunoblotting with actin specific antisera. M, monomer; D, dimer; T, trimer.
After confirming an effect on yeast growth, the ACDVc was amplified by PCR using Taq DNA polymerase with an inherently low fidelity of nucleotide incorporation. Sc was then transformed with the amplified product and linearized plasmid. Once transformed with both DNA molecules, yeast homologous recombination can occur between the >30bp of homology on the plasmid and the PCR product, thereby resulting in a replication competent plasmid with ACDVc expression under control of the GAL1 promoter. The transformations were plated onto Ura+gal to select for Sc carrying the plasmid (Ura) and to induce expression from pYC-ACD (gal). Inducing ACDVc expression only allowed the growth of Sc containing pYC-ACD with a non-functional copy of ACDVc, but selected against Sc carrying a plasmid expressing a functional ACDVc, which prevents colony formation. 80 individual colonies arose following the transformation of PCR product and plasmid, while no colonies grew from transformation of either PCR product or linearized plasmid alone.
Using this approach, 16 individual aa changes, 9 multiple residue changes, and 21 frame-shift truncations were identified (Fig. 3B). The individual mutants were primarily distributed across the first 285 aa of the ACDVc, with the exception of one, R2315H. This distribution of mutants supports the high degree of aa conservation between the five ACD homologs, as well as the results from the LSM, that the N-terminal ~300 residues of ACDVc are less amenable to alteration than the C-terminal ~150 residues (Fig. 1). The residues identified in this yeast screen overlap with the LSM in many areas, most evidently in region I, where 3 of the 16 mutants are located. Notably, three of the yeast mutants (L2022P, L2076P, W2237R) are in aa identified in the LSM approach as deficient in actin cross-linking and five (E1992G, D2025N/G, Q2136R, A2140T, R2315H) were located within 3 aa of an LSM insertion suggesting the mutations could affect the same loop, strand, or helix (Fig. 1).
Due to the severity of the growth defect in yeast, it was difficult to confirm that ACDVc cross-links actin in yeast. Thus, to demonstrate that the residues identified in this approach represented defects in actin cross-linking activity, four of the mutations that allowed yeast colony formation (Fig. 3C) were transferred to pEGFP-ACD. Upon transfection into HeLa cells, all four mutants caused cell rounding but showed markedly reduced actin cross-linking activity compared to transfection of wild-type pEGFP-ACD (Fig. 3D). This result indicates that the yeast-based approach allowed the isolation of partially defective ACD mutants.
Investigation of the type of base pair changes selected by this mutagenesis strategy showed that this method was biased towards particular nucleotide changes. Of the 60 base pair changes identified, 23 were A→G and 20 were T→C (data not shown). This nucleotide bias favored the mutation of Leu, Trp, and Glu residues as 60% of the total aa changes identified (individual and multiple) involved one of these three residues (Fig. 3B). There was also a bias for transition to Pro or Gly, as half of the single mutations resulted in conversion to either a Pro or Gly. This bias and the overlap with the LSM mutants suggests that this screen identified mutations that induced conformational changes in the overall protein structure, which could disrupt actin cross-linking activity, in addition to the identification of potential essential catalytic residues.
Targeted Ala-Substitution Mutagenesis of the ACD Reveals Residues Essential for Full Activity in HeLa cells
Based on the cumulative data from LSM and PCR mutagenesis and the five ACD proteins (Fig. 1), a list of candidate essential residues was generated. Our selection of residues was limited to aa in close proximity to those identified in either the LSM or yeast mutagenesis strategies as essential for activity and those that were 100% identical or highly conserved between the five ACDs. In addition, several residues in a previously predicted Walker box consensus ATP binding site, SIDAQGKT, at residues 2120–2127 were selected (Cordero et al., 2006). Finally, since many peptide ligases require serine and histidine in their active site, highly conserved Ser and His residues were selected for mutagenesis.
