Abstract
Multiple genetic linkage studies support the hypothesis that the 15q13–14 chromosomal region contributes to the etiology of schizophrenia. Among the putative candidate genes in this area are the α7 nicotinic acetylcholine receptor gene (CHRNA7) and its partial duplication, CHRFAM7A. A large chromosomal segment including the CHRFAM7A gene locus, but not the CHRNA7 locus, is deleted in some individuals. The CHRFAM7A gene contains a polymorphism consisting of a 2 base pair (2 bp) deletion at position 497–498 bp of exon 6. We employed PCR-based methods to quantify the copy number of CHRFAM7A and the presence of the 2 bp polymorphism in a large, multi-ethnic population. The 2 bp polymorphism was associated with schizophrenia in African Americans (genotype p=0.005, allele p=0.015), and in Caucasians (genotype p=0.015, allele p=0.009). We conclude that the presence of the 2 bp polymorphism at the CHRFAM7A locus may have a functional significance in schizophrenia.
Keywords: nicotinic acetylcholine receptor, CHRFAM7A, CHRNA7, association study, deletion, schizophrenia, duplication, P50
1. Introduction
Understanding the genetic determinants of schizophrenia will lead to more promising treatments for this devastating psychiatric disorder, which afflicts approximately 1% of the world population. This disease has demonstrated a substantial genetic component in twin, family and adoption studies, raising hopes that genetic linkage studies could identify predisposing genes of large to moderate effect (Cardno et al., 1999; Cardno and Gottesman, 2000; Leo, 2006; Wynne et al., 2006). To date, definitive linkage results have been difficult to achieve. However, chromosomal rearrangements on 15q11–14 are associated with neurodevelopmental syndromes. Intrachromosomal recombination events that map to 15q11–14 have been implicated in Prader-Willi/Angelman syndrome (Christian et al., 1998; Robinson et al., 1998). Sharp et al. recently reported a recurrent microdeletion at 15q13.3 that is associated with mental retardation and seizures (Sharp et al., 2008) and large recurrent, but rare, microdeletions at 15q11.2 and 15q13.3 were associated with schizophrenia (Stefansson et al., 2008). Rare chromosomal deletions and duplications in several locations in the genome, including chromosomes 15, 22, and 1 have also been found to be associated with schizophrenia (International Schizophrenia Consortium, 2008). As these rearrangements are associated with neurocognitive dysfunction, the region is likely to be important in brain development. The α7 nicotinic cholinergic receptor gene (CHRNA7) and its partial duplication (CHRFAM7A) map within this dynamic region (Baron, 2001; Freedman and Leonard, 2001; Linthorst and Reul, 2008). Within the large deletion and duplication events described above, this segmental duplication containing CHRNA7 also has complex rearrangements. The purpose of the current study was to determine whether the partial duplication, CHRFAM7A, is associated with schizophrenia or its physiological correlate, the P50 gating deficit.
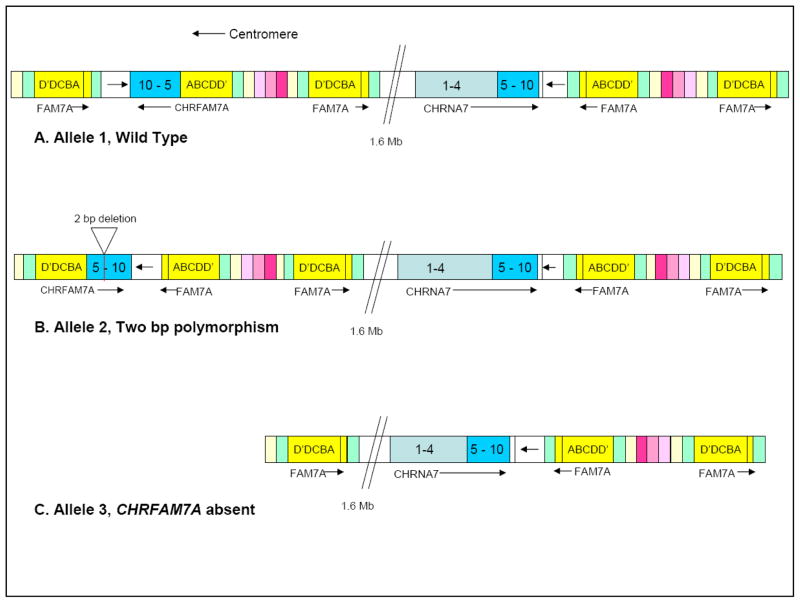
Genetic analysis of CHRNA7 has been complicated by a partial duplication, now designated CHRFAM7A, which maps 1.6 Mb centromeric to the gene (Gault et al., 1998; Riley et al., 2000b) (see Figure 1). This appears to be a relatively recent event unique to humans; it is not carried by closely related primates (Locke et al., 2003). In this duplication, exons 5–10, intervening introns, and the 3′ untranslated region of CHRNA7 are conserved (> 99% nucleotide identity). In place of exons 1–4, novel exons termed D′, D, C, B and A are fused 700 bp upstream of CHRNA7 exon 5 (GenBank AF029838) (Gault et al., 1998). The breakpoint occurs in an alu sequence within intron 4. Riley et al. (2002) also described another exon, E, lying between exons A and B. Exons C-A are the result of a partial duplication of a putative kinase-like gene (ULK4) on chromosome 3p21. Axons C-A also map to several additional loci in the chromosome 15q13–14 region. Exons D′ and D map to no fewer than 5 distinct loci on chromosome 15, but not to chromosome 3. Their origin is unknown.
Figure 1. Genomic map of CHRNA7 and its partial duplication, CHRFAM7A.
(A) Allele 1, wild-type CHRFAM7A allele. (B) Allele 2, two bp polymorphism and inversion of CHRFAM7A (C) Allele 3, duplicated gene absent.
FAM7A=duplication of sequences from gene ULK4 on chromosome 3
Mutation screening in human brain mRNA for both CHRNA7 and CHRFAM7A identified 33 polymorphisms (Gault et al, 2003). Thirty of these polymorphisms are single nucleotide polymorphisms (SNPs). Three insertion/deletion polymorphisms were identified, including a 2 bp deletion polymorphism (2 bp polymorphism) in exon 6 that maps exclusively to CHRFAM7A. In addition, some chromosomes are missing the CHRFAM7A locus and rare individuals are missing both copies.
Riley et al. (2001) suggested that the CHRFAM7A locus is the result of a primary duplication of the full-length CHRNA7 gene, followed by a non-homologous deletion of sequences contained in the more centromeric copy of CHRNA7. The absence of the CHRFAM7A gene on some chromosomes would represent a later deletion of the primary duplication event due to further recombination. An alternative explanation, suggested by Flomen et al. (2008) is that the absence of CHRFAM7A represents an ancestral sequence that did not undergo duplication of CHRNA7. Further, CHRFAM7A exists in two orientations with reference to CHRNA7 (Flomen et al., 2008). The 2 bp polymorphism in exon 6 of CHRFAM7A is in strong linkage disequilibrium with this polymorphic inversion of CHRFAM7A and surrounding loci. On chromosomes containing the 2 bp polymorphism, CHRFAM7A is almost always in the same orientation as CHRNA7; at loci that do not contain the 2 bp polymorphism, CHRFAM7A is almost always in the opposite orientation. Thus, numerous recombination events over time have created the complex and fragmented region observed today.
