Abstract
Drosophila melanogaster is a widely used model organism for the molecular dissection of mitosis in animals. However, despite the popularity of this system, no studies have been published on the ultrastructure of Drosophila kinetochores and kinetochore fibers (K-fibers) in somatic cells. To amend this situation, we used correlative light (LM) and electron microscopy (EM) to study kinetochores in cultured Drosophila S2 cells during metaphase, and after colchicine treatment to depolymerize all microtubules (MTs). We find that the structure of attached kinetochores in S2 cells is indistinct, consisting of an amorphous inner zone associated with a more electron-dense peripheral surface layer that is approximately 40–50 nm thick. On average, each S2 kinetochore binds 11±2 MTs, in contrast to the 4–6 MTs per kinetochore reported for Drosophila spermatocytes. Importantly, nearly all of the kinetochore MT plus ends terminate in the peripheral surface layer, which we argue is analogous to the outer plate in vertebrate kinetochores. Our structural observations provide important data for assessing the results of RNAi studies of mitosis, as well as for the development of mathematical modelling and computer simulation studies in Drosophila and related organisms.
Introduction
For more than a century, researchers have viewed kinetochores as specialized regions on centromeres responsible for the movement of chromosomes during mitosis (e.g., Metzner 1894). The kinetochore’s critical role in chromo-some segregation has led extensive investigations of kinetochore function in a wide range of organisms and model systems (Rieder and Salmon 1998; Cleveland et al. 2003; Maiato et al. 2004a; Chan et al. 2005). Classical EM studies led to the view that the kinetochore in vertebrates resembles a trilaminar disk (Brinkley and Stubblefield 1966; Jokelainen 1967; Roos 1973, 1977; Rieder 1982). In this model, an electron-dense inner plate is located on the surface of the centromeric heterochromatin and it is separated from an electron-dense outer plate by a lighter middle layer. This inner plate contains nucleosomes, a specialized histone, CENP-A, and other proteins, including CENP-C (Cleveland et al. 2003; Chan et al. 2005). Thin-section EM studies of high-pressure frozen specimens suggest that the electron-lucent middle layer is largely a fixation/dehydration artefact and that the outer plate is an extended fibrous mat (McEwen et al. 1998).
The plus ends of kinetochore microtubules (kMTs) terminate in the outer plate (Rieder 1990; Vandenbeldt et al. 2006). This region contains the four-subunit Ndc80 complex, which has been shown to play a critical role in attaching MTs to kinetochores in a wide range of organisms, including Saccharomyces cerevisiae, Schizosac-charomyces pombe, Caenorhabditis elegans, Xenopus, Gallis gallus, and humans (reviewed by Maiato et al. 2004a; DeLuca et al. 2005; Kline-Smith et al. 2005). On kinetochores that lack MTs, the distal surface of the outer plate appears as a ribosome-free exclusion zone in high-pressure frozen/freeze-substituted specimens, and as a compact fibrous corona in conventionally fixed specimens (McEwen et al. 1998). The corona material contains a dynamic network of mainly transient proteins involved in MT capture, spindle checkpoint response, and the regulation of MT dynamics. They include motor proteins such as cytoplasmic dynein and the plus-end directed kinesin CENP-E, as well asMT stabilizing proteins such as CLASPs (Maiato et al. 2004a). Upon binding of MTs, many corona proteins partially or fully dissociate from the outer plate, and the corona structure becomes correspondingly more difficult to detect by EM (Rieder 1982, 1990; Cassimeris et al. 1990; Hoffman et al. 2001; Howell et al. 2001).
Due to its powerful genetics, the Drosophila melanogaster system has been widely used as a model for studying mitosis in animals. Many important and widely conserved genes, including the Polo and Aurora kinases and the ZW10/Rod complex, were originally discovered through mutagenesis screenings of this organism (reviewed by Blagden and Glover 2003; Karess 2005). The combination of a fully sequenced genome and efficient RNA interference (RNAi) in cultures of Drosophila S2 cells has proven particularly useful both for high-throughput genome-wide screens and for the molecular dissection of mitosis and cytokinesis (e.g., Goshima and Vale 2003; Bettencourt-Dias et al. 2004; Somma et al. 2002; Echard et al. 2004). Importantly, these tools can be used to study several mammalian kinetochore components that have conserved counterparts in Drosophila, including CENP-A (CID in Drosophila), CENP-C, CENP-E (CENP-meta), dynein, ZW10/Rod and CLASPs (MAST/Orbit) (Henikoff et al. 2000; Lemos et al. 2000; Inoue et al. 2000; Yucel et al. 2000; Heeger et al. 2005; Karess 2005).
Despite the popularity of the Drosophila system for work on mitosis, there are no EM studies on the kinetochore structure in Drosophila somatic cell lines such as S2. Therefore, we have conducted a thorough EM analysis of the S2 kinetochore, focusing on three fundamental aspects: 1) the structure of the kinetochore and how it changes with the acquisition of MTs; 2) the number of MTs that become attached to each kinetochore; and 3) the interface between the K-fiber and the kinetochore.
Materials and methods
Cells and immunofluorescence
For immunofluorescence, S2R+ cells or S2 cells stably expressing CID-GFP were grown overnight at 25°C on uncoated coverslips in Schneider’s medium (Invitrogen) supplemented with 10% FBS, or on concanavalin A (Calbiochem)-treated coverslips, and were immediately fixed for 10 min in 4% paraformaldehyde in cytoskeleton buffer (137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 0.4 mM KH2PO4, 2 mM MgCl2, 2 mM EGTA, 5 mM PIPES, 5.5 mM glucose, pH 6.9). Cells were then extracted in cytoskeleton buffer, supplemented with 0.3% Triton X-100 for 10 min at room temperature, and rinsed in PBS with 0.1% Triton X-100 (PBS-T). Cultures were subsequently blocked in PBS-T with 10% FBS for 30 min. Primary and secondary antibody incubations were carried out in blocking solution for 1 h at room temperature, with three 5-min washes in PBS-T after each incubation. Rabbit antibodies against CENP-meta and Cnn (kind gifts of M. Goldberg, Cornell University, Ithaca, NY, USA and T. Kaufman, Indiana University, Bloomington, IN, USA) were used diluted 1:500. For α-tubulin staining, a mouse monoclonal antibody B512 (Sigma, St. Louis, MO, USA) was used at 1:2,000. For CENP-C and CID immunostaining, rabbit and chicken antibodies (kind gifts from C. Lehner, University of Bayreuth, Bayreuth, Germany and Gary Karpen, University of California at Berkeley, CA, USA) were used at 1:5,000 and 1:1,000, respectively. All secondary antibodies were used at 1:2,000 dilution (Invitrogen—Molecular Probes). DNA was counterstained with DAPI (4 µg/ml), which was added to the secondary antibody incubation, and coverslips were mounted on slides with 90% glycerol in Tris–HCl pH 8.5 supplemented with 0.5% n-propylgalate as an anti-fading agent.