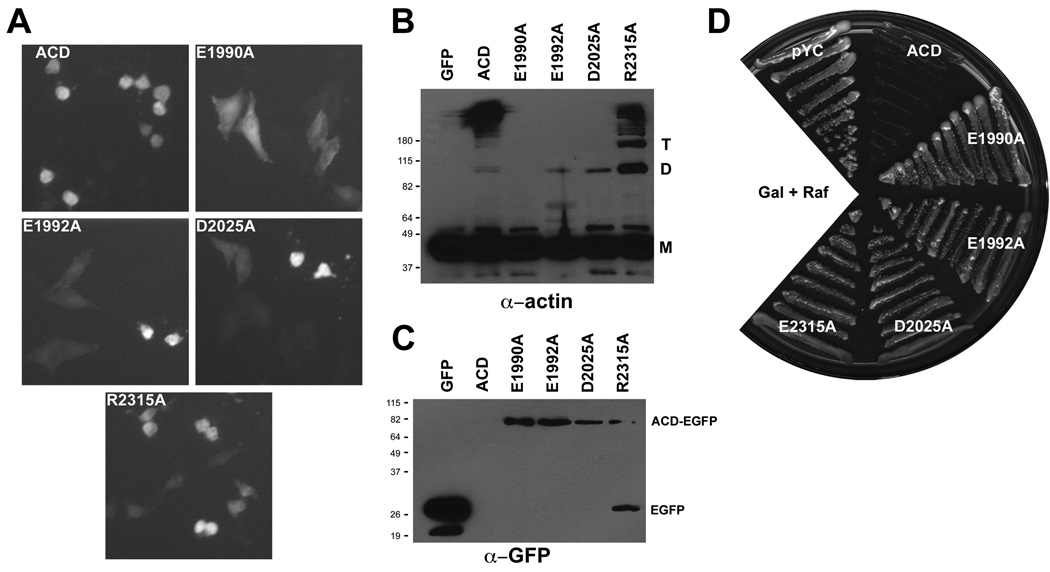
Each of these selected residues was changed to Ala within pEGFP-ACD. The resulting plasmids were transfected with an optimized strategy using HeLa cells compared to the COS-7 cells used for the LSM. Following transfection of pEGFP-ACD plasmids, each mutant was tested for expression by anti-GFP immunoblot, cell rounding by microscopy, and actin cross-linking by immunoblot. 34 of the 38 transiently-expressed mutant proteins caused cell rounding and cross-linked actin indistinguishably from wild-type ACDVc (Fig. 1 inverted filled arrows, Table S3, and data not shown).
By contrast, the E1990A mutant neither rounded cells nor cross-linked actin (Fig. 4A and 4B) indicating that E1990 is absolutely essential for ACDVc activity. Transfection of pEGFP-ACD E1992A or D2025A resulted in a mixture of round and normal shaped GFP-positive HeLa cells (Fig. 4A), and immunoblots showed small amounts of cross-linked actin upon extended exposure of the blot (Fig. 4B). pEGFP-ACD R2315A transfected cells were mostly rounded (Fig. 4A); however, there was a decrease in the average length of cross-linked actin oligomers compared to cells expressing wild-type ACDVc (Fig. 4B). The decreased amount of cross-linking suggests that E1992A, D2025A, and R2315A mutants are partially defective. A defect due to these three mutations is consistent with isolation of these residues in the yeast screen, as well as identification of disruptive LSM insertions within three residues (Fig. 1).
Fig. 4.
Ala substitution mutations in ACDVc decrease cell rounding, actin cross-linking activity and abrogate ACDVc-mediated yeast growth arrest. (A–C) HeLa cells were transfected with the indicated pEGFP-ACD plasmid and, after 24 hrs, cells were photographed (A) or processed for immunoblotting with actin (B) or GFP (C) specific antisera. (D) Sc containing either pYC, or pYC-ACD with the indicated mutations were grown on SC-Ura gal+raf for ~48 hrs at 30°C.
Targeted Ala-Substitution Mutagenesis Identifies Amino Acids Essential for ACD-mediated Yeast Growth Inhibition
Since yeast proved to be a valid approach for assessment of mutations in ACD function, nine additional residues were substituted for Ala in pYC-ACD and tested for their ability to prevent Sc colony formation. Sc expressing all nine new mutants did not grow on gal plates indicating that ACDVc is functional and the mutation did not significantly affect activity (Table S3).
