Abstract
Transcriptional regulation of human iNOS (hiNOS) gene is highly complex and requires an orchestrated flow of positive and negative transcription factors that bind to specific cis-acting upstream response elements. Very little specific information exists about the far upstream region of the hiNOS gene. Oct-1 protein belongs to the POU domain transcription factor family and is constitutively expressed in all dividing cells. It's essential for proliferation, differentiation and other key cell processes. However, the role of Oct-1 in regulating human iNOS gene expression has not been reported. In this work, the octamer sequence 5′-ATGCAAAT-3′ at -10.2 kb in the hiNOS promoter was identified as high-affinity Oct-1 binding by electrophoretic mobility shift assay (EMSA) in vitro and chromatin immunoprecipitation (ChIP) assay in vivo. Mutation of Oct-1 motif at -10.2 kb in the hiNOS promoter decreased cytokine-induced hiNOS promoter activity by 40%. Cytokine-induced hiNOS promoter activity was also significantly reduced by Oct-1 siRNA targeting. Overexpression of Oct-1 increased cytokine-induced hiNOS protein expression in primary human hepatocytes. Furthermore, the Oct-1 motif at -10.2 kb of the hiNOS promoter conferred increased transcriptional activity to the heterologous thymidine kinase (TK) promoter irrespective of cytokine induction. Taken together, this work identifies a far-upstream functional Oct-1 enhancer motif at -10.2 kb in the hiNOS promoter that regulates cytokine-induced hiNOS gene transcription, and further underscores the tight control mechanisms regulating expression of the human iNOS gene.
Keywords: Nitric oxide (NO), iNOS (NOS2), transcription factor, Oct-1, POU
Introduction
Nitric oxide (NO) has diverse roles as a vasodilator, neurotransmitter, antimicrobial effector molecule, and immunomodulator. Nitric oxide (NO) is produced at high levels by inducible nitric oxide synthase (iNOS) during septic and inflammatory conditions. It can result in either protective or damaging effects in clinical diseases depending on the physiological or pathophysiological state.1,2
Expression of iNOS gene was initially reported in murine macrophages,3 and the murine iNOS cDNA was cloned from lipopolysaccharide (LPS) and IFNγ-stimulated macrophages.4,5,6 The iNOS gene is typically not expressed in resting cells, but activated by various infectious agents, microbial products, or cytokines.7,8 Human iNOS (hiNOS) expression in primary human hepatocytes was originally identified by stimulating with a cytokine mixture (CM) of TNF-α,IL-1β, IFN-γ, and LPS,9,10 and the hiNOS cDNA was cloned from LPS and cytokine-stimulated primary human hepatocytes.11
Regulation of hiNOS gene expression is mainly at the transcriptional level, although post-transcriptional mechanisms are also active.10,12,13,14,15 We reported that the 5′-flanking promoter region of the hiNOS gene as far as -16 kb upstream was necessary for maximal cytokine inducibility.10,16 This contrasts with the murine iNOS gene where the LPS and cytokine-inducible most transcription factors are acting within the proximal -1.0 kb region in the murine iNOS promoters.17,18