Cells destined for chromosome spreads were grown in six-well plates, treated with 30 µM colchicine (Sigma) for 16 h, counted, centrifuged, and resuspended at 105 cells/ml in Ephrussi–Beadle Ringers solution (EBR; 129 mM NaCl, 4.7 mM KCl, 1.9 mM CaCl2, 10 mM HEPES, pH 6.9) for non-hypotonic treatment (Vass et al. 2003). A total of 500 µl of the cell suspension was spun onto slides at 1,000 rpm for 5 min in a Cytospin 2 (Shandon), and was immediately fixed and processed as above. Quantitative three-dimensional data sets of representative cells were collected using a DeltaVision microscope (Applied Precision, Issaquah, WA, USA), based on an Olympus IX-70 inverted microscope equipped with a cooled Photometrics CH350L CCD camera, and subsequently deconvoluted and projected onto a single plane using SoftWorx (Applied Precision). Alternatively, a Zeiss Axiovert 200M driven by Axiovision software and equipped with a Zeiss Axiocam MRm was used to collect three-dimensional data sets of representative cells, which were subsequently blind deconvoluted with Autoquant X (Autoquant, Troy, NY, USA). Adobe Photoshop 6.0 (Adobe Systems) was used to process all images.
Correlative light and electron microscopy
S2R+ cells were grown at 25°C as described above. Coverslip cultures were subsequently mounted in Rose chambers (Khodjakov and Rieder 2006) filled with Schneider’s medium and 10% FBS. For time-lapse recordings, cells were visualized by DIC as they entered mitosis, and images were collected every 30 s on the DeltaVision microscope described previously. After nuclear envelope breakdown (NEB), when most chromosomes were positioned at the metaphase plate, the area of the coverslip containing the cell of interest was marked with a diamond scribe and the culture immediately fixed with 2.5% gluteraldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) for 30 min. For MT depolymerization, S2R+ cells were incubated for 16 h with 30 µM colchicine and fixed as described above. All cells were processed for correlative electron microscopy as previously described (Rieder and Cassels 1999).
Electron microscopy
Images of 100-nm-thick serial sections of Epon-embedded S2 and S2+ cells were recorded on film on a Zeiss 910 electron microscope at 5,000–10,000× magnification. Three-dimensional serial-section reconstructions were created by tracing chromosomes, kinetochores, and MTs on digitally scanned images using the Sterecon software (Marko and Leith 1996). The number of MTs bound to each kinetochore was determined using duplicate counting trials on film negatives from a complete serial-section series through each kinetochore (McEwen et al. 1997). Counting was facilitated by digital scanning of the negatives for enlargement and contrast adjustment. A total of 31 kinetochores from three different cells were analyzed. Microsoft Excel (Microsoft, Redmond, WA, USA) was used to compute statistical parameters and to plot bar graphs.
Dual-axis EM tomography was performed as described previously (McEwen and Marko 1999). Tilt series images were recorded on a Zeiss 910 electron microscope at 100 kV using a Gatan (TVIPS) 1K × 1K CCD camera with a pixel size of 1.6 nm. The tilt angle increment was varied according to the cosine of the tilt angle, over an angular range of ±60°, and with an angular increment at the untilted image of 2.0°. Ten-nanometer colloidal gold particles, attached to a single surface of the section, served as fiducial markers, and both SPIDER and IMOD were used for image alignment and tomographic computations (Frank et al. 1996; Kremer et al. 1996).
Results
The structure of the S2 kinetochore as revealed by immunofluorescence
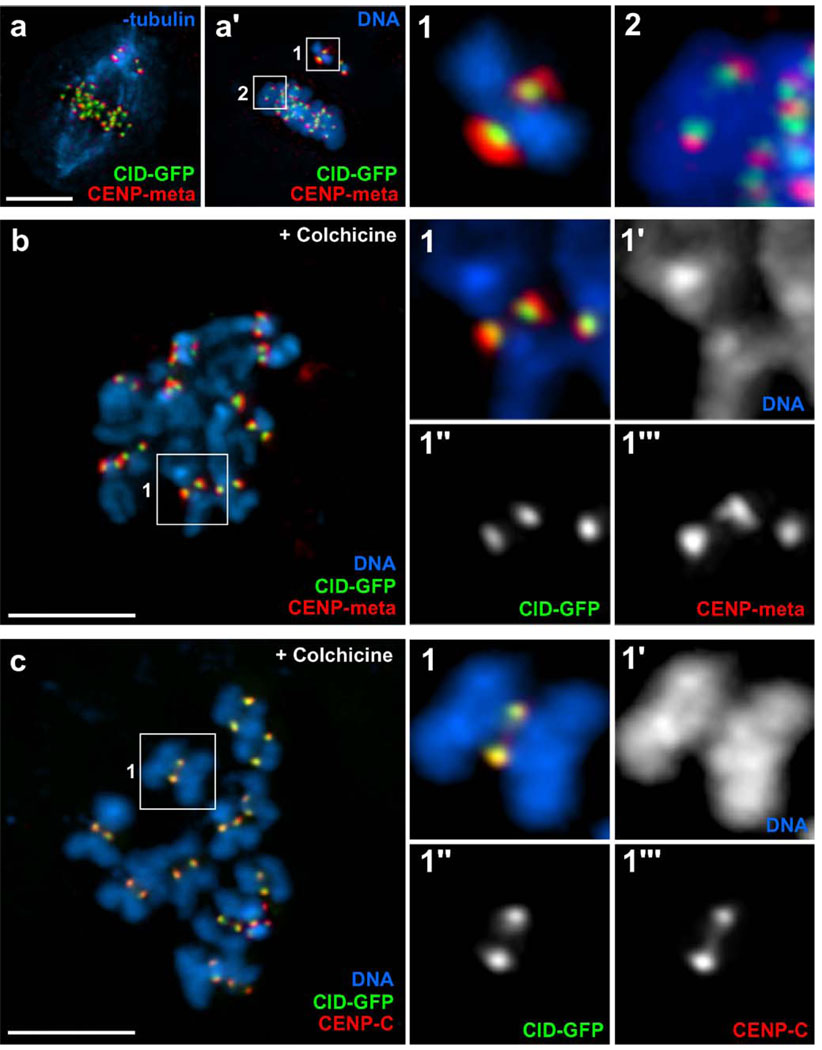
We used indirect immunofluorescence methods to define the general location of several structural proteins in Drosophila S2 kinetochores, including CENP-A (CID), CENP-C, and CENP-E (Cenp-meta). For this purpose, we used cells stably expressing CID-GFP (Heun et al. 2006), an inner kinetochore protein, and we compared the distribution of kinetochore proteins among unattached, mono-oriented, and bi-oriented kinetochores (Fig. 1). These studies revealed that while the CID signal remained indistinguishable between mono-oriented and bi-oriented chromosomes, the Cenp-meta signal showed a significant change in distribution between these two attachment states (Fig. 1a′1 and a′2). Accordingly, Cenp-meta staining on mono-oriented