The CHRNA7 gene is genetically linked to schizophrenia and its endophenotype, the P50 auditory gating deficit. The P50 endophenotype, a neurophysiological auditory gating deficit, is commonly seen in schizophrenics and their first degree relatives (Freedman et al., 1991). This trait involves inhibition of response to repetitive auditory stimuli and can be measured by means of auditory evoked potentials in a paired pulse paradigm. Scalp electrodes record waves with a 50 millisecond latency (P50) following paired auditory stimuli delivered 0.5 seconds apart. In a normal response, the subject decreases the amplitude of the second response (test response) compared to the response to the first stimulus (conditioning response) through the action of an inhibitory neuronal pathway. The results are reported as the P50 test-conditioning (T/C) ratio (Freedman et al., 1991). The endophenotype is normalized by smoking in schizophrenic patients (Leonard et al., 1998a) and is strongly linked to the D15S1360 dinucleotide repeat marker within intron 2 of the CHRNA7 gene on chromosome 15q14 (Freedman et al., 1997). Other markers in this region have shown weaker linkage to schizophrenia as a disease. (Freedman et al., 2001; Gejman et al., 2001; Leonard et al., 1998b; Liu et al., 2001; Neves-Pereira et al., 1998; Riley et al., 2000a; Tsuang et al., 2001; Xu et al., 2001)
A role for the α7 nicotinic receptor in mediating the P50 response is strongly supported by pharmacological and biochemical data, both in humans and in animals (Adler et al., 1998; Leonard et al., 2001). The snake toxin α-Bungarotoxin (α-BTX) preferentially binds to the α7 receptor in the CNS. α-BTX binding levels are decreased in schizophrenic post-mortem hippocampus, where the P50 response is mediated (Freedman et al., 1995). α-BTX binding is also decreased in cingulate cortex and in the reticular nucleus of the thalamus in schizophrenia, while CHRNA7 protein levels are reduced in prefrontal cortex (Court et al., 1999; Guan et al., 1999; Marutle et al., 2001). The proximal promoter of CHRNA7 contains functional SNPs that decrease transcription and are associated with schizophrenia (Leonard et al., 2002). This is consistent with the observation that CHRNA7 expression is aberrant in schizophrenia.
Multiple lines of evidence indicate the involvement of genes in the 15q13–14 locus in schizophrenia, and the CHRFAM7A locus itself is polymorphic. We determined the copy number and 2 bp polymorphism status of CHRFAM7A quantitatively and evaluated them as risk factors for schizophrenia and for increased P50 T/C ratio in a large multi-ethnic population of Caucasian, African American and Hispanic descent.
2. Results
Southern blot was used on an initial set of samples and fluorescent in situ hybridization (FISH) was used on a subset of these to confirm and visualize the findings on Southern blot. Both Southern blot and FISH are highly labor intensive and time consuming. Southern blot results were used for comparison to empirically determined genotype results utilizing dHPLC and RT-PCR, which more efficiently and rapidly detected CHRFAM7A copy number and the 2 bp polymorphism.
2.1. Southern Blot Analysis
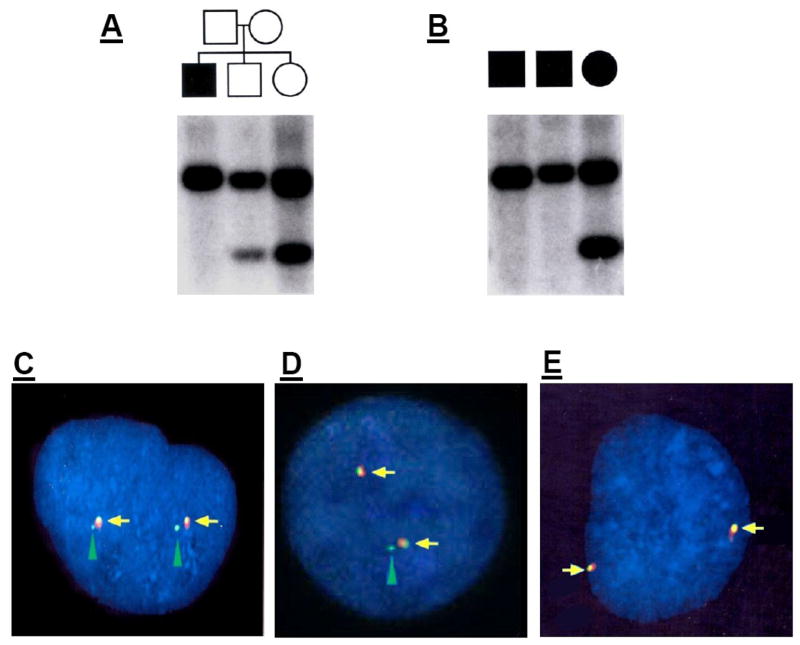
An RsaI polymorphism in intron 4 results in a 768 bp target for CHRFAM7A and a 1094 bp fragment for CHRNA7. RsaI-digested genomic DNA from 3 siblings was used for a Southern blot (Figure 2A). In this pedigree, the 768 bp target is missing from the affected individual on the left, a schizophrenic male, indicating that this individual has two full-length copies of CHRNA7 but is missing both copies of CHRFAM7A. The other two siblings, who are not diagnosed with schizophrenia, have at least one copy of the CHRFAM7A locus as well as the CHRNA7 locus. The sibling on the right appears to have two copies of CHRFAM7A, while the sibling in the center appears to have only one copy. Figure 2B shows 3 other unrelated schizophrenic individuals, two of whom are homozygous deleted for the CHRFAM7A locus. The third individual has two copies of CHRFAM7A (Lane 3, Figure 2B).
Figure 2. Southern Blot Analysis of CHRFAM7A Copy Number.
(A) Southern blot of RsaI digested genomic DNA from a family with one affected male offspring (dark box) and two unaffected offspring (white boxes). The probe hybridizes to both CHRNA7 and CHRFAM7A in exon 5. The upper band corresponds to a predicted fragment digested from the CHRNA7 locus (1094 bp), while the lower hybridization band is the smaller fragment (768 bp) from CHRFAM7A. The affected offspring is missing the CHRFAM7A band, indicating that he has no copies of CHRFAM7A. The offspring in the center carries one copy of CHRFAM7A, while the offspring on the left carries 2 copies.
(B) Three other unrelated affected subjects. Two are missing the CHRFAM7A fragment but retain the CHRNA7 band. The third individual is a control for the blot/hybridization and contains both bands.
(C, D, E) FISH analysis of interphase chromosomes from individuals with 0, 1 or 2 copies of CHRFAM7A. Two PAC clones were fluorescently labeled. PAC 10743 contains sequence for exon 6–10 and is labeled with Spectrum Green. PAC 10905 contains sequence for exons 1–3 and is labeled with Spectrum Orange. Both PACs hybridize to the full length gene, where the overlapping yellow signal is evident (yellow arrows). CHRFAM7A hybridizes only with PAC 10743, resulting in an additional green signal (green arrowheads).
(C)Two distinct loci are labeled on each chromosome in this individual who has two copies of CHRFAM7A
(D) An individual with one copy of CHRFAM7A.
(E) Both copies of the CHRFAM7A gene are missing.