In addition, four defective and four fully active mutants first characterized in pEGFP-ACD were also transferred to pYC-ACD to confirm that their affect on actin cross-linking in HeLa cells correlated with an affect on yeast colony formation. As expected, Sc expressing the four fully active mutants from pEGFP-ACD were unable to form colonies when plated on gal agar (Table S3) and the four mutants with decreased cross-linking activity when expressed from pEGFP-ACD (E1990A, E1992A, D2025A, and R2315A) showed growth in Sc that was similar to empty vector alone on gal agar (Fig. 4D). Notably, non-Ala substitutions in E1992, D2025, and R2315 were all isolated in the yeast random approach (Fig. 4B), emphasizing the utility of the epPCR screen to identify essential residues. Interestingly, despite their reduced actin cross-linking in HeLa cells, yeast expressing various mutant ACDs showed no difference in viability compared to empty vector alone, suggesting that full activity is required to prevent yeast colony formation (Fig. S1). Altogether, out of 46 individual Ala-substitution mutants constructed using both site-directed methods, 91% were functional, including mutation of the proposed ATP contact residues in a putative Walker box and all of the selected Ser and His residues indicating that these common ligase residues are not essential. Only 1 residue, E1990, was identified that was absolutely essential while 3, E1992A, D2025A, and R2315A, were partially defective.
Sequence Alignment Reveals ACDs have Active Sites Related to the Glutamine Synthetase Family of Peptide Ligases
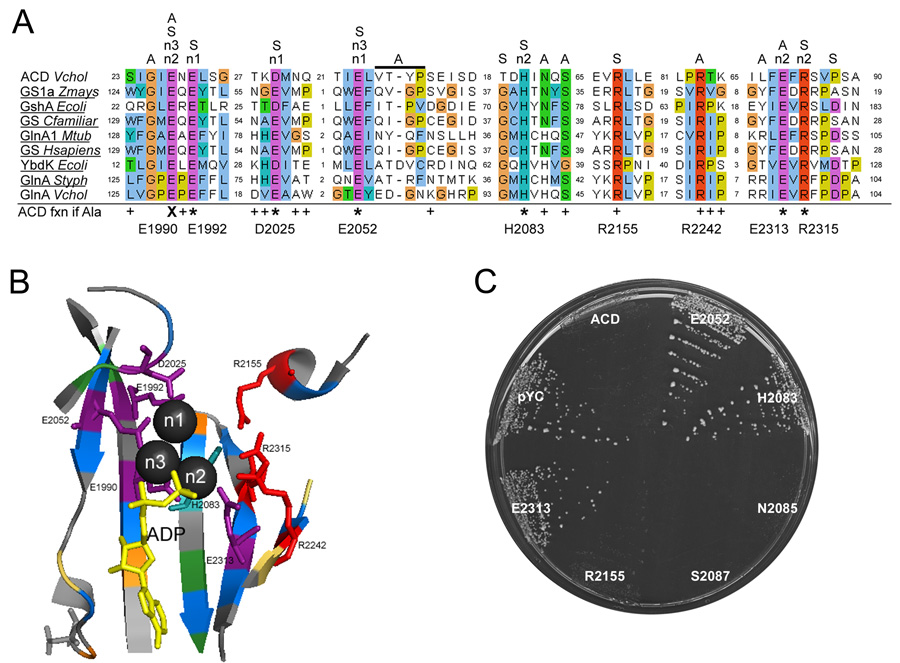
With knowledge of essential regions and important residues, we reinitiated a bioinformatics approach to identify distant functional homologs of ACDVc in order to further characterize its mechanism of action. HHpred (Soding et al., 2005) returned 10 potential short homology regions to various proteins. Of particular interest, homology to a GCS family member, YbdK, indicated that residues aligned to E1990, E1992, and D2025 were 100% conserved between the two proteins. Analogous to ACDVc, the GCS and related GS enzyme families both carry out ATP-dependent ligation reactions (Kudryashov et al., 2008a, Kudryashov et al., 2008b, Abbott et al., 2001). The GS enzymes generate glutamine from glutamate and ammonia whereas GCS synthesizes γ-glutamylcysteine from glutamate and cysteine. The active sites of both GS and GCS enzymes are nearly identical and are composed of a binding pocket for three Mg/Mn2+ ions, ATP, glutamate, and ammonia (GS) or cysteine (GCS) (Abbott et al., 2001, Eisenberg et al., 2000, Krajewski et al., 2008, Hibi et al., 2004, Lehmann et al., 2004).