chromosomes revealed an expanded crescent area, especially evident in the unattached kinetochore from the mono-oriented pair, while in bi-oriented chromosomes the signal was confined to a smaller, well-defined spot. Next, we compared the inter-kinetochore distance between attached (bi-oriented) and unattached kinetochores by measuring the distances between CID-GFP signals and Cenp-meta signals in metaphase cells and on chromosome spreads obtained from colchicine-treated cells (Fig. 1b). On unattached chromosomes, the CID-GFP and Cenp-meta signals between sister kinetochores were separated by 0.63±0.1 µm (N=33) and 0.88±0.13 µm (N=30), respectively. On bi-oriented chromosomes positioned at the metaphase plate, the CID-GFP and Cenp-meta signals between sister kinetochores were separated by 0.84±0.08 µm (N=30) and 1.12±0.08 µm (N=30), respectively. The distance difference between unattached and attached kinetochores is statistically significant (p<0.05, Student’s t test).
Fig. 1.
Characterization of the structural domains of the Drosophila kinetochore at the light microscope level. S2 cells stably expressing CID-GFP (green) were immunostained for kinetochore-associated proteins and were observed by immunofluorescence microscopy in the presence and absence of MTs. a Prometaphase cell immunostained for CENP-meta (red) and α-tubulin (blue). a′ The same cell, counter-stained with DAPI (blue). A well-defined metaphase plate and two misaligned chromosomes can be observed near one pole. One of the misaligned chromosomes is shown at higher magnification (window 1), as well as a properly bi-oriented aligned chromosome (window 2). Two immediate conclusions are evident. The first is that while the CID signal does not change perceptibly between the two situations, the CENP-meta signal on the misaligned chromosome reveals a hemispheric, crescent-like structure that is significantly larger than in bi-oriented chromosomes. The second conclusion is that inter-kinetochore distance in a bi-oriented chromosome is higher than in misaligned chromosomes, which are likely to be mono-oriented. b Chromosome spread showing the localization pattern of CID-GFP (green) and CENP-meta (red) at kinetochores in the absence of MTs, which were induced to depolymerize by incubation with colchicine. Windows 1–1''' show a higher magnification of an individual chromosome from b; note that the two proteins only partially co-localize, with CENP-meta being more external and enlarged than CID (as in the case of misaligned chromosomes). c Co-immunostaining of CID-GFP (green) and CENP-C (red) revealed significant co-localization of the two proteins. Scale bar=5 µm
As an additional control for these assays, we compared the locations of CID-GFP and CENP-C, two inner kinetochore proteins (Fig. 1c). As expected, each protein localized as a pair of well-defined spots on the centromeric region, and any difference in their relative positions was beyond the resolution limit of light microscopy (Fig. 1c1). From these data we conclude that, as in vertebrates, the centromere regions and kinetochores on Drosophila S2 chromosomes become distorted as chromosomes attach to the spindle.
Correlative light and electron microscopy and the structure of the S2 cell spindle
Same-cell correlative light and electron microscopy is used to define the ultrastructural correlates that underlie dynamic cellular events (reviewed in Rieder and Cassels 1999). This method involves fixation of a cell during or immediately after an event of interest, for a subsequent 3D electron microscopic analysis. Many types of questions, especially those related to mitosis, require that the cell remains flat, so that the event can be clearly followed. Furthermore, cells that remain flat throughout mitosis while cultured as a monolayer can be easily sectioned parallel to the long spindle axis, thus greatly facilitating EM studies of the spindle. However, S2 cells are generally grown in suspension, or in a semi-adherent way, which promotes rounding. In the past, this has been overcome by one of two minimally invasive techniques. In one, the cells are cultured on concanavalin A-coated coverslips, which promotes cell adhesion and spreading (Rogers et al. 2002). In the other, an agar overlay is used to flatten cells via surface tension (Maiato et al. 2004b). The first method often inhibits the cytokinesis part of mitosis, while the second is complex and time-consuming.