A few studies have elucidated mechanisms of how some transcription factors regulate the hiNOS promoter. TNF-α or IL-1β signal through NF-κB binding to −5.5, −5.8, and −6.1 kb cis-acting DNA elements, while IFN-γ signals through Stat-1 by binding to motifs at −5.2 and −5.8 kb in the hiNOS promoter.16,19,20 Another group showed that cytokine-responsiveness required 5′-flanking DNA region extending to −8 kb and demonstrated CM-inducible activating protein (AP)-1 binding sites at −5.1 and −5.3 as well as a functional role for a NF-κB element located at −8.2 kb in the hiNOS promoter.21 However, NF-κB repressing factor (NRF) protein binding to a novel negative response element (NRE) at −6.7 kb in the hiNOS promoter was recognized to mediate constitutive silencing of hiNOS transcription.22 AABS response element exhibited a dual role in regulating hiNOS gene expression. The AABS site at -0.19 kb in the hiNOS promoter region mediated basal hiNOS transcription in a tissue-specific pattern in liver cells, and it also functioned as a “switch point” for cytokine inducibility where C/EBPβ (LAP) binding to an intact AABS was necessary, but not sufficient, for inducible promoter activity.23 Additional chromatin-based mechanisms in the control of hiNOS gene transcription include DNA methylation of CpG dinucleotides.24 Recently, β-catenin/TCF-4 binding in vivo to two TCF-4 binding element (TBE) motifs at −3.8 and −6.1 kb in the hiNOS promoter has been reported.25
The octamer motif has been reported to be an important regulatory sequence in many promoters. The octamer-binding transcription factors Oct-1 and Oct-2 belong to the POU (Pit-Oct-Unc) domain family of transcription factors,26 exerting their effects through binding to sequences related to the octamer element (5′-ATGCAAAT-3′).27 While Oct-1 is ubiquitously expressed in all dividing cells, a smaller protein, Oct-2, is found predominantly in B lymphocytes, macrophages, and other hematopoietic cells, as well as cells of the central nervous system.28,29,30 Even though Oct-1 and Oct-2 recognize the same sequence with equal affinity in vitro,31 they regulate different sets of genes.32 Target genes controlled by Oct-1 include those encoding snRNAs,33 histone H2B,34 Pit-1,35 Oct-1,36 and p15INK4b.37 Oct-2 activates B cell specific mRNA promoters38 with a B-cell-specific cofactor (OCA-B/Bob-1/OBF-1).39,40,41
Since the role of Oct-1 in regulating hiNOS gene transcription was unknown, in the present work we examined the 5′-flanking sequence for potential cis-acting Oct-1 binding elements. We identify a far upstream Oct-1 motif at -10.2 kb in the hiNOS promoter that exerts an important role for cytokine-induced hiNOS gene expression.
Results and Discussion
The potential Oct-1 binding elements on the hiNOS promoter are located
The human iNOS promoter sequence (GenBank Accession #: AF049872) was analyzed for binding sites of transcription factors including Oct-1 using Transcription Element Search Software (TESS) from the University of Pennsylvania. We identified one perfectly matched 8/8 nucleotide Oct-1 consensus sequence (5′-ATGCAAAT-3′) at -10,239 bp (Figure 1). Allowing for one mismatched nucleotide (7/8), there were an additional eight potential consensus Oct-1 response elements at -13,147, -12,914, -11,644, -10,611, -10,134, -9,103, -5,606, and -671 bp, respectively, from the transcription initiation site (Figure 1).
Figure 1.
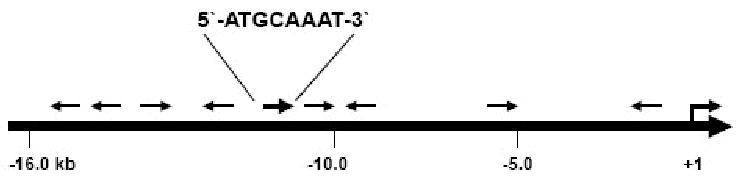
The location of potential Oct-1 binding elements on the hiNOS promoter. The hiNOS promoter sequence (GenBank Accession #: AF049872) was analyzed for binding sites of transcription factors including Oct-1 using Transcription Element Search Software from the University of Pennsylvania. Oct-1 consensus (8/8) sequence (ATGCAAAT) is shown at −10.2 kb and all the other sites contain one mismatched base out of 8 nucleotide consensus Oct-1 response element. The arrow signs stand for the predicted Oct-1 binding sites with their corresponding orientation.