2.2. Fluorescence In Situ Hybridization
Interphase chromosomes from individuals with 0, 1 or 2 copies of CHRFAM7A, as determined by Southern blot, were processed for in situ hybridization (Christian et al., 1998). P1-derived artificial chromosome (PAC) 10905 is labeled with Spectrum Orange and hybridizes to exons 1–4; PAC 10743 is labeled with Spectrum Green and hybridizes to exons 6–10. Both PACs hybridize to the full length gene, where the overlapping yellow signal is evident (Figure 2C–E, yellow arrows). CHRFAM7A (exons 5–10) hybridizes only with PAC 10743, resulting in an additional green signal that corresponds to CHRFAM7A (green arrowheads). (C)Two distinct loci are labeled on each chromosome in this individual who has two copies of CHRFAM7A. (D) An individual with one copy of CHRFAM7A. Only one green signal from PAC 10743 is present. (E) An individual missing both copies of CHRFAM7A by Southern blot analysis shows cohybridization of both PAC probes at the CHRNA7 locus (yellow arrows), but the second distinct signal from PAC 10743 is not observed.
2.2. Genetic Analysis
2.2.1. CHRFAM7A copy number and frequency of the 2 bp polymorphism: differences among the three ethnicities
Genotypes for CHRFAM7A copy number and the 2 bp polymorphism were determined by dHPLC, as described in Methods. A chromosome that contains a copy of CHRFAM7A and does not carry the 2 bp polymorphism in exon 6 of CHRFAM7A was designated the wild-type allele (allele 1); the 2 bp polymorphism is designated allele 2, and a missing CHRFAM7A gene was designated allele 3 (See Figure 1). Allele frequencies for CHRFAM7A copy number and for the 2 bp polymorphism in controls for each ethnic group are shown in Table 1. In order to determine if there are significant differences in CHRFAM7A allele frequencies between the three ethnic groups, population allele frequency comparisons were made by performing Chi Square tests for proportions in the control subjects. Alleles were in Hardy-Weinberg equilibrium for African Americans (χ2HWE = 3.57, 3 df, p = 0.31), Caucasians (χ2HWE = 2.71, 3 df, p = 0.44), and Hispanics (χ2HWE = 4.13, 3 df, p = 0.25)
Table 1.
Allele frequencies for the 2 bp polymorphism and for CHRFAM7A copy number in controls of the three ethnicities
| Allele | African American | Caucasian | Hispanic |
|---|---|---|---|
| Allele 1 | 0.74* (89) | 0.50 (194) | 0.34# (25) |
| Allele 2 | 0.14* (17) | 0.42 (161) | 0.59# (44) |
| Allele 3 | 0.12 (14) | 0.08 (29) | 0.07 (5) |
African Americans were significantly more likely to carry allele 1 (χ2= 30.87, 2df, p = 5.20×10−7, OR 2.81, 95% CI 2.03–3.89), but less likely to carry allele 2 (OR 0.23, 95% CI 0.13–0.39) compared to Caucasians.
Hispanics were significantly more likely to carry allele 2 (χ2= 7.95, 2df, p = 0.017, OR 2.04, 95% CI 1.35–3.09), and less likely to carry allele 1 (OR 0.50, 95% CI 0.31–0.81) compared to Caucasians. Allele 1= wild type. Allele 2 = 2 bp polymorphism. Allele 3 = CHRFAM7A absent.
All p values presented are empirical p values derived after a minimum of 104 Monte Carlo simulations.
Numbers in parentheses represent the number of alleles in each group.
The CHRFAM7A allele frequencies in the controls of the three ethnicities differed significantly (Table 1). The African American subjects were more likely than Caucasians to be carrying the wild-type allele (χ2= 30.87, 2df, p = 5.20×10−7, OR 2.81, 95% CI 2.04–3.89; all p values presented in this study are empirically derived via the Monte Carlo simulation method), and less likely to be carrying the 2 bp polymorphism (χ2= 30.87, 2df, p = 5.20×10−7, OR 0.23, 95% CI 0.13–0.39) compared to Caucasians. African Americans were more likely to be missing a copy of CHRFAM7A, but this did not reach significance (OR 1.59, 95% CI 0.82–3.10). The African American subjects were also more likely than Hispanics to be carrying the wild-type allele (χ2= 43.69, 2df, p = 7.09×10−10, OR 5.63, 95% CI 3.31–9.56), and less likely to be carrying the 2 bp polymorphism (χ2= 43.69, 2df, p = 7.09×10−10, OR 0.11, 95% CI 0.06–0.21). Hispanics were significantly more likely than Caucasians to carry the 2 bp polymorphism. (χ2= 7.95, 2df, p = 0.017, OR 2.03, 95% CI 1.34–3.08) and less likely to be carrying the wild type allele (χ2= 7.95, 2df, p = 0.017, OR 0.50, 95% CI 0.31–0.81).
2.2.2. Association with Schizophrenia
Ethnically matched, unrelated controls were used for assessment of preferential association of either CHRFAM7A copy number or the 2 bp polymorphism with schizophrenia. Controls were ethnically matched to patients as determined by admixture analysis (Stephens et al., 2009). Association was assessed by case-control comparisons using Chi-square tests. If sample sizes were small, Fisher-Freeman-Halton exact tests for proportions were used. Allele frequencies for CHRFAM7A copy number and for the 2 bp polymorphism in schizophrenics vs. controls for each ethnic group are shown in Table 2. Genotype frequencies in schizophrenics vs. controls are shown in Table 3. In Caucasians, the 2 bp polymorphism (Allele 2) was significantly associated with schizophrenia (χ2= 9.462, 2df, empirical p= 0.009). Subjects with the 2 bp polymorphism carried significantly higher odds for schizophrenia compared to those with the wild-type allele (OR 1.49, 95% CI 1.14–2.42). In genotype tests, both genotype 2/2 and genotype 2/3 were significantly associated with schizophrenia (χ2= 13.309, 5df, p= 0.015). Subjects with genotype 2/2 (2 copies of CHRFAM7A, both of which contain the 2 bp polymorphism) carried higher odds for schizophrenia compared with genotype 1/1 (wild-type) (OR= 2.07, 95% CI 1.16–3.71). Individuals with genotype 2/3, a single copy of CHRFAM7A that contains the 2 bp polymorphism, had the highest odds for schizophrenia (OR= 2.64, CI= 1.19–5.84).
Table 2.
Allele Frequencies in Cases and Controls
| African Americans | |||
| Allele: | Allele 1 | Allele 2 | Allele 3 |
| Cases | 0.58 (77) | 0.28* (37) | 0.14 (18) |
| Controls | 0.74 (89) | 0.14 (17) | 0.12 (14) |
| Caucasians | |||
| Allele: | Allele 1 | Allele 2 | Allele 3 |
| Cases | 0.41 (258) | 0.50* (319) | 0.09 (57) |
| Controls | 0.50 (194) | 0.42 (161) | 0.08 (29) |
| Hispanics | |||
| Allele: | Allele 1 | Allele 2 | Allele 3 |
| Cases | 0.31 (8) | 0.54 (14) | 0.15 (4) |
| Controls | 0.34 (25) | 0.59 (44) | 0.07 (5) |
Subjects carrying Allele 2 had significantly higher odds of schizophrenia in African Americans (OR 2.52, CI 1.31–6.21, χ2= 8.222, 2df, p= 0.015) and in Caucasians (OR 1.49, CI 1.14–2.42, χ2= 9.462, 2df, p= 0.009).
Allele 1= wild type. Allele 2 = 2 bp polymorphism at 497–498. Allele 3= duplicate gene absent.
Numbers in parentheses represent the number of alleles in each group.
All p values presented are empirical p values derived after 104 Monte Carlo simulations.