To facilitate our EM studies, we searched the literature for S2 clones that remain flat during mitosis. We found that the original S2 cells isolated by Schneider (1972) were epithelial-like in appearance and grew as a loose monolayer. This contrasts sharply with the morphology of S2 lines commonly used today. In general, these latter lines do not respond to wingless signaling, because they lack the wingless receptor, Dfrizzled-2. However, a relatively recent paper describes another S2 clone that expresses both Dfrizzled-1 and Dfrizzled-2; it was named S2 receptor plus (S2R+; Yanagawa et al. 1998). Interestingly, the morphology of S2R+ cells closely resembles that originally described by Schneider for S2 cells, i.e., growth as an epithelial cell-like monolayer. As these cells remain relatively flat during mitosis, they have the physical characteristics required for high-resolution EM studies on the spindle. As in other S2 clones, S2R+ cells typically double in culture over 21–23 h, are highly aneuploid (nearly tetraploid), and take ~20 min from NEB to anaphase (data not shown). As seen in Fig. 2, they remain flat throughout mitosis and form typical astral spindles containing prominent K-fibers (see also Supplementary Figure 1S).
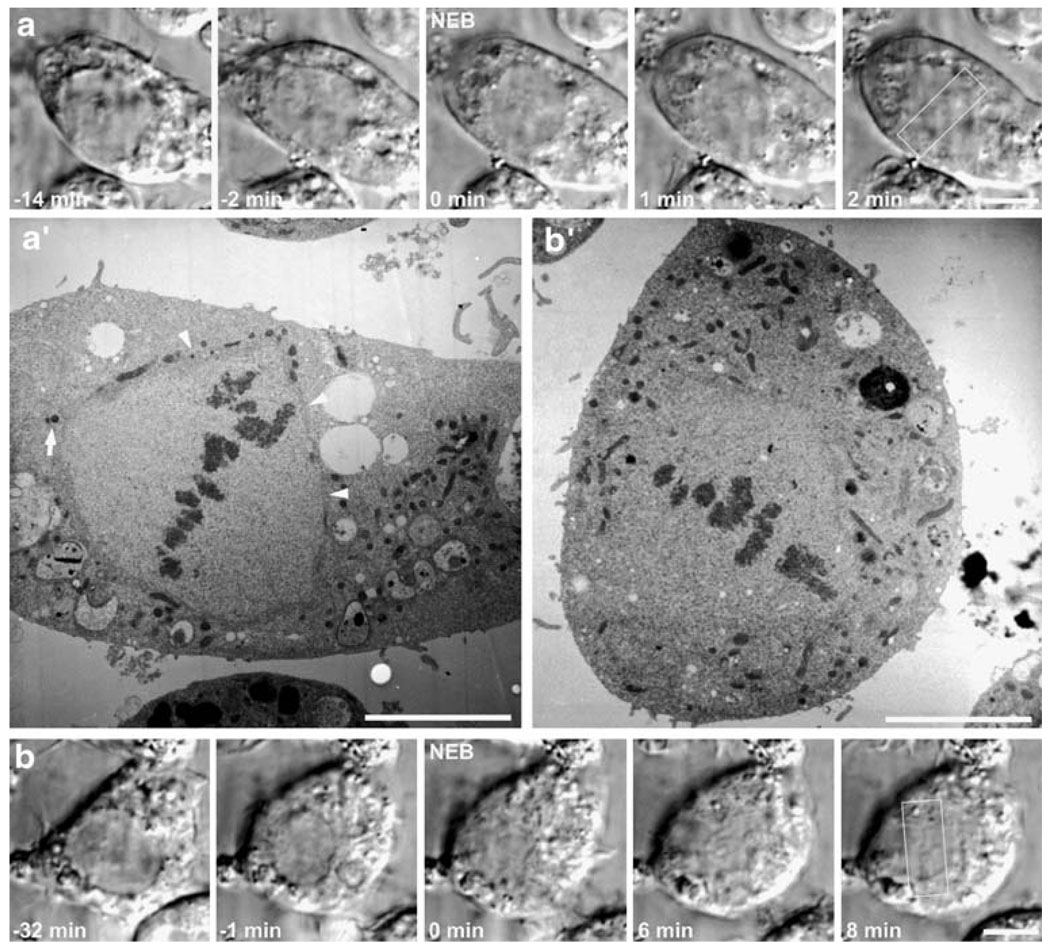
Fig. 2.
Correlative light and electron microscopy analyses of Drosophila S2R+ cells. a, b Selected frames from time-lapse sequences of two cells visualized by DIC microscopy as they entered mitosis. Note the presence of the respective nuclear envelopes and their disassembly (0 min). Cells were fixed when a metaphase plate became apparent (white rectangle). a′, b′. Corresponding cells from a and b visualized by transmission electron microscopy. Note that in a′ the cell was fixed early after NEB; consequently, it is possible to observe remnants of the nuclear envelope surrounding the spindle region (arrowheads). One of the centrosomes with two centrioles is also indicated (arrows). Scale bars=5 µm
We used high-resolution time-lapse video-LM to observe individual S2R+ cells during mitosis, and then fixed them at selected times for subsequent serial-section EM analyses. The example shown in Fig. 2a was followed for 2 min after NEB before fixation, while the one depicted in Fig. 2b was followed for 8 min after NEB. In each case, the cell of interest was fixed when a metaphase plate became evident, and relocated after plastic embedding using standard correlative LM/EM techniques (e.g., Rieder and Cassels 1999). Low-magnification EM views confirmed that the cells were in metaphase at the time of fixation (Fig. 2a′,b′). However, despite the alignment of all chromosomes on the cell equator, remnants of the nuclear envelope could still be observed in the cell fixed shortly after NEB (Fig. 2a′, arrowheads). By contrast, in the cell fixed 8 min after NEB, we found a well-formed metaphase plate with no vestiges of the nuclear envelope (Fig. 2b′). These data underscore the speed at which the spindle forms in S2 cells, and demonstrate that correlative LM/EM can be used on Drosophila cells in culture.