Oct-1 specifically binds to oligonucleotides containing the Oct-1 motif at -10.2 kb of hiNOS promoter in vitro
To examine whether the Oct-1 protein can interact with the potential Oct-1 binding site at -10.2 kb of hiNOS in vitro, we carried out EMSAs with nuclear extracts from HepG2 cells. We compared Oct-1 protein/DNA-binding using a consensus Oct-1 oligonucleotide (Santa Cruz, CA) with the human iNOS-specific Oct-1 motif at -10.2 kb. We also synthesized mutant oligonucleotides for the human iNOS-specific -10.2 kb Oct-1 element as shown in Table 1. Strong Oct-1 protein/DNA binding was seen using the wild type Oct-1 consensus oligonucleotide as well as the human iNOS-specific Oct-1 motif at -10.2 kb (Figure 2A, lanes 2 and 9). Specificity of DNA binding was shown by cold competition with excess unlabelled oligonucleotides (lanes 3 and 10), and binding was restored when mutant Oct-1 oligonucleotide was used as competitor (lanes 4 and 11). The identity of the protein in the DNA complexes was confirmed by supershifts with two different Oct-1 antibodies (lanes 5, 6, 12, & 13). As negative control, antibodies against NF-κB p50 or C/EBPβ did not elicit a supershift. Finally, radiolabeled probe of mutant Oct-1 -10.2 kb oligo didn't show any distinct protein/DNA complexes, also confirming specificity of binding. We had similar gel shift results with nuclear extracts from several other human cell lines such as A549, HCT116, and DLD-1 (data not shown).
Table 1.
Consensus Oct-1 sequence and Oct-1 motif at -10.2 kb of hiNOS promoter for EMSA. Oct-1 binding element is shown in bold type and mutated nucleotides are underlined.
| Consensus Oct-1: | 5′ TGTCGAATGCAAATCACTAGAA 3′ |
| Mutant Oct-1: | 5′ TGTCGAATGCAAGCCACTAGAA 3′ |
| Oct-1 at −10.2kb: | 5′ GCTTGGGTATGCAAATGGCTGG 3′ |
| Mutant Oct-1: at −10.2kb | 5′ GCTTGGGTGCGCAAGCGGCTGG 3′ |
Figure 2.

Oct-1 binds to oligonucleotides containing the Oct-1 motif at −10.2 kb of hiNOS promoter. A) Nuclear extracts from HepG2 cells were incubated with 32P-labeled oligonucleotides from consensus Oct-1 oligonucleotides, oligonucleotides containing Oct-1 motif at -10.2 kb, or mutant Oct-1 motif at −10.2 kb of hiNOS promoter, followed by EMSA as described under Materials and Methods. B) ChIP assay was performed as described under Materials and Methods. Lane C, no DNA; lane I, input of genomic DNA; lane G, IgG; lane O, Oct-1 antibody; lane P, NF-κB p50 antibody.
Chromatin immunoprecipitation (ChIP) assay confirms Oct-1 specifically binds to the motif at -10.2 kb in the hiNOS promoter in vivo
To verify that the Oct-1 protein/DNA binding observed by EMSAs at -10.2 kb in the hiNOS promoter was also occurring in living cells, we performed ChIP assay in human A549 cells. A PCR band of expected 212 bp size was strongly seen with input genomic DNA (Figure 2B, lane I) and with immunoprecipitation by Oct-1 antibody (lane O), but not with NF-κB p50 antibody (lane P) or with control IgG antibody (lane G, weak band). As negative control, ChIP assay did not show Oct-1 binding when GAPDH was amplified (data not shown). These results confirm that the transcription factor Oct-1 associates with the Oct-1 motif at -10.2 kb of hiNOS promoter in vivo and is consistent with the in vitro EMSAs results.