The residues that form the active sites of GS and GCS enzymes are highly conserved amongst enzymes from both prokaryotes and eukaryotes (Abbott et al., 2001). Manual alignment of the ACDVc sequence to the GS and GCS proteins that have been crystallized showed overlap in the GS/GCS active site with all of the aa essential for full ACDVc activity (Fig. 5A). Specifically, a β-strand in the Mg/ATP binding site of all GS/GCS enzymes consists of an absolutely conserved G×E×E motif (Iyer et al., 2008). This motif corresponds to GIENEL (aa 1988–1992) of ACDVc (Fig. 5A and 5B). This motif includes residues E1990 and E1992 and is 100% conserved throughout all 5 ACD proteins (Fig. 1 and 4). E1990 corresponds to a GS/GCS residue that interacts directly with ATP and contacts two Mg/Mn2+ ions (n2 and n3). These metal ions coordinate ATP into the GCS/GS active site and are implicated in transfer of the γ-phosphate from ATP to the OH group of glutamate. The residue corresponding to E1992 on GS/GCS binds glutamate as well as coordinates the n1 metal that interacts with the bound glutamate (Abbott et al., 2001, Krajewski et al., 2008).
Fig. 5.
ACDVc shares structural similarity with GCS and GS enzymes. (A) Alignment of ACDVc with portions of the active site from various GCS and GS species. The crystal structure has been solved for those underlined. The activity each residue plays in GCS/GS activity is shown above: A, ATP-binding/hydrolysis; n#, Mg/Mn2+ ion contact; S, substrate binding. Residue color key: blue, hydrophobic; orange, glycine; purple, charged; yellow, proline; teal, histidine; green, polar; red, arginine. The residues colored in (A) correspond to the residues highlighted in (B). The affect of the Ala-substitution mutations on ACD activity is indicated below the alignment: +, indistinguishable from wild-type; *, decreased actin cross-linking and allowed yeast colony formation; X, no actin cross-linking and allowed yeast colony formation. (B) A portion of the GCS active site from E. coli GshA (PDB 1VA6) showing ADP (yellow stick) and Mg (spheres). The homologous residues required for full ACD activity are represented as sticks; Glu, Asp (purple) and Arg (red). (C) Colony growth of Sc containing pYC-ACD expressing the indicated structural alignment based Ala substitution on SC–Ura gal+raf for ~48 hrs at 30°C.
The other two residues that decrease activity when mutated to Ala correspond to GS/GCS residues involved in Mg/Mn2+ binding as well; D2025 binds n1 and R2315 binds n2 (Fig. 5A and 5B). Fitting the other functional regions of ACDVc identified in this study shows overlap with other regions of the GS/GCS active site as well, indicating a similar reaction mechanism despite the high degree of sequence divergence away from the active site (Fig. 5A and data not shown).
Based on the alignment of ACD to the GS/GCS active site, seven more ACDVc residues were chosen for mutation. Each residue (E2052, H2083, N2085, S2087, R2155, R2242, and E2313) was replaced by Ala in pYC-ACD and/or pEGFP-ACD then tested for its ability to prevent yeast colony formation when grown on gal or cross-link actin upon transfection. Residues shown to be involved in GS/GCS binding to ATP and Mg2+ (E2313), or glutamate and Mg2+ (E2052, H2083) were essential for ACDVc activity and allowed yeast colony formation on gal (Fig. 5C and Table S4). However, mutation in conserved residues exclusively involved in the GS/GCS enzymes substrate binding (R2155) or ATP alone (N2085, S2087, R2242) did not relieve the growth inhibition of ACDVc expression and did not permit growth on gal agar or affect actin cross-linking in HeLa cells (Fig. 5C, Table S4, and data not shown). These results indicate that the residues responsible for ACDVc to bind the substrate (actin) are likely different than those required for GS/GCS, but that the enzymes share a similar Mg/ATP binding site and catalytic mechanism for iso-peptide bond formation.
Discussion
ACDs are enzyme ligases that introduce iso-peptide bonds between side chains glutamic acid 270 and lysine 50 of actin. In this study, multiple mutation strategies, either random or targeted, were used to modify over 25% of the ACDVc protein sequence thereby revealing regions of the protein that are important for actin cross-linking in vivo, as well as identifying specific residues essential for catalysis.