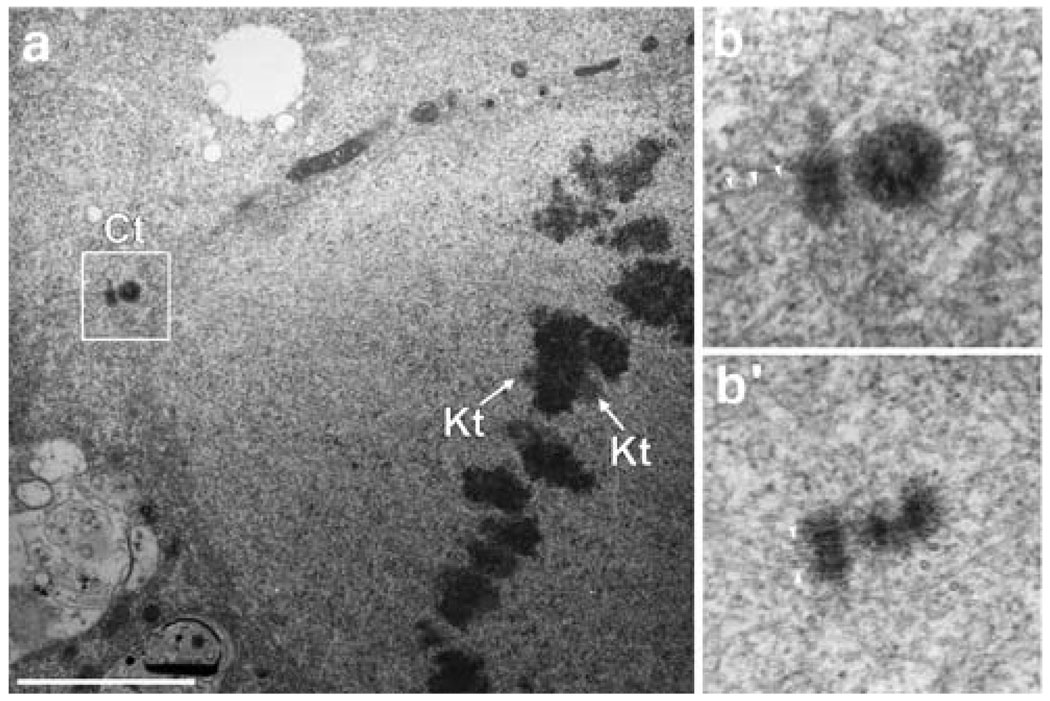
An overview of spindle ultrastructure in one of these cells is shown in Fig. 3a. One conspicuous feature of S2 cells is that the orthogonally oriented centrioles are extremely short (about 0.25 µm; Fig. 3b), in agreement with previous observations (Szollozi et al. 1986; Moritz et al. 1995). As in other cell types, the centrioles define the centrosome and are surrounded by MTs that can be seen in cross-sections and in longitudinal sections. In some views, several MTs are seen to extend directly from the MT triplet blades of the centrioles (arrows in Fig. 3b and b′), and not from the pericentriolar material. It has recently been suggested that in the absence of γ-tubulin, the elongation of centriolar MTs functions as an alternative mechanism for centrosomal MT assembly in Drosophila cells (Raynaud-Messina et al. 2004). Our ultrastructural data reveal that this mechanism also operates normally in cells containing γ-tubulin. In low-magnification overviews, sister kinetochores can be recognized as two protrusions that extend from opposite sides of the chromosomes (indicated in Fig. 3a).
Fig. 3.
Ultrastructural characterization of the mitotic apparatus of Drosophila S2R+ cells. a. Visualization of a half-spindle region from S2R+ cells showing one centrosome (Ct) composed of two centrioles, plus a kinetochore pair (Kt) with attached MTs. Several K-fibers can be identified. b Higher-magnification view of the centrosome visualized in a. b′. Similar view of the other centrosome. Note that both centrioles have MTs directly attached (arrowheads). Scale bar=2.5 µm
Structure of the metaphase S2 kinetochore and kinetochore fiber
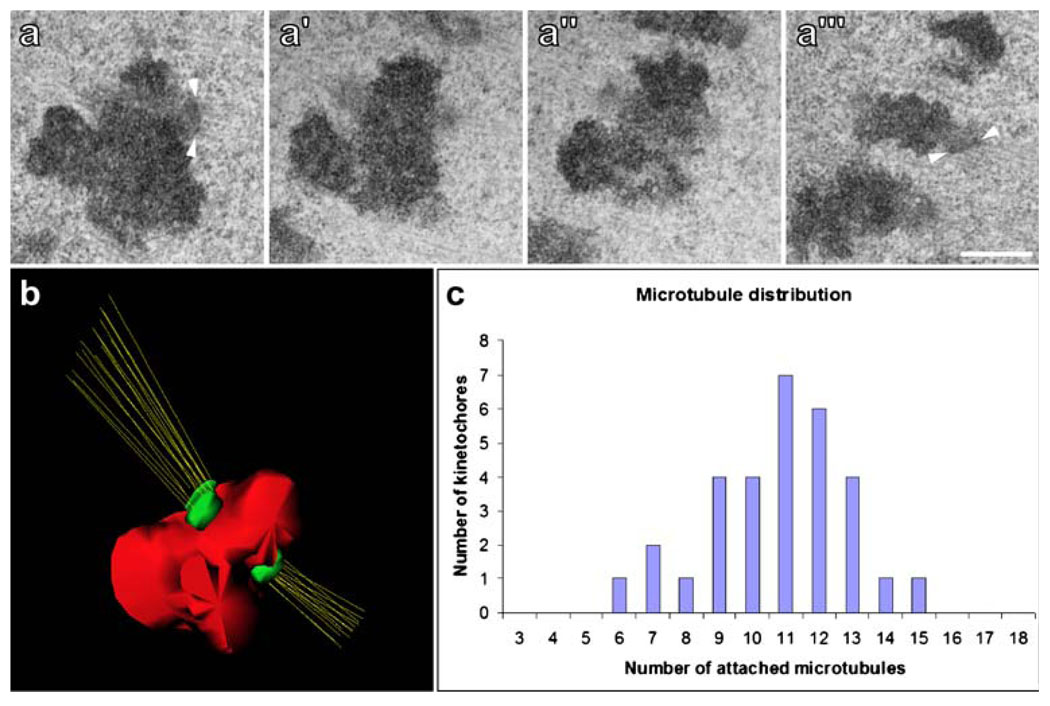
We examined kinetochores from metaphase S2R+ cells in serial sections (Fig. 4). Generally, the kinetochores appeared as poorly organized structures consisting of an amorphous material that is more electron-opaque than the surrounding cytoplasm but less electron-opaque than the chromatin (Fig. 4a–a′; see also Fig. 5, Fig 6). In some serial sections, notably those at the beginning and end of the series, the kinetochore region closely associated with the heterochromatin showed an additional thin, more electron-dense layer (Fig. 4a, a''', arrowheads and Fig. 5, Fig 6). To facilitate the visualization of kinetochores and their associated MTs, we generated 3D-reconstructions from serial sections (Fig. 4b). We also used serial-section EM to quantify the number of MTs bound to each kinetochore. Analyzing 31 kinetochores from three different metaphase cells, we found that S2R+ kinetochores bind 6–15 MTs. The histogram of kMT binding shows a near Gaussian distribution (Fig. 4c). On average, metaphase kinetochores bind 11±2 MTs per kinetochore, which is twice the number observed for kinetochores in Drosophila spermatocytes (Lin et al. 1981).