Mutation of the Oct-1 motif at -10.2 kb of the hiNOS promoter decreases cytokine-inducible hiNOS promoter activity
Previously we have shown that maximal cytokine-induced hiNOS gene expression requires transcriptional elements extending to -16 kb in the far upstream region of the human iNOS promoter.10,16 Functional cytokine-induced NF-κB, Stat1, and AP-1 sites have been reported in the enhancer region from -5 to -6 kb in the hiNOS promoter.16,19,20 However, the role of the -10.2 kb Oct-1 element in regulating hiNOS transcription is completely unknown. To examine the effect of the -10.2 kb hiNOS-specific Oct-1 element on cytokine-induced hiNOS promoter activity, we transfected human A549 cells with hiNOS promoter – luciferase reporter constructs, and then stimulated with cytokines. As previously demonstrated, the -7.2 kb hiNOS promoter exhibited a ∼ 4-5 fold induction after cytokine stimulation (Figure 3), while the -16 kb construct containing the -10.2 kb Oct-1 element showed a 10-fold induction in cytokine-stimulated hiNOS promoter activity. Mutation of the Oct-1 motif at -10.2 kb in the context of the -16 kb promoter construct showed a significant 40% decrease in cytokine inducibility compared to the wild type promoter activity. This is important, as previously the crucial cytokine-mediated regulatory elements were shown to reside in the -5 to -6 kb enhancer region, and indicates that an intact far upstream Oct-1 response element is required for maximal hiNOS gene transcription. Interestingly, mutation of the -10.2 kb Oct-1 element in the -16kb hiNOS promoter construct did not influence basal promoter activity. Taken together with the gel shift and ChIP data, these results suggest that this Oct-1 motif is required for maximal cytokine induction, but is not essential for basal transcriptional activity.
Figure 3.

Mutation of Oct-1 motif at -10.2 kb of hiNOS promoter decreases the promoter activities by CM. A549 cells were transiently co-transfected with luciferase reporter plasmid containing -7.2kb, -16kb, or -16kb mutant (mutant Oct-1 at -10.2 kb) of hiNOS promoter. Following 48 hours of incubation, cell lysates were prepared for luciferase assay after 8-hour treatment of CM. (* indicates p < 0.05 vs. -16kb.)
Oct-1 silencing decreases hiNOS promoter activity and also down-regulates hiNOS mRNA and protein expression
To further demonstrate a functional role for Oct-1 binding in regulating hiNOS mRNA and protein expression, we performed siRNA experiments targeting Oct-1 itself. Transient transfection of A549 cells with Oct-1 siRNA (Santa Cruz) decreased Oct-1 mRNA (Figure 4B) and Oct-1 protein (Figure 4C) in a cytokine-independent manner. Control (scrambled) siRNA did not affect Oct-1 mRNA levels. Co-transfection of Oct-1 siRNA with the -16kb hiNOS promoter reporter plasmid decreased cytokine-induced hiNOS promoter activity by ∼40% compared to control siRNA (Figure 4A), indicating that Oct-1 silencing significantly decreased hiNOS transcriptional activity to the same magnitude that mutation of the -10.2 kb Oct-1 element elicited. Oct-1 siRNA silencing did not affect hiNOS promoter activity with the -7.2 kb hiNOS promoter construct, which is consistent with the notion that the critical Oct-1 binding occurs further upstream at -10.2 kb. In addition to decreasing cytokine-stimulated hiNOS promoter activity, Oct-1 silencing also decreased hiNOS mRNA and protein levels (Figure 4B and 4C), confirming a functional role for Oct-1 in the regulation of cytokine-induced hiNOS gene expression.
Figure 4.

Oct-1 silencing down-regulates hiNOS mRNA and protein expression. A) A549 cells were transiently co-transfected with luciferase reporter plasmid containing -7.2 or -16 kb of hiNOS promoter and either control (scrambled) siRNA or Oct-1 siRNA. Following 48 hours of incubation, cell lysates were prepared for luciferase assay as control (open bars) or after 8-hour of CM treatment (closed bars). B/C) A549 cells were transfected with either control (scrambled) siRNA, Oct-1 siRNA, or no transfection. After 48 hour incubation, total RNA and protein extract were prepared after 8-hour CM treatment. (* indicates p<0.05 vs. -16 kb Control.)