The LSM technique generated insertions throughout the ACDVc coding sequence that disrupted the actin cross-linking ability of the ACDVc. Based on homology with the GS/GCS enzymes, six of these insertions are found within five residues from the predicted ACDVc active site, suggesting that these disrupt functional regions of the ACDVc. Other insertions that are outside of the proposed active site may disrupt actin binding or simply effect overall protein structure. As validation for initially utilizing this technique, the LSM approach correctly predicted regions I and III that correlate to two anti-parrallel β-strands that likely form part of the active site based on alignment to GS/GCS enzymes. A single disruptive insertion in region VI’ also correctly highlighted the importance of another β-strand within the proposed ACDVc active site.
Together, the site-directed studies conducted in either cell culture or Sc identified seven residues as critical for full ACDVc function in addition to 46 non-essential residues. Importantly, the residues that abrogated toxicity in yeast were also impaired in cross-linking within tissue culture cells when transfected as GFP-fusions. The E1990A mutation completely blocked the HeLa cell rounding and actin cross-linking ability of ACDVc, whereas the E1992A, D2025A, E2052A, and R2315A constructs showed decreased amounts of cross-linked actin by immunoblotting. These data suggest that although these four partial mutants are able to cross-link actin, they do so much less efficiently than wild-type ACDVc, thereby leaving enough actin monomer for yeast to survive and divide and to maintain proper HeLa cell shape.
Our alignment to other ATP-dependent peptide ligases revealed the potential mechanism of Mg/ATP binding by ACDVc. By using information from mutagenesis to fit the ACDVc sequence to the GS/GCS active site (Abbott et al., 2001), we were able to predict the role of identified functional residues. All seven essential residues in ACDVc correspond to residues within the GS/GCS active site that are directly involved in GS/GCS Mg/ATP binding and/or transfer of the γ-phosphate from ATP to glutamate (Abbott et al., 2001). We hypothesize that ACDVc Mg/ATP binding or PO4 transfer is significantly affected by single residue changes, similar to the GCS active site (Abbott et al., 2001), and these residues were therefore readily identified using our approaches.
Similarly, according to their roles in the GS/GCS active site, we can generate a hypothesis for the basis of partial versus complete inactivation of the ACDVc by the Ala-substitution mutants. Despite their roles in binding glutamate, ATP, or Mg/Mn2+, among the defective Ala-substitution mutants, only E1990 is predicted to contact all three ligands; glutamate, two metal ions, and ATP. This indicates that E1990 coordinates ATP and substrate binding in order to facilitate PO4 transfer, thereby accounting for the complete inactivity of the enzyme. Correspondingly, mutation of the equivalent residue in T. brucei GCS (E53) results in a completely inactive enzyme (Abbott et al., 2001). E2052 is predicted to contact two metal ions (n1 and n3) similar to E1990, however no direct contact with ATP is suggested. Conversely, E2313 likely binds ATP and n2, but not n3. E1992, D2025, H2083, and R2315 are not predicted to contact ATP directly, but a decreased metal binding may reduce the affinity for ATP or actin compared to wild-type ACDVc.
While our data imply that Mg/ATP coordination is functionally conserved between ACDVc and the GS/GCS proteins, substrate binding appears to be divergent. The manual alignment of ACDVc to the GS/GCS proteins showed conservation of 8 of the ~15 residues believed to be involved in GS/GCS substrate binding, compared to complete overlap between the ACDVc and GS/GCS residues required for Mg/ATP binding and PO4 transfer. Indeed, mutation of ACDVc R2155, which corresponds to a residue of GS/GCS exclusively involved in binding of glutamate, had no discernable effect on ACDVc activity, nor did inclusion of glutamate or ammonia in an in vitro ACDVc actin cross-linking assay (data not shown). The substrate for ACDVc is two monomers of actin, a 42 kD protein, compared to the relatively small GS and GCS substrates, glutamic acid and ammonia or cysteine. Because of this substrate size difference, the ACDVc-actin association likely requires a more complex binding interaction than the GS-glutamate/ammonia or GCS-glutamate/cysteine surfaces.
The GS/GCS enzymes consist of large order multimers (≥10 subunits) with active sites formed between neighboring subunits. The GS/GCS active site resembles a bifunnel, with ATP and glutamate entering the active site on opposite sides of the funnel (Eisenberg et al., 2000). The conservation between the active site residues of ACDVc suggests that the ACDVc active site may be similar to that of the GC/GCS, but there is no evidence at this time of ACDVc multimer formation. Formation of an ACDVc multimer might help coordinate the two actin molecules during iso-peptide bond formation or potentially form an actin binding pocket. In addition, a large space would be necessary to accommodate monomer and the large actin multimers that are formed as cross-linking progresses over time (Cordero et al., 2006). Structural changes caused by multiple residue insertions or disruptive mutations could block coordination of multiple actin molecules, thereby preventing cross-linking altogether. Thus, the inactive LSM insertions and epPCR mutants may have affected a number of newly considered putative ACDVc activities including multimer formation, actin binding and protomer positioning.