Table 3.
Genotype Frequencies in Cases and Controls.
| African Americans | ||||||
| Genotype | 1/1 | 1/2 | 1/3 | 2/2 | 2/3 | 3/3 |
| Cases | 0.29 (19) | 0.42* (28) | 0.17 (11) | 0.03 (2) | 0.08 (5) | 0.02 (2) |
| Controls | 0.58 (35) | 0.20 (12) | 0.12 (7) | 0.03 (2) | 0.02 (1) | 0.05 (3) |
| Caucasians | ||||||
| Genotype | 1/1 | 1/2 | 1/3 | 2/2 | 2/3 | 3/3 |
| Cases | 0.16 (50) | 0.44 (140) | 0.06 (18) | 0.23# (73) | 0.10# (33) | 0.01 (3) |
| Controls | 0.23 (44) | 0.46 (88) | 0.10 (18) | 0.16 (31) | 0.06 (11) | 0.00 (0) |
| Hispanics | ||||||
| Genotype | 1/1 | 1/2 | 1/3 | 2/2 | 2/3 | 3/3 |
| Cases | 0.08 (3) | 0.46 (8) | 0.00 (0) | 0.15 (4) | 0.31 (4) | 0.00 (0) |
| Controls | 0.11 (4) | 0.43 (18) | 0.03 (1) | 0.32 (12) | 0.11 (4) | 0.00 (0) |
In African Americans, subjects with genotype 1/2 had significantly higher odds of schizophrenia (OR 4.30, CI 1.79–10.33, Exact test, p=0.005)
In Caucasians, subjects with genotype 2/2 (OR 2.07, CI=1.16–3.71) and 2/3 (OR=2.64, CI=1.19–5.84) had significantly higher odds of schizophrenia (χ2= 13.309, 5df, p= 0.015).
Allele 1= wild type. Allele 2 = 2 bp polymorphism at 497–498. Allele 3= duplicate gene absent.
Numbers in parentheses represent the number of subjects in each cohort.
All p values presented are empirical p values derived after 104 Monte Carlo simulations.
In African Americans, Allele 2 was significantly associated with schizophrenia (χ2= 8.222, 2df, empirical p= 0.015). Subjects with the 2 bp polymorphism were significantly more likely to have schizophrenia compared to those with the wild-type allele (χ2= 8.222, 2df, p= 0.015, OR= 2.52, 95% CI 1.31–6.21). In genotype tests, individuals with genotype 1/2 carried higher odds for schizophrenia compared with those with genotype 1/1 (Exact test, p= 0.005, OR= 4.30, CI= 1.79–10.33). Individuals with genotype 2/3 had the highest odds for schizophrenia (OR 9.21) but this group was small and confidence intervals were large (95% CI 0.87–22.14).
In Hispanics, neither the allele (Exact test, empirical p= 0.447) nor genotype (Exact test, p= 0.420) frequencies reached significance for association with schizophrenia. The small group size limits the conclusions that can be made for this ethnic group.
For allele 3, the absence if CHRFAM7A, no statistically significant association with schizophrenia was found for allele frequencies. However, within each ethnicity, the frequency of allele 3 was higher in schizophrenics vs. controls. The odds ratios for schizophrenia for allele 3 were 1.49, 1.48 and 2.50 for the African American, Caucasian and Hispanic populations respectively. This allele is relatively rare.
2.2.3. Association with P50 Gating Ratios
P50 gating ratios were analyzed with respect to age, sex, ethnicity, 2 bp polymorphism, and CHRFAM7A copy number in a subset of 169 controls. There was no effect of sex (p=0.788, MANOVA) or ethnic group (p= 0.630) on P50, and linear regression showed no correlation between age and P50 (R2= 0.0, p= 0.865). For the 2 bp polymorphism and copy number, logistic regression analysis was performed to determine appropriate groupings for the P50 data, as was done in a previous study (Leonard et al., 2002). This analysis indicated that the best fit for the data was to divide the control subjects (n=169) into three groups: Subjects with P50 T/C ratios less than 0.25 (n=102), those with p50 between 0.25 and 0.55 (n=56), and those with P50 greater then 0.55 (n=11). Chi square analysis of these groups for the CHRFAM7A alleles indicated no statistically significant differences in allele frequencies among the three P50 groups for either the 2 bp polymorphism (p=0.082) or for CHRFAM7A copy number (p=0.731). Association with P50 in the schizophrenic cohort was not analyzed; all of the schizophrenic subjects had abnormally high P50 values, with no value falling below 0.50.
3. Discussion
3.1. Functional Significance of CHRFAM7A
A 2 bp polymorphism at the CHRFAM7A locus was significantly associated with schizophrenia in both the Caucasian and African American populations. There was no statistically significant association in Hispanics. However, given the small sample size, it would be premature to draw conclusions about the Hispanic population. Several studies have examined the association of the 2 bp polymorphism in CHRFAM7A with schizophrenia and other psychiatric disorders (Flomen et al., 2006; Lai et al., 2001; Liou et al., 2001; Raux et al., 2002). None have reported significant association of this polymorphism with schizophrenia. The current study was larger, with a total of 396 cases and 289 controls altogether. This may have provided more statistical power to detect an association with a small effect size. Techniques used in the current study relied primarily on the probes for exon 5, exon 6, and the intron 4/intron A breakpoint of the known duplicate gene for detection of copy number. If other partial duplications exist that do not contain these sequences, for example a duplication containing only sequences from exons 1–4 or 8–10, they would not have been detected. Therefore given the unstable nature of this region of the genome, the possibility remains that there are other duplications present that have not been included in the current analysis.
It is not known whether the 2 bp polymorphism causes functional changes in CHRFAM7A. CHRFAM7A is expressed as mRNA in the brain and contains exons 5–10 of CHRNA7 sequence, which account for 76% of the coding sequence for the gene. Exon 1 of CHRNA7, which is not duplicated in CHRFAM7A, encodes the signal peptide sequence for targeting and entry of the a7 peptide into the endoplasmic reticulum for assembly and transport. Exons 2–4 contribute to the formation of the extracellular ligand binding domain as well as glycosylation sites, which have been shown to be important for the functional α7* homomeric receptors expressed in vitro (Chen et al., 1998). In contrast, exons D′-A have no known sequence motifs that confer protein function, and no known signal translocation sequence (Goder and Spiess, 2001; Kozak, 1999). The 2 bp polymorphism creates three premature UGA translation stop codons in exon 6 (Gault et al., 2003). However, these stop codons can often be read through (MacBeath and Kast, 1998). If translation occurs, a foreshortened peptide may have different properties, either for a regulatory function, or in assembly with the full length subunit.
The orientation of CHRFAM7A on the chromosome may also have functional consequences. The 2 bp polymorphism is in strong linkage disequilibrium with an inversion involving a 320 kb region that contains CHRFAM7A. On chromosomes with the 2 bp polymorphism, CHRFAM7A almost always occurs in the same orientation as CHRNA7, in contrast to the “wild type” allele, which is almost always in the opposite orientation to CHRNA7 (Flomen et al., 2008). There are no known promoter elements within CHRFAM7A and the exact point of transcription start has not been identified. However, upstream elements for the wild type and inverted alleles would be different. This may be important in determining whether wild type or inverted copies of CHRFAM7A are transcribed.