Overexpression of Oct-1 enhances hiNOS protein expression
Since Oct-1 was found to have an important role in regulating cytokine-induction of hiNOS gene expression, we addressed the question of whether Oct-1 over-expression could “super-drive” hiNOS gene expression and subsequent hiNOS protein levels. In these experiments, we utilized fresh primary human hepatocytes as we noted a low basal level of Oct-1 mRNA (Figure 5A) compared to the A549 cell line (Figure 4B). Infection of the primary human hepatocytes with adenovirus expressing Oct-1 (Ad-Oct-1) (Vector Biolabs, Philadelphia, PA), but not empty vector Ad-Ψ5, resulted in high levels of Oct-1 mRNA (Figure 5A). The human hepatocytes transduced to over-express Oct-1 showed greater cytokine-induced hiNOS protein compared to control transduction with empty adenovirus (Figure 5A and 5B, lanes 5 & 6 vs lanes 2 & 3).
Figure 5.
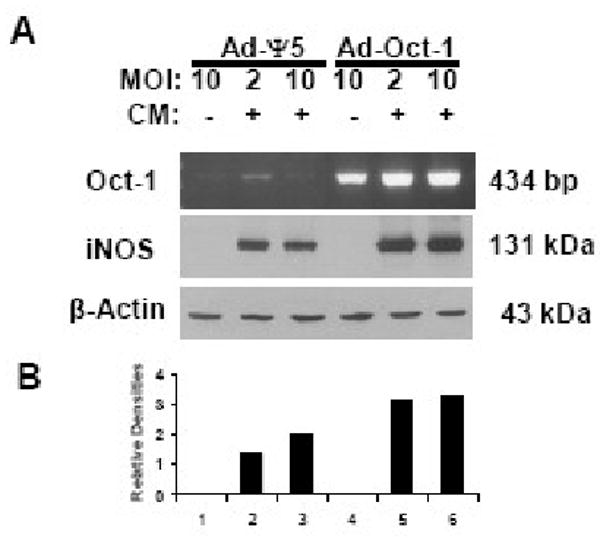
Overexpression of Oct-1 increases hiNOS protein expression in human hepatocytes. A) Human hepatocytes were infected with control virus Ad-Ψ5, or Ad-Oct-1 with different MOIs for 1 day and then treated with CM for 6 (RT-PCR) or 8 hours (Western blot). Total RNAs were prepared for RT-PCR and the whole cell extracts for western blot with anti-iNOS antibody or anti-Actin antibody. B) Relative densities of iNOS expression are corrected against Actin levels.
In resting human hepatocytes, basal hiNOS protein was not detectable by western blot (Figure 4A), and Ad-Oct-1 over-expression did not trigger basal hiNOS protein detection, consistent with our earlier results showing a lack of effect of mutation of the endogenous -10.2 kb Oct-1 element on basal hiNOS promoter activity or the inability of Oct-1 siRNA silencing to decrease basal hiNOS promoter activity.
Oct-1 motif at -10.2 kb of hiNOS promoter confers transcriptional activation to a heterologous thymidine kinase (TK) promoter
Since the -10.2 kb Oct-1 hiNOS promoter element had a functional role in hiNOS gene expression, we next sought to determine if this Oct-1 response element could confer transcriptional activation to a heterologous promoter. We fused 3 copies of the wild-type or mutant -10.2 kb hiNOS promoter Oct-1 element in front of the minimal herpes thymidine kinase (TK) promoter. Transfection of HepG2 or A549 cells with the wild-type pTK3XOct-1 significantly increased transcriptional activation compared to the minimal TK vector (Figure 6), while transfection of 3 copies of the mutant Oct-1 motif was not different than the minimal TK construct. These results indicate that the -10.2 kb Oct-1 hiNOS promoter element can indeed confer transcriptional activation to a heterologous promoter.
Figure 6.

Oct-1 motif at -10.2 kb confers transcriptional activation to a heterologous thymidine kinase (TK) promoter. HepG2 or A549 cells were transiently transfected with a luciferase reporter gene construct driven by three copies of Oct-1 motif (pTK3XOct-1) or mutant Oct-1 motif at -10.2 kb (pTK3XMutOct-1) of hiNOS promoter. Luciferase activities were determined 1 day after transfection. (* indicates p<0.02 vs. empty vector. † indicates p<0.01 vs. pTK3XOct-1.)