In all, using a combination of genetic approaches, we have defined both broad regions and specific residues of the ACDVc required for actin cross-linking revealing functional regions of the protein that previously had no known homologues. Based on these essential aa, we were able to use minimal sequence conservation to reveal that the ACDVc is functionally homologous to ligases involved in aa metabolism, further highlighting the novelty of this unique mechanism of toxin action. Notably, the identified critical residues are 100% conserved in 4 other ACDs identified within protein toxins of A. hydrophila and other Vibrios, suggesting these ACDs may have similar catalytic function and may be important to evade innate immunity by similar mechanisms.
Experimental procedures
Cell lines, Reagents and DNA manipulation
HeLa and COS-7 fibroblast cells (ATCC) were grown in DMEM (Invitrogen) containing 10% FBS (Invitrogen) and 50 units/ml penicillin and 50 units/ml streptomycin at 37°C with 5% CO2. All restriction enzymes and DNA ligases were purchased from New England Biolabs (NEB) or Invitrogen. All chemicals were purchased from Sigma unless noted otherwise. Plasmid DNA was isolated using Econospin spin columns (Epoch biolabs) and sequencing was performed in the Northwestern University Genomics core facility.
Cloning for linker scanning mutagenesis
The acd within the MARTX rtxA gene was amplified from a colony of V. cholerae N16961 using primers ACDentr UP and ACDentr DOWN representing codons for MARTX residues 1964–2374 according to the annotation of Lin et al (Lin et al., 1999). The forward primer introduces an ATG for methionine followed a novel BglII restriction site. The amplified product was cloned into pENTR/D-TOPO (Invitrogen) by manufacturer’s protocols creating plasmid pENTR-ACD. To test if the cloned acd was functional in mammalian cells as described below, it was recombined into the Gateway® EGFP fusion destination vector pDEST47 (Invitrogen) using the LR recombination reaction according to manufacturer’s protocols creating plasmid pDEST-ACD.
Linker Scanning Mutagenesis
Introduction of 5 codon insertions in the acd sequence was done using the NEB GPS-LS Linker-Scanning System. Due to its small size (3.8 kb), pENTR-ACD was used as the target for mutagenesis to reduce the frequency of transposon insertions in the vector. pENTR-ACD was subjected to in vitro transposition using the chloramphenicol-resistant Tn7 derived transposon on plasmid pGPS4 followed by electroporation to E. coli DH5α and plating on L agar with 34 µg/ml chloramphenicol and 50 µg/ml kanamycin. Plasmid DNA was purified from individual colonies and insertions within the acd gene were identified by restriction digestion with BsrG1 and oriented by digestion with BglII. All total, 738 plasmids were screened and 110 plasmids were identified with transposon insertions in the acd gene. The insertion site was identified by sequencing with NEB primer N or primer S that anneal to the transposon sequence. 28 of the 110 insertions created an out of frame mutation, 11 were duplicates, and 1 had a plasmid rearrangement. The remaining 70 plasmids were digested with PmeI, re-circularized using T4 ligase, and electroporated to E. coli. For analysis of function, the modified acd genes were transferred to pDEST47 using the Gateway® LR recombination reaction. Two methods were used to transfer 40 of the LSM mutations into pEGFP-ACD, a eukaryotic expression vector that expresses ACD from aa 1963–2419 fused to EGFP (Sheahan et al., 2004). pENTR-ACD LSM plasmids were digested with BstXI and BstEII and ligated to similarly digested pEGFP-ACD. Insertions occurring before the BstXI-BstEII fragment were amplified by PCR from pDEST-ACD templates with primers VgrG1963 and CΔ45 (Sheahan et al., 2004), products were digested with BglII and HindIII and ligated to similarly digested pEGFP-ACD. Plasmids were transfected then assayed for actin cross-linking by immunoblot (see below).