Fig. 4.
The Drosophila mitotic kinetochore and K-fiber. a–a''' Serial-sections through a kinetochore from a metaphase S2R+ cell. Note the highly amorphous structure that protrudes at the centromeric region. In some favorable views, such as a and a''', it is possible to visualize a thin electron-dense layer immediately adjacent to the amorphous material where MTs are bound (arrowheads). Scale bar=0.5 µm. b Three-dimensional reconstruction from serial sections of the chromosome observed in a. In this particular case, the sister kinetochores are bound to 15 and 12 MTs. c Direct MT counts from 31 kinetochores. The average number of kMTs bound is 10.8 MTs/kinetochore, with a standard deviation of 2.1, and a range of 6–15
Fig. 5.
The ultrastructure of the colchicine-treated Drosophila mitotic kinetochore. a, b Serial sections through two individual kinetochores from S2R+ cells treated with colchicine to depolymerize MTs. In the absence of MTs, the Drosophila kinetochore reveals a well-defined electron-dense outer layer with a hemispherical shape (brackets and arrowheads) that extends from a poorly defined internal region. Scale bar=200 nm
Fig. 6.
Comparison between the ultrastructures of Drosophila, PtK1, and HeLa cell kinetochores in the absence and presence of MTs. The side-by-side comparison with vertebrate kinetochores indicates that the electron-dense outer layer observed in unbound Drosophila kinetochores (brackets and arrowheads), although it is less distinct, clearly corresponds to the classical outer plate in unbound vertebrate kinetochores. The presence of MTs distorts the kinetochore structure in all three species. Nevertheless, the putative outer plate (arrowheads) is still visible in favourable views of Drosophila kinetochores. Scale bar=200 nm
In vertebrates, the kinetochore undergoes a structural change as it becomes associated with MTs (Roos 1973; Rieder 1982, 1990). To characterize the structure of non-attached kinetochores in S2R+ cells, we treated cultures with colchicine to disassemble MTs. In the absence of MTs, S2R+ kinetochores appeared as hemispheres of material that is less electron opaque than the underlying centromeric heterochromatin (Fig. 5). In some views, a 40- to 50-nm thick layer of more densely stained material was seen on the peripheral surface of the kinetochore (Fig. 5 arrowheads). As has been observed for vertebrates, the Drosophila kinetochore upon MT attachment becomes more disorganized, and the peripheral layer becomes less distinctive (Fig. 6).
EM tomographic analysis of the Drosophila kinetochore-microtubule interface
Thus far, our results indicate that, after conventional fixation and embedding, the S2R+ kinetochore is a less defined structure than the vertebrate kinetochore (Fig. 6). However, the peripheral layer does resemble the outer plate in vertebrate kinetochores in terms of thickness, staining properties, and overall shape (Fig. 6, arrowheads). In vertebrates, the majority of kMT plus ends terminate in the outer plate (Rieder 1982, 1990; Maiato et al. 2004a; Vandenbeldt et al. 2006). To determine whether the peripheral layer of S2R+ kinetochores has a similar function, we used EM tomography to compute 3-D reconstructions of individual kinetochore profiles in 100-nm-thick plastic sections (Fig. 7). We examined 26 kMT plus ends in 1.6-nm-thick slices viewed from tomographic 3D reconstructions of eight different kinetochores, and we found that all of the kMT plus ends terminated within 100 nm of the peripheral edge of the kinetochore (Fig. 7a–c). Often, this edge was marked by a distinctive fibrous band, as indicated by the brackets in Figs. 7a–c, and 21 of the kMTs examined (81%) terminate in the region roughly delineated by this band. The other five penetrate a little deeper, but never more than 100 nm.
Fig. 7.
Electron tomography of the Drosophila kinetochore-microtubule interface: a–c 1.6-nm thick slices from tomographic reconstructions of three different S2R+ kinetochores. Boxed areas indicate selected kMTs, and the brackets indicate a plate-like kinetochore structure. Scale bar=200 nm. a′–c′. Individual kMTs corresponding to the boxed areas in a–c. Note that the distortion of the microtubule wall is a consequence of chemical fixation that is particularly noticeable because of the higher resolution provided by electron tomography. Conformations detected for kMT plus-ends of S2R+ cells by electron tomography were manually classified as curved or straight according to criteria used by Vandenbeldt et al. (2006). According to these criteria, a′ and c′ were classified as curved, whereas b′ was classified as straight. Scale bar=200 nm
Discussion
With the discovery of RNAi technology, Drosophila S2 cells have been widely used for the molecular dissection of mitotic spindle formation, function, and architecture (e.g., Goshima and Vale 2003; Kwon and Scholey 2004; Goshima et al. 2005a,b; Morales-Mulia and Scholey 2005; Mahoney et al. 2006). Although these molecular depletion studies constitute an inherently powerful approach for studying protein function, assessment of phenotypic results in S2 cells has thus far been limited to LM resolutions. In the current work, we have characterized the prototypic ultrastructure for S2 kinetochores. We have also demonstrated the feasibility of applying correlative LM/EM techniques to S2 cells imaged live by video LM.