3.2. Association with P50
Although the 2 bp polymorphism in CHRFAM7A increases risk for schizophrenia, it was not significantly associated with P50 gating in the controls (p=0.082). Most of our control subjects had low T/C ratios. Logistic regression analysis indicated that a three group model including subjects with P50 ratios below 0.25, between 0.25 and 0.55, and over 0.55 best fit the data. We found no association in our control population, suggesting that the 2 bp polymorphism does not directly mediate the P50 response.
Our findings do not replicate a previous finding of association of CHRFAM7A with the P50 auditory gating deficit. Raux et al. (2002) noted that the 2 bp polymorphism appeared to associate with higher P50 levels in a French cohort of controls, as measured by the percentage of individuals who fall above a T/C of 0.50 or below a T/C ratio of 0.45. Classifying our data as in Raux et al. did not alter our results. Stimuli used by Raux et al. were presented at an intensity of 75dB measured at the ear of the subject. In the current study, subjects were tested with a stimulus at 50dB above threshold (average threshold is approximately 32 dB). It is possible that these procedures are not equivalent.
Previous work has demonstrated that promoter polymorphisms in the full-length CHRNA7 gene are associated with the P50 gating deficit in control subjects (Leonard et al., 2002). Approximately 20% of non-mentally ill subjects exhibited abnormal gating deficits and most of these subjects carried a CHRNA7 promoter polymorphism. Almost all schizophrenic patients, however, have an auditory sensory gating deficit (Freedman et al., 1991; Adler et al., 1998). In the current study, the schizophrenic subjects who had P50 recordings displayed this deficit with test to conditioning ratios that were greater than 0.50 and were not, therefore, examined for association of CHRFAM7A with auditory gating.
3.3. Evidence for the Sequence of Recombination Events
Looking at the relative allele frequencies in different populations may provide information about the sequence of recombination events that lead to the current arrangement of the 15q13–14 region. Since the 2 bp polymorphism is only seen in CHRFAM7A, it must have occurred after the partial duplication of CHRNA7. If this polymorphism appeared later in human history as migrations to Europe and the Middle East progressed, we would expect that populations of African descent might have lower frequencies of this allele. The allele frequencies in our African American and Caucasian controls differed significantly (p =1.98×10−7, see Table 3). Compared to Caucasians, the African American subjects are almost 3 times more likely to be carrying Allele 1 (wild type), and they were also significantly less likely than Caucasians to be carrying Allele 2. These data suggest that Allele 1 has an earlier origin, and that the 2 bp polymorphism occurred later as human populations migrated into Europe. This also supports the hypothesis put forth by Flomen et al. that the original orientation of the partial duplication of CHRNA7 may have been with CHRNA7 and CHRFAM7A in opposite directions. The inversion event reoriented them so that they are in the same direction. This inversion would have been accompanied by, or shortly followed by the emergence of the 2 bp polymorphism.
It has been hypothesized that chromosomes that are missing CHRFAM7A (allele 3) represent an ancestral structure that existed before the partial duplication of CHRNA7 (Flomen et al., 2008). Populations of African descent should have higher frequencies of allele 3 if this hypothesis is correct. The other alternative is that the absence of CHRFAM7A represents a later occurring deletion of CHRFAM7A. In this scenario, European populations would have higher frequencies of this allele. Since deletions in the 15q14 region are often associated with neuropsychiatric disorders, allele 3 would more likely be associated with schizophrenia. We did not find an increased risk of schizophrenia for subjects who were missing one or both copies of CHRFAM7A. African Americans were, however, more likely to be carrying this allele. These findings support the hypothesis that Allele 3 is likely to represent an ancestral allele rather than the deletion of CHRFAM7A.
In summary, we found an association of the 2 bp polymorphism in the CHRFAM7A gene in the 15q13 region with schizophrenia, but not with the P50 gating deficit endophenotype. Thus the presence of the 2 bp polymorphism may be a modest genetic risk factor for the development of schizophrenia. The relative allele frequencies in African Americans vs. Europeans suggest that the 2 bp polymorphism occurred after populations had migrated from Africa. In addition, absence of CHRFAM7A is likely to represent an ancestral allele that existed before the partial duplication of CHRNA7 and therefore may not represent a deletion event.
4. Experimental Procedures
4.1. Subjects
Samples from a total of 687 subjects were used in this study (See Table 4). There were 317 Caucasian subjects with schizophrenia and 192 control subjects, 66 African American subjects with schizophrenia and 60 controls, and 13 Hispanic subjects with schizophrenia and 37 controls. A consensus diagnosis of schizophrenia based on DSM-IIIR or DSM-IV criteria was made following a systematic and comprehensive examination of multiple sources of available information obtained from relatives, medical records, clinicians, and direct assessment using one or more diagnostic interviews including the Diagnostic Interview for Genetic Studies (DIGS), the Structured Clinical Interview for Axis I DSM Disorders (SCID), or the Kiddie-Schedule for Affective Disorders and Schizophrenia (K-SADS). Subjects with a diagnosis of schizotypal personality disorder, as well as those with schizoaffective, schizophreniform, and bipolar disorders were excluded from this sample. Control subjects were evaluated using the SCID-Non-patient Edition. All subjects in the study were unrelated to each other.
Table 4.
Demographic characteristics of the study population
| African American | Schizophrenia | 66 |
| Control | 60 | |
| Total | 126 | |
| Caucasian | Schizophrenia | 317 |
| Control | 192 | |
| Total | 509 | |
| Hispanic | Schizophrenia | 13 |
| Control | 37 | |
| Total | 50 |
Numbers of subjects with schizophrenia and controls for the three ethnic groups in the study.
4.2. Genomic DNA Isolation
Genomic DNA used in this study was isolated from brain, fibroblasts or lymphoblasts. Lymphoblast and fibroblast DNA was extracted using a salting-out method as described previously. DNA from human brain was isolated using a standard phenol extraction protocol (Sambrook and Russell, 2001). One hundred and ten individual DNA samples were obtained from the NIMH Genetics in Schizophrenia Initiative collection (http://nimhgenetics.org).
4.3. Genotype Detection
In order to determine genotypes, it was necessary to evaluate both copy number and 2 bp polymorphism status. Three amplicons were designed to determine the copy number of CHRFAM7A and infer the presence or absence of the 2 bp polymorphism. Primers were synthesized in the laboratory using a DNA Synthesizer (ABI, Foster City, CA). All primers and thermal profiles for PCR are shown in Table 5.
Table 5.
Amplicons used in the current study
| Amplicon | Size (bp) | Primers | Thermal Profile |
|---|---|---|---|
| Amplicon 1 | 165 or 167 bp | 5′-ttcaaggagttcctgctaca-3′ 5′-cattctccattggggatat-3′ |
95°C/10 min, (94°C 20 sec, 56°C/1 min 72°C/45 sec)×35, 72°C/10 min, and then 4°C |
| Amplicon 2 | 155 bp | 5′-tgtcaccttcacagtgacca-3′ 5′-agacactggggcgctta-3′ |
95°C/10 min, (94°C 20 sec, 56°C/1 min 72°C/45 sec)×35, 72°C/10 min, and then 4°C |
| Amplicon 3 | 117 bp | 5′-agtaatagtgtaatactgtaactttaaaatgtgttacttgt-3′ 5′-agccgggatggtctcgat-3′ |
95°C/1.5 min, (94°C/15 sec, 58°/30 sec, 72°/30 sec)×40, 95×/1 min, 60°/1 min |
| β2 microglobulin | 82 bp | 5′-tgggtttcatccatccgacatt-3′ 5′-agacaagtctgaatgctccactttt-3′ |
95°C/1.5 min, (94°C/15 sec, 58°/30 sec, 72°/30 sec)×40, 95°/1 min, 60°/1 min |
4.4. Detection of the 2 bp polymorphism
The WAVE dHPLC instrument (Trangenomic, Inc., Omaha, NE) was utilized for detection of the 2 bp polymorphism. Amplicon 1 was generated to determine the status of this polymorphism. PCR primers were designed to amplify a product from exon 6 spanning the 2 bp polymorphism. A 167 bp product was obtained from genomic targets that did not contain the 2 bp polymorphism. If the 2 bp polymorphism was present, 165 bp products were generated.