Construction of pYC-ACD random mutants by recombination
The DNA encoding ACDVc (aa 1963–2419) was amplified from V. cholerae N16961 ΔhlyA ΔhapA (KFV119) (Sheahan et al., 2004) by PCR using Pfx50 polymerase (Invitrogen) with pYC-ACD UP and pYC-ACD DOWN (Table S1). This fragment was then digested with KpnI and XbaI and ligated into similarly digested pYC to create pYC-ACD.
The effect of ACDVc on yeast was determined by transforming either pYC or pYC-ACD into Sc strain INVSc1 using the PLATE method (Gietz et al., 1992) and plated onto SC-Ura agar containing glu (SC-Ura+glu)(MBL). Fresh transformants were grown overnight in SC-Ura+glu broth at 30°C. 5 µl of 10-fold serial dilutions of each culture was spotted onto either SC-Ura+glu or SC-Ura containing gal and raffinose plates (SC-Ura+gal) and incubated for 2 days at 30°C. Raf was added as a carbon source to allow comparable growth rates as glu.
For yeast gap repair cloning (Ma, 1987), pYC-ACD was used as template to amplify acd as described above. Amplification was performed with 4U goTaq polymerase (Fisher Scientific) and buffer, 200 µM dNTPs, and 1 µM of each primer for 40 cycles. pYC-ACD was prepared for transformation by digestion with BstEII and SphI. This digestion removed the DNA encoding for aa 2017 through 2403. ~1 µg of purified PCR product and ~0.1 µg BstEII-SphI digested plasmid were transformed into INVSc1 as above. Following transformation, cells were plated onto SC-Ura+gal to induce expression from pYC-ACD and incubated at 30°C. Colonies arising after ~3 days were restreaked and maintained on SC-Ura+gal plates. Plasmid DNA was then isolated from each yeast colony, transformed into E.coli, re-isolated, and sequenced.
Construction of pYC-ACD specific mutants by recombination
Ala-substitution mutants were constructed in Sc using homologous recombination between pYC-ACD and PCR products containing the substitutions (Ma, 1987). Ala-substitutions were incorporated into the 3’-end of primers with at least 30 nucleotides of homology 5’ to the targeted residue (Table S1). Pfx50 polymerase was used to amplify the ACDVc either upstream or downstream of the residue using N- or C-terminal ACDVc specific primers. pYC-ACD cleaved with BamHI was transformed along with PCR product using the PLATE method into INVSc1. Transformations were plated onto SC-Ura+glu at 30°C and allowed to grow for ~3 days. Plasmid DNA was then isolated from yeast, transformed into E.coli, re-isolated, and sequenced.
Construction of Ala substitutions in pEGFP-ACD
Ala-substitution mutants were constructed with specific primers (Table S1) using the Stratagene Quikchange XL II site-directed mutagenesis kit or by Bionexus, Inc. (Oakland, CA) with pEGFP-ACD (Sheahan et al., 2004) as the template for the reactions. All mutations were confirmed by DNA sequencing.
In vivo Actin Cross-linking and Microscopy
HeLa or COS-7 cells were grown in 6-well dishes for ~24 hrs then transiently transfected using Lipofectamine Plus (Invitrogen) or FuGene HD (Roche) following the manufacturer’s protocols. Transfections were allowed to proceed for 18–24 hours before the cells were washed, collected, and boiled in 2× SDS-PAGE loading buffer. Cells were imaged in the dish using an inverted Leica DMIRE2 microscope with a CCD camera and processed using OPENLAB (Improvision) and Adobe Photoshop.
Western blotting
Cell lysates from transiently transfected tissue culture cells were separated by SDS-PAGE and transferred to Hybond-C Extra nitrocellulose membranes (Amersham Pharmacia). Membranes were subjected to western blotting with anti-actin antibody (Sigma) as previously described (Sheahan et al., 2004) or with anti-GFP-HRP (Miltenyi Biotec) for confirmation of ACD-GFP expression. Membranes were developed with Supersignal West Pico chemiluminescent substrate (Pierce).
Protein alignment and Structure prediction
Amino acids 1963–2419 of V. cholerae rtxA (GI:4455065) was used as query for BLAST analysis against the 988 partial and complete genome sequences currently available through NCBI (as of 02/09). Alignments were performed using CLUSTAL W and highlighted using TEXTSHADE through the Biology Workbench 3.2 server (http://workbench.sdsc.edu). HHpred (Soding et al., 2005) was used to identify structural homologues of ACDVc using aa1963–2419 as input. All of the sequences from the GS and GCS enzymes deposited in PDB were aligned in Kalign (Lassmann & Sonnhammer, 2005) then ACDVc was manually aligned based on residues involved in activity. These aligned regions were then inserted back into Kalign to determine sequence relatedness. A cartoon depicting a portion of the active site from E. coli GCS (GshA, PDB ID: 1VA6) was prepared with MacPyMol (Delano Scientific).