The immunofluorescence studies illustrated in Fig. 1 reveal that the Drosophila homologs of CENP-A (CID) and CENP-C are stably located at the core of S2 kinetochores. As in vertebrate kinetochores, fluorescence of the CENP-E (Cenp-meta) was more extensive in the absence of MT binding (Hoffman et al. 2001; Howell et al. 2001; Thrower et al. 1996). Kinetochores in S2 cells showed distortion and increased separation of sister kinetochores upon MT binding, a clear indication that they experience tension forces. We then presented results from correlative time-lapse video LM-EM studies in S2 cells, which revealed that the overall ultrastructure of the spindle in S2 cells is similar to that in vertebrate cells (Fig. 2 and Fig 3).
In general, the ultrastructure of kinetochores in Drosophila somatic cells is similar to that reported for early prometaphase I bivalent kinetochores in Drosophila spermatocytes (Goldstein 1981; Church and Lin 1982). Those early structural studies described unattached Drosophila kinetochores as a “single bilaminar hemisphere” ~560 nm in diameter, with an inner layer ~280 nm thick and a more electron-dense outer layer ~60 nm thick (Goldstein 1981; Lin et al. 1981; Church and Lin 1982). We also find the S2 kinetochore to be a bilaminar hemisphere, with a thin, darker-staining peripheral layer on the surface of a hemisphere. The material of the hemisphere has a distinctly different texture and different staining properties than the underlying chromatin (Fig. 4 and Fig 5). After MT attachment, the kinetochore becomes more disorganized, in the sense that the thin peripheral layer is often not apparent (Fig. 4a and Fig 6; Goldstein 1981).
Despite the similarities in ultrastructure between spermatocyte and S2 kinetochores, we found that the latter, on average, bind twice as many kMTs during metaphase as do spermatocyte kinetochores. The number of MTs that make up a K-fiber is variable according to cell type, and is presumably limited by the surface area of the kinetochore (reviewed by Rieder 1982). Vertebrate kinetochores typically bind 15–30 MTs during metaphase, while S. cerevisiae kinetochores attach to a single MT (Peterson and Ris 1976). Based on the classic studies with meiosis II spermatocytes, it is widely assumed that the Drosophila somatic kinetochore binds to 4–6 MTs (Lin et al. 1981; Maiato and Sunkel 2004). However, our results reveal that Drosophila mitotic kinetochores on average bind 11 MTs and have the capacity to bind up to 15 MTs. These numbers not only provide fundamental information for assessing the results of molecular depletion studies but also are particularly important for mathematical modeling and computer simulation studies in Drosophila (Brust-Mascher et al. 2004; Mogilner et al. 2006; Civelekoglu-Scholey et al. 2006).
We do not think that the difference in the number of MTs bound per kinetochore, between Drosophila S2 cells and spermatocytes, arises from differences in fixation protocols, because we used basically the same specimen preparation protocol, of glutaraldehyde fixation, osmium post fixation, and uranyl acetate en bloc staining, as had been employed in the previous studies (Lin et al. 1981; Goldstein 1981; Church and Lin 1982). An interesting possibility is that the difference that we observe in numbers of attached kMTs arises from differences in kMT turnover between spermatocytes and somatic cells. KMT turnover is relatively rapid in Drosophila embryos (Brust-Mascher et al. 2004; Civelekoglu-Scholey et al. 2006). In contrast, kMTs appear to be relatively stable in S2 cells (Maiato et al. 2005). Consequently, at a given time point, the number of kMTs bound per kinetochore is expected to be closer to the total binding capacity in S2 cells than in Drosophila embryos (see the models of Zhai et al. 1995 and Civelekoglu-Scholey et al. 2006). The kMT turnover rate in Drosophila spermatocytes is unknown, but our hypothesis predicts that it will be similar to the turnover rate in embryos. An easier way to test the hypothesis would be to count the number of kMTs attached to kinetochores in embryos. The number should be significantly lower in embryos than S2 cells, if kMT turnover is the major factor in determining the number of kMTs attached.
Another difference between S2 and spermatocyte kinetochores is that we do not observe any kMTs penetrating into the inner kinetochore and ending in the chromosome (e.g., compare Fig. 7a–c to Fig. 3–Fig. 7 and Table 1 in Church and Lin 1982). Rather, in electron tomographic images we find that 81% of the kMTs terminate in a band of fibers at the kinetochore periphery that most likely corresponds to the peripheral layer seen in projection images. This is strikingly similar to our finding in PtK1 cells that 86% of kMT plus ends terminate in the kinetochore outer plate (Vandenbeldt et al. 2006).
Although the two structural features differ in appearance, we argue that there are several strong parallels between the peripheral layer of S2 kinetochores and the outer plate of vertebrate kinetochores. First, both are about 40–50 nm wide in specimens prepared by conventional methods (Fig. 4, Fig 5 and Fig 6; Roos 1973; McEwen et al. 1993). Second, both the vertebrate outer plate and the peripheral layer undergo a transition to a more disorganized structure, upon MT attachment (e.g., Fig. 6). Third, the fact that most kMTs terminate in the outer plate of vertebrate kinetochores and the peripheral layer of S2 kinetochores strongly implies that the two have similar functions. Fourth, in electron-tomographic reconstructions, the peripheral layer is similar in appearance to the outer plate, despite differences in specimen preparation (compare Fig. 7 to Fig. 4 in Vandenbeldt et al. 2006). Finally, using electron tomography to distinguish straight (Fig. 7b) and curved (Figs. 7a′,c′) conformations of the kMT plus ends, we previously found the surprising result that two-thirds of the kMT plus ends in both PtK1 outer plate and S2R+ peripheral layer have the curved conformation (Vandenbeldt et al. 2006). The curved conformation is associated with the depolymerization state of MTs (Mandelkow et al. 1991; Chretien et al. 1995; Arnal et al. 2000). Hence, our structural data indicate strong parallels between the peripheral layer of kinetochores in S2 cells and the outer plate of kinetochores in vertebrate cells.