PCR reaction conditions were as follows: 500 nM of each primer, 150μM dNTPs, 2 mM MgCl2, 1X Taq Gold Buffer and 2.5 U Taq Gold DNA polymerase (Perkin Elmer, Branchburg, NJ). 150–200 ng of genomic DNA was added and the tubes were briefly spun at 1000×g and then placed in a GeneAmp 9600 thermal cycler (ABI). Inspection of PCR products on a 1.5–2% agarose gel containing ethidium bromide revealed only one visible major band of the correct size for Amplicons 1 and 2. PCR reactions lacking genomic DNA were run as a negative control.
PCR products were eluted from the WAVE using a buffer gradient of increasing concentrations of acetonitrile. Initial WAVE mobile phase buffer concentrations were 52% Buffer A (0.1M triethylammonium acetate (TEAA), pH 7.0) and 48% Buffer B (0.1M TEAA, 25% acetonitrile, pH 7.0). At 0.5 minutes after injection, a mobile phase gradient was initiated with 47% Buffer A and 53% Buffer B. The final gradient was 37.5% Buffer A and 62.5% Buffer B at 5 minutes. All Amplicon 1 samples were run at 50°C and 0.9 ml/minute. The presence of the 2 bp polymorphism was detected by the appearance of two major peaks at approximately 165 and 167 bp, as referenced to a PUC DNA sizing standard (Transgenomic). Quantification of CHRFAM7A alleles was calculated as a ratio of the 165 bp UV absorbance peak to the 167 bp absorbance peak.
4.5. Detection of CHRFAM7A copy number
Four Methods were used to determine CHRFAM7A copy number.
4.5.1. Southern Blotting
Southern blot analysis was performed as described previously (Gault et al., 1998). Briefly, the deletion of theCHRFAM7A locus was visualized by hybridization of a [32P]-dCTP radiolabeled exon 5 probe (Amersham) to RsaI restriction enzyme fragments (Biolabs, Beverly, MA). An RsaI polymorphism in intron 4 results in exon 5 restriction fragments of 1094 bp for the CHRNA7 gene, and 768 bp for CHRFAM7A. The exon 5 probe is completely contained within each restriction fragment. The relative abundance of each band was used to score the individual for CHRFAM7A deletion genotype.
4.5.2. Fluorescent In Situ Hybridization (FISH)
FISH was used to confirm the deletion of the CHRFAM7A locus. FISH was performed using standard techniques. Cultured lymphoblasts were arrested in interphase and hybridized with P1 artificial chromosome (PAC) clones obtained from Genome Systems, Incyte Corp., Palo Alto, CA. PAC 10905 hybridizes with exons 1–4 and was labeled with Spectrum Orange fluorescent dye using nick translation. PAC 10743 hybridizes with exons 6–10 and was labeled with Spectrum Green. The labeled PAC clones were mixed with Cot-1 DNA to block repetitive sequences and placed on a slide containing an interphase chromosome spread from cultured lymphoblasts. The slides were simultaneously denatured and hybridized overnight using the Vysis HYBrite instrument (Vysis Inc., Downer’s Grove, IL). Following high stringency washes the slides were counterstained with DAPI to identify the chromosomes in the background. Analysis of the slides was performed using a Zeiss Axiophot microscope equipped with a cooled CCD camera (KAF 1400, Photometrics, Tucson, AZ) and IP Lab Spectrum (Signal Analytics, Vienna, VA) or Quips mFISH Software (Vysis). A minimum of 30 interphase nuclei were analyzed per sample.
4.5.3. dHPLC analysis of copy number
Amplicon 2 was designed to determine the copy number of CHRFAM7A via dHPLC. For this amplicon, we utilized a highly prevalent and informative polymorphism, a G to A transition at position 690 located in exon 7 that is polymorphic in both CHRFAM7A and CHRNA7. The 690 A allele is found at a higher frequency in CHRFAM7A but is very rare in CHRNA7 (Gault et al., 2003). PCR reaction conditions were the same as for amplicon 1.
Amplicon 2 spans the intron 6/exon 7 junction of both genes. Primers for amplicon 2 amplify 155 bp products from both gene loci. Amplicon 2 samples were heated to 95°C for 5 minutes and allowed to cool to 25°C over a 45 minutes period to generate both homo- and heteroduplexes prior to loading on the WAVE™. The conditions used to resolve distinct genotype patterns were empirically derived based on initial recommendations obtained from WAVEMAKER™ (Transgenomic) software. Total run time was 7.2 minutes at 64.0°C at 0.9 ml/minute. Start gradient was initiated at 0.5 minutes with 47% Buffer B. Run stop time was 5.5 minutes at 56% Buffer B. WAVEMAKER™ software was used to analyze all WAVE™ chromatograms. Genotype was determined by qualitative assessment of the WAVE™ chromatograms based on internal control genomic DNA. The presence of the A allele indicates a copy of CHRFAM7A.
4.5.4. RT-PCR analysis of copy number
Approximately 10–15% of individuals could not be genotyped using amplicons 1 and 2 due to ambiguities in the WAVE chromatograms. In these individuals, copy number of CHRFAM7A was determined with RT-PCR using amplicon 3. Amplicon 3 spans the 5′ breakpoint of CHRFAM7A where intron 4 sequence diverges into intron A. Amplicon 3 was also used to confirm that all homozygous deleted individuals were detected.
A SYBR green assay was used. Ct values were determined in triplicate for the CHRFAM7A specific assay and compared with Ct values for an endogenous control, the beta-2-microglobulin gene. SYBR green results were interpreted using values corresponding to 0, 1, or 2 copies of CHRFAM7A which were determined empirically using 107 samples of known copy number previously determined by Southern Blot. The PCR efficiency of amplicon 3 was 100.5% and the correlation coefficient for the standard curve was 0.989. For the beta-2-microglobulin amplicon, PCR efficiency was 101.6% and the correlation coefficient was 0.998. All samples were within the range of the standard curve.
4.5.5. P50 Recordings
Measurement of the P50 ratio of the test to conditioning amplitudes was previously described (Freedman et al., 1991; Leonard et al., 2002). One hundred and sixty nine unrelated control subjects with no history of mental illness, recorded in this previous study, were genotyped for CHRFAM7A copy number and the 2 bp polymorphism.
4.5.6. Statistical Analysis
For the purpose of analysis, we designated the absence of CHRFAM7A and the 2 bp polymorphism each as alleles of CHRFAM7A, giving rise to a genetic model with 3 alleles and 6 genotypes. The wild-type gene was designated Allele 1, the 2 bp polymorphism was designated as Allele 2, and an absent CHRFAM7A locus was designated as Allele 3.