Oct-1 binding and transcriptional activation are not affected by cytokines
Since the Oct-1 binding at -10.2 kb in the hiNOS promoter was required for maximal cytokine induction of the human iNOS gene, we next addressed whether cytokine stimulation actually influenced the degree of Oct-1 DNA-binding. EMSAs with nuclear extracts from resting or 6-hr cytokine (CM)-stimulated DLD-1 cells did not show a difference in the intensity of the protein-DNA complexes (Figure 7A), suggesting that Oct-1 DNA binding was independent of cytokine stimulation. Similar findings were observed for Oct-1 mediated transcriptional activation on the minimal heterologous TK reporter which occurred in a cytokine-independent manner (Figure 7B).
Figure 7.

Oct-1 binding and transcription control is not affected by CM. A) Nuclear extracts from resting or 6 hr CM-stimulated DLD-1 cells were incubated with 32P-labeled oligonucleotides from Oct-1 motif at -10.2 kb of hiNOS promoter, followed by EMSA as described under Materials and Methods. B)A549 cells were transiently transfected with the same plasmids as those of Figure 6. Luciferase activities were determined 1 day after transfection in resting or 8 hr CM-stimulated A549 cells. (* indicates p<0.02 vs. empty vector. † indicates p<0.01 vs. pTK3XOct-1.)
Previously we reported that promoter reporter plasmids containing -16 kb of the 5′- flanking upstream region of the hiNOS gene showed greater cytokine-induced transcriptional activity compared to smaller promoter constructs containing -7.2 or -5.8 kb regions.10,16 We and others have reported that hiNOS promoter has functionally active binding elements for NF-κB, Stat1, AP-1, NRE, KLF6, AABS, TBE1/TBE2, and other cis-acting DNA elements, which are mostly located within -7 kb from the transcription initiation site. The identification of active Oct-1 binding motif located at -10.2 kb in this study further expands the binding region of transcription factors that regulate hiNOS expression, and explains in part the increased cytokine inducibility seen when the promoter region from -7.2 kb to -16 kb is included in the cytokine-stimulated transfection experiments (Figure 3).
Several groups reported that the downstream octamer (Oct) motif in the murine iNOS promoter is required for iNOS activation by lipopolysaccharide (LPS),42 IL-6,43 IFNγ plus LPS,44 Brn-3,45 and with NF-κB and HMG-I(Y).46 The Oct-1 containing downstream murine promoter region was also implicated in repression by exogenous NO,47 triptolide,48 or alcohol.49 This work in the human iNOS promoter underscores the complexities and species-specific regulation of the human iNOS gene, which has been shown by several groups to require far upstream promoter binding regions for maximal transcriptional activation.
In this work, we showed for the first time that a canonical Oct-1 motif at -10.2 kb of the hiNOS promoter is binding with the Oct-1 protein in vitro and in vivo and mutation of the motif decreases the promoter activity by CM. This Oct-1 binding motif at -10.2 kb is one of the farthest functional upstream elements from the transcription initiation site. Silencing of Oct-1 by transfecting Oct-1 siRNA decreased cytokine-induced (but not basal) hiNOS promoter activity and hiNOS mRNA and protein levels. These data support that the Oct-1 motif at -10.2 kb of hiNOS promoter is functional and Oct-1 works as an enhancer in the hiNOS promoter with cytokine stimulation in human cell lines as well as primary human hepatocytes. The findings suggest that intact Oct-1 binding is necessary, but not sufficient for maximal cytokine-induced hiNOS transcription. Interestingly, Oct-1 motif at -10.2 kb inserted into the minimal herpes TK promoter conferred transcriptional activation independent of CM stimulation, suggesting that the Oct-1 binding may work directly to enhance transcription of a minimal promoter element, however it may also function cooperatively with other cytokine-induced transcription factor binding, as in the case with human iNOS.