Supplementary Material
Acknowledgements
We dedicate this paper to Dr. Bill Brinkley for his pioneering work on mammalian kinetochore ultrastructure. We also thank Drs. Shin-ichi Yanagawa (Kyoto University, Japan) and Gary Karpen (University of California at Berkeley, CA) for the generous gift of the S2R+ and S2-GFP-CID cell lines, respectively. We are also indebted to Drs. Mike Goldberg (Cornell University, Ithaca, NY), Christian Lehner (University of Bayreuth, Germany) and T. Kaufman (Indiana University, Bloomingdale, IN) for antibodies used in this paper. Finally, we would like to thank Dr. Alexey Khodjakov for many insightful discussions about the Drosophila kinetochore. This work was supported by: POCI/SAU-MMO/58353/2004 to HM; NIH R01 GM40198 to CLR; and NIH R01 GM06627 and NSF MCB 0110821 to BFM. SMP holds a studentship from Fundação para a Ciência e a Tecnologia of Portugal (SFRH/BD/15244/2004). We are grateful for technical support from the Wadsworth Center’s Core Facility for Electron Microscopy.
Footnotes
Electronic supplementary material Supplementary material is available for this article at http://dx.doi.org/10.1007/s00412-006-0076-2 and is accessible to authorized users.
Contributor Information
Helder Maiato, Email: maiato@ibmc.up.pt, Institute for Molecular and Cell Biology, Rua do Campo Alegre 823, 4150-180 Porto, Portugal; Laboratory of Cell and Molecular Biology, Faculty of Medicine, University of Porto, 4200-319 Porto, Portugal.
Polla J. Hergert, Wadsworth Center, New York State Department of Health, Albany, NY 12201-0509, USA
Sara Moutinho-Pereira, Institute for Molecular and Cell Biology, Rua do Campo Alegre 823, 4150-180 Porto, Portugal.
Yimin Dong, Wadsworth Center, New York State Department of Health, Albany, NY 12201-0509, USA.
Kristin J. Vandenbeldt, Wadsworth Center, New York State Department of Health, Albany, NY 12201-0509, USA
Conly L. Rieder, Wadsworth Center, New York State Department of Health, Albany, NY 12201-0509, USA Department Biomedical Sciences, School of Public Health, State University of New York at Albany, Albany, NY 12222, USA.
Bruce F. McEwen, Wadsworth Center, New York State Department of Health, Albany, NY 12201-0509, USA Department Biomedical Sciences, School of Public Health, State University of New York at Albany, Albany, NY 12222, USA.
References
- Arnal I, Karsenti E, Hyman AA. Structural transitions at microtubule ends correlate with their dynamic properties in Xenopus egg extracts. J Cell Biol. 2000;149:767–774. doi: 10.1083/jcb.149.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Blagden SP, Glover DM. Polar expeditions-provisioning the centrosome for mitosis. Nat Cell Biol. 2003;5:505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci USA. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Rupp G, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96(Pt 1):9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- Chan GK, Liu ST, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church K, Lin HP. Meiosis in Drosophila melanogaster. II. The prometaphase-I kinetochore microtubule bundle and kinetochore orientation in males. J Cell Biol. 1982;93:365–373. doi: 10.1083/jcb.93.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O’Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005a;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005b;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 2005;19:2041–2053. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, et al. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Imaging the division process in living tissue culture cells. Methods. 2006;38:2–16. doi: 10.1016/j.ymeth.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Sandall S, Desai A. Kinetochore-spindle microtubule interactions during mitosis. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kwon M, Scholey JM. Spindle mechanics and dynamics during mitosis in Drosophila. Trends Cell Biol. 2004;14:194–205. doi: 10.1016/j.tcb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Lemos CL, Sampaio P, Maiato H, Costa M, Omel’yanchuk LV, Liberal V, et al. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J. 2000;19:3668–3682. doi: 10.1093/emboj/19.14.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HP, Ault JG, Church K. Meiosis in Drosophila melanogaster. I. Chromosome identification and kinetochore microtubule numbers during the first and second meiotic divisions in males. Chromosoma. 1981;83:507–521. doi: 10.1007/BF00328276. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Maiato H, Sunkel CE. Kinetochore-microtubule interactions during cell division. Chromosome Res. 2004;12:585–597. doi: 10.1023/B:CHRO.0000036587.26566.81. [DOI] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004a;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004b;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko M, Leith A. Sterecon-three-dimensional reconstructions from stereoscopic contouring. J Struct Biol. 1996;116:93–98. doi: 10.1006/jsbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Marko M. Three-dimensional transmission electron microscopy and its application to mitosis research. Methods Cell Biol. 1999;61:81–111. doi: 10.1016/s0091-679x(08)61976-7. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J Cell Biol. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzner R. Beitrage zur granulalehre. I. Kern und kerntheilung. Arch Anat Physiol. 1894:309–348. [Google Scholar]
- Mogilner A, Wollman R, Civelekoglu-Scholey G, Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B, Mazzolini L, Moisand A, Cirinesi AM, Wright M. Elongation of centriolar microtubule triplets contributes to the formation of the mitotic spindle in gamma-tubulin-depleted cells. J Cell Sci. 2004;117:5497–5507. doi: 10.1242/jcs.01401. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL. Formation of the astral mitotic spindle: ultrastructural basis for the centrosome-kinetochore interaction. Electron Microsc Rev. 1990;3:269–300. doi: 10.1016/0892-0354(90)90005-d. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- Roos UP. The fibrillar organization of kinetochore and the kinetochore region of mammalian chromosomes. Cytobiologie. 1977;16:82–90. [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Somma MP, Fasulo B, Cenci G, Cundari E, Gatti M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol Biol Cell. 2002;13:2448–2460. doi: 10.1091/mbc.01-12-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollozi A, Ris H, Szollosi D, Debec A. A centrioles-free Drosophila cell line. A high voltage EM study. Eur J Cell Biol. 1986;40:100–104. [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil Cytoskeleton. 1996;35:121–133. doi: 10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Vandenbeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules within a kinetochore fiber. Curr Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, et al. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–218. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- Yucel JK, Marszalek JD, McIntosh JR, Goldstein LS, Cleveland DW, Philp AV. CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J Cell Biol. 2000;150:1–11. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.