Supplementary Material
Supplemental Information includes Fig. S1 and Tables S1–4 and can be found with this article online.
Acknowledgements
We thank C. Cordero and L. Rogers for early contributions to this project, M. Feda and I. Antic for construction of plasmids used herein, and R. Tungekar for technical assistance. Special thanks to K. Prochazkova and G. Minosav for helpful discussions. This work was supported by National Institutes of Health Grant AI051490 (to K.J.F.S.) and an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (to K.J.F.S.). B.G. was supported by an Institutional NRSA Post-doctoral Research Fellowship in Immunology and Molecular Pathogenesis, T32-AI007476-11.
References
- Abbott JJ, Pei J, Ford JL, Qi Y, Grishin VN, Pitcher LA, Phillips MA, Grishin NV. Structure prediction and active site analysis of the metal binding determinants in gamma-glutamylcysteine synthetase. J Biol Chem. 2001;45:42099–42107. doi: 10.1074/jbc.M104672200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero CL, Kudryashov DS, Reisler E, Satchell KJ. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J Biol Chem. 2006;281:32366–32374. doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero CL, Sozhamannan S, Satchell KJ. RTX toxin actin cross-linking activity in clinical and environmental isolates of Vibrio cholerae. J Clin Microbiol. 2007;45:2289–2292. doi: 10.1128/JCM.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. Structure-Function relationships of glutamine synthetases. Biochim Biophys Acta. 2000;1477:122–145. doi: 10.1016/s0167-4838(99)00270-8. [DOI] [PubMed] [Google Scholar]
- Fullner KJ, Mekalanos JJ. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 2000;19:5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi T, Nii H, Nakatsu T, Kimura A, Kato H, Hiratake J, Oda J. Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc Natl Acad Sci U S A. 2004;101:15052–15057. doi: 10.1073/pnas.0403277101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua SB, Qiu M, Chan E, Zhu L, Luo Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3 doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and opportunities for drug and herbicide design. J Mol Biol. 2008;375:217–228. doi: 10.1016/j.jmb.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Kudryashov DS, Cordero CL, Reisler E, Satchell KJ. Characterization of the enzymatic activity of the actin cross-linking domain from the Vibrio cholerae MARTXVc toxin. J Biol Chem. 2008a;283:445–452. doi: 10.1074/jbc.M703910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashov DS, Durer ZA, Ytterberg AJ, Sawaya MR, Pashkov I, Prochazkova K, Yeates TO, Loo RR, Loo JA, Satchell KJ, Reisler E. Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin. Proc Natl Acad Sci U S A. 2008b;105:18537–18542. doi: 10.1073/pnas.0808082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, Amaro C, Wu KM, Valiente E, Chang YF, Tsai SF, Chang CH, Hor LI. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J Bacteriol. 2008;190:1638–1648. doi: 10.1128/JB.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann C, Doseeva V, Pullalarevu S, Krajewski W, Howard A, Herzberg O. YbdK Is a carboxylate-amine ligase with a gamma-glutamyl:cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–383. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]
- Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci U S A. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host & Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;55:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Olivier V, Salzman NH, Satchell KJ. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect Immun. 2007;75:5043–5051. doi: 10.1128/IAI.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MH, Biswas K, Hossain MA, Sack RB, Mekalanos JJ, Faruque SM. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 2008;27:347–355. doi: 10.1089/dna.2008.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell KJ. MARTX: Multifunctional-Autoprocessing RTX Toxins. Infect Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell KJF, Geissler B. The Multifunctional-Autoprocessing RTX toxins of Vibrios. In: Proft T, editor. Microbial Toxins: Current Research and Future Trends. Auckland: Caister Academic Press; 2009. [Google Scholar]
- Sheahan KL, Cordero CL, Satchell KJ. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc Natl Acad Sci U S A. 2004;101:9798–9803. doi: 10.1073/pnas.0401104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information includes Fig. S1 and Tables S1–4 and can be found with this article online.