Our in vivo ChIP data and in vitro mutagenesis studies clearly show that Oct-1 proteins directly interact with this far upstream site to transcriptionally activate the hiNOS gene. But we also cannot rule out that Oct-1 may regulate other genes to have an indirect effect on hiNOS gene expression in the Oct-1 overexpression or silencing experiments. From the chromatin structure analysis and in vivo footprinting for the human iNOS gene, CM inducible DNAse I hypersensitive sites were also identified in the far upstream region between -10.0 and -12.0 kb50, which correlates well with our Oct-1 motif at -10.2 kb. This far upstream 5′-flanking region might confer cytokine-inducibility and function as a cytokine-responsive enhancer to regulate human iNOS gene transcription from a relative distance. It is possible that Oct-1 located at these remote hypersensitive sites might form a long range DNA loop51 with essential regulatory elements in the downstream proximal hiNOS core promoter.
Materials and Methods
Cell lines and treatment
The human colorectal cancer cell lines DLD1 and HCT116, lung cancer cell line A549, and hepatoblastoma cell line HepG2 were obtained from the American Type Culture Collection (Manassas, VA) and cultured at 37°C in 5% CO2 in medium as recommended, respectively. A549 media contain 10% fetal bovine serum (HyClone, Logan, UT), 100 units/ml penicillin (invitrogen, Carlsbad, CA), 100 μg/ml streptomycin(invitrogen) and 15 mM of l M HEPES (LONZA, Walkersville, MD) in F-12 Kaighn's Modification, 1 × (invitrogen). HepG2 cells were cultured in DMEM medium (LONZA) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine (invitrogen), and 15 mM of l M HEPES. DLD-1 cells were cultured in RPMI-1640 medium (LONZA) with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM of l M HEPES. HCT116 cells were cultured in McCoy's 5A medium (invitrogen) with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM of l M HEPES. If necessary, cells were stimulated with a cytokine mixture (CM) consisting of 1,000 units/ml human tumor necrosis factor(TNF)-α (R&D Systems, Minneapolis, MN), 100 units/ml interleukin (IL)-1β (provided by C. Reynolds, National Cancer Institute, NIH, Bethesda, MD), and 250 units/ml human interferon (IFN)-γ (R&D Systems or Roche Pharmaceuticals, Nutley, NJ), which were purified recombinant proteins.
Hepatocyte isolation and culture
Human hepatocytes were isolated from histologically normal liver and were kindly provided by Drs. Steven Strom and Ken Dorko (University of Pittsburgh Core Pathology Facility, Pittsburgh, PA) according to an Institutional Review Board–approved protocol. Human hepatocytes were prepared by a three-step collagenase perfusion technique and cultured in William's medium E (invitrogen) supplemented with 5 % calf serum (HyClone), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 15 mM of l M HEPES.
Plasmids
The human iNOS promoter reporter plasmid piNOS(7.2)luc and piNOS(16.0)luc contain −7.2 kb and −16.0 kb, respectively, of upstream 5′-flanking DNA linked to the luciferase reporter gene and have been described earlier.10,22 Mutation of the putative −10.2 kb Oct-1 binding element was generated from the piNOS(16.0)luc reporter plasmid by using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's recommendation. The primer sequences are as followed. Forward: 5′-GAGTGGTGCTTGGGTGCGCAAGCGGCTGGGAGGATATCC-3′. Reverse: 5′GGATATCCTCCCAGCCGCTTGCGCACCCAAGCACCACTC-3′. The Oct-1 motif at -10.2 kb of hiNOS promoter is underlined and the mutated nucleotides are bold and Italic. Three copies of Oct-1 motif at −10.2 kb or mutant Oct-1 were inserted into pTK-luc, a plasmid carrying a minimal promoter fragment of the herpes thymidine kinase gene with luciferase reporter gene at a HindIII site to create different heterologous constructs in either orientation. The sequences are as followed.
WT forward: 5′-AGCTTATGCAAATGGCTGGGTATGCAAATGGCTGGGTATGCAAATGGA-3′.
WT reverse: 5′-AGCTTCCATTTGCATACCCAGCCATTTGCATACCCAGCCATTTGCATA-3′.