In order to determine if there are significant differences in CHRFAM7A allele frequencies between the three ethnic groups, population allele frequency comparisons were made by performing Chi Square tests for proportions among unrelated controls of the three groups.
Associations of CHRFAM7A copy number and the 2 bp polymorphism with schizophrenia were done by case-control comparisons using Chi-square tests. If sample sizes were small, Fisher-Freeman-Halton exact tests for proportions were used.
Significant nominal p values were further evaluated by the Monte Carlo simulation method and empirical p values were generated following a minimum of 10,000 permutations. Monte Carlo simulations were performed using the CLUMP software program (Sham and Curtis, 1995). All p values presented in this study represent empirical p-values derived from the Monte Carlo simulations. Ethnically matched, unrelated controls were used for assessment of preferential association of either CHRFAM7A copy number or the 2 bp polymorphism with schizophrenia. Probands were unrelated and family members, with or without schizophrenia, were excluded from the study. To evaluate association with the P50 gating deficit, P50 data from unrelated control individuals were compared. Logistic regression analysis was performed to determine appropriate groupings for the P50 data, and Chi-square tests were used to determine if there were significant differences in CHRFAM7A allele frequencies among the P50 groups. Analyses were performed using SAS software (SAS, Cary, NC).
Acknowledgments
T32 MH15442 to MLS; AA013973 to MLL; NIDA DA09457, NIMH MH81177, and The Veterans Affairs Medical Research Service to SL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler LE, Olincy A, Waldo MC, Harris JG, Griffith J, Stevens K, Flach K, Nagamoto H, Bickford P, Leonard S, Freedman R. Schizophrenia, sensory gating, and nicotinic receptors. Schiz Bull. 1998;24:189–202. doi: 10.1093/oxfordjournals.schbul.a033320. [DOI] [PubMed] [Google Scholar]
- Baron M. Genetics of schizophrenia and the new millennium: Progress and pitfalls. Am J Hum Gen. 2001;68:299–312. doi: 10.1086/318212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman II, Farmer AE, McGuffin P, Reveley AM, Murray RM. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Archives of General Psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- Chen D, Dang H, Patrick JW. Contributions of N-linked glycosylation to the expression of a functional alpha7-nicotinic receptor in Xenopus oocytes. J Neurochem. 1998;70:349–357. doi: 10.1046/j.1471-4159.1998.70010349.x. [DOI] [PubMed] [Google Scholar]
- Christian S, Bhatt NK, Martin SA, Sutcliffe JS, Kubota T, Huang B, Mutirangura A, Chinault AC, Beaudet AL, Ledbetter DH. Integrated YAC contig map of the Prader-Willi/Angelman region on chromosome 15q11–q13 with average STS spacing of 35 kb. Genome Res. 1998;8:146–157. doi: 10.1101/gr.8.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court J, Spurden D, Lloyd S, McKeith I, Ballard C, Cairns N, Kerwin R, Perry R, Perry E. Neuronal nicotinic receptors in dementia with Lewy bodies and schizophrenia: alpha-bungarotoxin and nicotine binding in the thalamus. J Neurochem. 1999;73:1590–1597. doi: 10.1046/j.1471-4159.1999.0731590.x. [DOI] [PubMed] [Google Scholar]
- Flomen RH, Collier DA, Osborne S, Munro J, Breen G, St Clair D, Makoff AJ. Association study of CHRFAM7A copy number and 2 bp deletion polymorphisms with schizophrenia and bipolar affective disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2006;141B:571–575. doi: 10.1002/ajmg.b.30306. [DOI] [PubMed] [Google Scholar]
- Flomen RH, Davies AF, Di Forti M, Cascia CL, Mackie-Ogilvie C, Murray R, Makoff AJ. The copy number variant involving part of the alpha7 nicotinic receptor gene contains a polymorphic inversion. Eur J Hum Genet. 2008;16:1364–1371. doi: 10.1038/ejhg.2008.112. [DOI] [PubMed] [Google Scholar]
- Freedman R, Coon H, Myles-Worsley M, Orr-Urtreger A, Olincy A, Davis A, Polymeropoulos M, Holik J, Hopkins J, Hoff M, Rosenthal J, Waldo MC, Reimherr F, Wender P, Yaw J, Young DA, Breese CR, Adams C, Patterson D, Adler LE, Kruglyak L, Leonard S, Byerley W. Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci U S A. 1997;94:587–592. doi: 10.1073/pnas.94.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman R, Hall M, Adler LE, Leonard S. Evidence in postmortem brain tissue for decreased numbers of hippocampal nicotinic receptors in schizophrenia. Biol Psychiatr. 1995;38:22–33. doi: 10.1016/0006-3223(94)00252-X. [DOI] [PubMed] [Google Scholar]
- Freedman R, Leonard S. Genetic linkage to schizophrenia at chromosome 15q14. Am J Med Gen. 2001;105:655–657. doi: 10.1002/ajmg.1548. [DOI] [PubMed] [Google Scholar]
- Freedman R, Leonard S, Gault JM, Hopkins J, Cloninger CR, Kaufmann CA, Tsuang MT, Farone SV, Malaspina D, Svrakic DM, Sanders A, Gejman P. Linkage disequilibrium for schizophrenia at the chromosome 15q13–14 locus of the alpha 7-nicotinic acetylcholine receptor subunit gene (CHRNA7) American Journal of Medical Genetics. 2001;105:20–22. [PubMed] [Google Scholar]
- Freedman R, Waldo M, Bickford-Wimer P, Nagamoto H. Elementary neuronal dysfunctions in schizophrenia. Schiz Res. 1991;4:233–243. doi: 10.1016/0920-9964(91)90035-p. [DOI] [PubMed] [Google Scholar]
- Gault J, Hopkins J, Berger R, Drebing C, Logel J, Walton K, Short M, Vianzon R, Olincy A, Ross RG, Adler LE, Freedman R, Leonard S. Comparison of polymorphisms in the α7 nicotinic receptor gene and its partial duplication in schizophrenic and control subjects. Am J Med Gen. 2003;123B:39–49. doi: 10.1002/ajmg.b.20061. [DOI] [PubMed] [Google Scholar]
- Gault J, Robinson M, Berger R, Drebing C, Logel J, Hopkins J, Moore T, Jacobs S, Meriwether J, Choi MJ, Kim EJ, Walton K, Buiting K, Davis A, Breese C, Freedman R, Leonard S. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics. 1998;52:173–185. doi: 10.1006/geno.1998.5363. [DOI] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, Badner JA, Cao QH, Zhang J. Linkage analysis of schizophrenia to chromosome 15. Am J Med Gen. 2001;105:789–793. doi: 10.1002/ajmg.1552. [DOI] [PubMed] [Google Scholar]
- Goder V, Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Guan ZZ, Zhang X, Blennow K, Nordberg A. Decreased protein level of nicotinic receptor alpha7 subunit in the frontal cortex from schizophrenic brain. Neuroreport. 1999;10:1779–1782. doi: 10.1097/00001756-199906030-00028. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Lai IC, Hong CJ, Tsai SJ. Association study of nicotinic-receptor variants and major depressive disorder. Journal of Affective Disorders. 2001;66:79–82. doi: 10.1016/s0165-0327(00)00292-5. [DOI] [PubMed] [Google Scholar]
- Leo J. Schizophrenia adoption studies. PLoS Med. 2006;3:e366. doi: 10.1371/journal.pmed.0030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S, Adler LE, Benhammou K, Berger R, Breese CR, Drebing C, Gault J, Lee MJ, Logel J, Olincy A, Ross RG, Stevens K, Sullivan B, Vianzon R, Vernich DE, Waldo M, Walton K, Freedman R. Smoking and mental illness. Pharmacol Biochem Behav. 2001;70:561–570. doi: 10.1016/s0091-3057(01)00677-3. [DOI] [PubMed] [Google Scholar]
- Leonard S, Gault J, Adams C, Breese CR, Rollins Y, Adler LE, Olincy A, Freedman R. Nicotinic receptors, smoking and schizophrenia. Restor Neurol Neurosci. 1998a;12:195–201. [PubMed] [Google Scholar]
- Leonard S, Gault J, Hopkins J, Logel J, Vianzon R, Short M, Drebing C, Berger R, Venn D, Sirota P, Zerbe G, Olincy A, Ross RG, Adler LE, Freedman R. Association of promoter variants in the alpha 7 nicotinic acetylcholine receptor subunit gene with an inhibitory deficit found in schizophrenia. Arch Gen Psychiatry. 2002;59:1085–1096. doi: 10.1001/archpsyc.59.12.1085. [DOI] [PubMed] [Google Scholar]
- Leonard S, Gault J, Moore T, Hopkins J, Robinson M, Olincy A, Adler LE, Cloninger CR, Kaufmann CA, Tsuang MT, Faraone SV, Malaspina D, Svrakic DM, Freedman R. Further investigation of a chromosome 15 locus in schizophrenia: analysis of affected sibpairs from the NIMH Genetics Initiative. Am J Med Genet. 1998b;81:308–312. doi: 10.1002/(sici)1096-8628(19980710)81:4<308::aid-ajmg6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Linthorst AC, Reul JM. Stress and the brain: solving the puzzle using microdialysis. Pharmacol Biochem Behav. 2008;90:163–173. doi: 10.1016/j.pbb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Liou YJ, Lai IC, Hong CJ, Liu HC, Liu TY, Tsai SJ. Association analysis of the partially duplicated alpha7 nicotinic acetylcholine receptor genetic variant and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2001;12:301–304. doi: 10.1159/000051273. [DOI] [PubMed] [Google Scholar]
- Liu CM, Hwu HG, Lin MW, Ou-Yang WC, Lee SFC, Fann CSJ, Wong SH, Hsieh SH. Suggestive evidence for linkage of schizophrenia to markers at chromosome 15q13–14 in Taiwanese families. Am J Med Gen. 2001;105:658–661. doi: 10.1002/ajmg.1547. [DOI] [PubMed] [Google Scholar]
- Locke DP, Archidiacono N, Misceo D, Cardone MF, Deschamps S, Roe B, Rocchi M, Eichler EE. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 2003;4:R50. doi: 10.1186/gb-2003-4-8-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G, Kast P. UGA read-through artifacts--when popular gene expression systems need a pATCH. BioTechniques. 1998;24:789–794. doi: 10.2144/98245st02. [DOI] [PubMed] [Google Scholar]
- Marutle A, Zhang X, Court J, Piggott M, Johnson M, Perry R, Perry E, Nordberg A. Laminar distribution of nicotinic receptor subtypes in cortical regions in schizophrenia. J Chem Neuroanat 4. 2001;22:115–126. doi: 10.1016/s0891-0618(01)00117-x. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Bassett AS, Honer WG, Lang D, King NA, Kennedy JL. No evidence for linkage of the CHRNA7 gene region in Canadian schizophrenia families. Am J Med Gen. 1998;81:361–363. doi: 10.1002/(sici)1096-8628(19980907)81:5<361::aid-ajmg3>3.0.co;2-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux G, Bonnet-Brilhault F, Louchart S, Houy E, Gantier R, Levillain D, Allio G, Haouzir S, Petit M, Martinez M, Frebourg T, Thibaut F, Campion D. The-2 bp deletion in exon 6 of the ‘alpha 7-like’ nicotinic receptor subunit gene is a risk factor for the P50 sensory gating deficit. Molecular Psychiatry. 2002;7:1006–1011. doi: 10.1038/sj.mp.4001140. [DOI] [PubMed] [Google Scholar]
- Riley BP, Makoff AM, Magudi-Carter M, Jenkins TJ, Williamson R, Collier DA, Murray RM. Haplotype transmission disequilibrium and evidence for linkage of the CHRNA7 gene region to schizophrenia in Southern African Bantu families. Am J Med Gen. 2000a;96:196–201. doi: 10.1002/(sici)1096-8628(20000403)96:2<196::aid-ajmg15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Riley BP, Makoff AM, Mogudi-Carter M, Jenkins TJ, Williamson R, Collier DA, Murray RM. High marker-density analyses of the α7-nicotinic cholinergic receptor subunit (CHRNA7) gene region on chromosome 15q13–q14 and 5′ RACE cloning of fragments specific to CHRNA7 or its partial duplication. Schiz Res. 2000b;41:93. [Google Scholar]
- Robinson WP, Dutly F, Nicholls RD, Bernasconi F, Penaherrerra M, Michaelis RC, Abeliovich D, Schinzel AA. The mechanisms involved in formation of deletions and duplications of 15q11–q13. J Med Gen. 1998;35:130–136. doi: 10.1136/jmg.35.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press: Cold Spring Harbor, New York; 2001. [Google Scholar]
- Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De GM, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K, Kahn RS, Linszen DH, van Os J, Wiersma D, Bruggeman R, Cahn W, de Haan L, Krabbendam L, Myin-Germeys I. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SH, Logel J, Barton A, Franks A, Schultz J, Short M, Dickenson J, James B, Fingerlin TE, Wagner B, Hodgkinson C, Graw S, Ross RG, Freedman R, Leonard S. Association of the 5′-upstream regulatory region of the alpha7 nicotinic acetylcholine receptor subunit gene (CHRNA7) with schizophrenia. Schizophr Res. 2009;109:102–112. doi: 10.1016/j.schres.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang DW, Skol AD, Faraone SV, Bingham S, Young KA, Prabhudesai S, Haverstock SL, Mena F, Menon AS, Bisset D, Pepple J, Sauter F, Baldwin C, Weiss D, Collins J, Boehnke M, Schellenberg GD, Tsuang MT. Examination of genetic linkage of chromosome 15 to schizophrenia in a large veterans affairs cooperative study sample. Am J Med Gen. 2001;105:662–668. [PubMed] [Google Scholar]
- Wynne LC, Tienari P, Nieminen P, Sorri A, Lahti I, Moring J, Naarala M, Laksy K, Wahlberg KE, Miettunen J. I. Genotype-environment interaction in the schizophrenia spectrum: genetic liability and global family ratings in the Finnish Adoption Study. Fam Process. 2006;45:419–434. doi: 10.1111/j.1545-5300.2006.00180.x. [DOI] [PubMed] [Google Scholar]
- Xu JZ, Pato MT, Dalla Torre C, Medeiros H, Carvalho C, Basile VS, Bauer A, Dourado A, Valente J, Soares MJ, Macedo AA, Coelho I, Ferreira CP, Azevedo MH, Macciardi F, Kennedy JL, Pato CN. Evidence for linkage disequilibrium between the alpha 7-nicotinic receptor gene (CHRNA7) locus and schizophrenia in Azorean families. Am J Med Gen. 2001;105:669–674. doi: 10.1002/ajmg.1549. [DOI] [PubMed] [Google Scholar]