Mutant forward: 5′-AGCTTATATAAGTGGCTGGGTATATAAGTGGCTGGGTATATAAGTGGA-3′.
Mutant reverse: 5′-AGCTTCCACTTATATACCCAGCCACTTATATACCCAGCCACTTATATA-3′.
The Oct-1 motif at -10.2 kb of hiNOS promoter is underlined and the mutated nucleotides are bold and Italic. Confirmation of the recombinant constructs was accomplished with DNA sequencing analysis by the University of Pittsburgh Sequencing Facility.
Transient transfection and reporter activity assay
Transfections of cells were carried out in six-well plates (Corning, Corning, NY) by using Lipofectamine or Lipofectamine 2000 (invitrogen) as suggested. Cells were splitted the day before transfection and exposed to Opti-MEM-I (invitrogen) medium containing 1 μg of reporter DNA and 0.5 μg of pIEP-LacZ DNA which is a cytomegalovirus (CMV) promoter-driven β-galactosidase gene to control for transfection efficiency between groups, and three times volume (4.5 μl, volume (μl)/ mass(μg)) of Lipofectamine or Lipofectamine 2000 after cells were washed with 1 × PBS. After 5 hours in 37 °C, 5 % CO2 chamber, 2 × medium was added for overnight. Cells were provided with normal medium and CM for 8 hours if necessary. Cells were lysed with 200 μl/well of 1 × Reporter Lysis Buffer (Promega, Madison, WI) and luciferase activity was assayed with 20 μl each lysate using Luciferase Assay System(Promega) with AutoLumat LB 953 luminometer (Berthold; Nashua, NH, USA). β-galactosidase activity was determined as recommended (Promega), using a 96-well multiplate reader with SOFTMAX software (Molecular Devices, Sunnyvale, CA, USA). Luciferase activity was normalized to β-galactosidase activity.
Western blotting analysis
SDS-PAGE was done according to the method of Towbin et al.52 The following specific antibodies were used for immunodetection with appropriate dilutions. Oct-1 antibody was from Santa Cruz. Horseradish peroxidase-linked anti-mouse or rabbit IgG was used as a second antibody. Peroxidase activity was detected by enhanced chemiluminescence (Pierce, Rockford, IL).
EMSA
Preparation of nuclear extract from HepG2 cells and electrophoretic mobility shift assay (EMSA) were done as described earlier.19 Cells were grown in 10-cm dish and treated with CM for 2 hours. All steps were carried out at 4 °C or on ice. The nuclear protein extracts were frozen at −80 °C or directly used for EMSA after determining the protein concentration using BCA Protein Assay Reagent Kit (Pierce). Oct-1 consensus or mutant oligonucleotides were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA) and other oligonucleotides were synthesized from invitrogen in Table 1. Radioisotope-labeled probes were prepared by end labeling with [γ-32P]dATP (DuPont/NEN, Boston, MA) and T4 polynucleotide kinase (Roche, Indianapolis, IN) and purified in TEN (Tris-EDTA + NaCl) by using G-25 resin column (Amersham Biosciences, Little Chalfont, UK). Nuclear proteins (5 μg) were incubated with radiolabeled probe (∼50,000 cpm), buffer containing 10 mM TrisCl, pH 7.5, 10 % Glycerol, and 0.2% Nonidet P-40, 2 - 4 μg poly(dIdC) (Roche), and 100 molar excess of competitors or 2 μg of antibodies in 20 μl for 30 min at room temperature as indicated. The samples were electrophoresed on 5% polyacrylamide gel in 25 mM Tris, 25 mM boric acid, and 0.5 mM EDTA. After electrophoresis, gel was dried and exposed onto film for autoradiography.
RT-PCR analysis
Total RNAs were prepared with Trizol (invitrogen) solution from 10-cm culture dishes of A549 or DLD-1 cells. RT-PCR was performed using OneStep RT-PCR Kit from Qiagen Inc. (Valencia, CA) and PCR machine from Perkin-Elmer according to the manufacturer's instructions. Primer pairs for detecting hiNOS level were as followed: 5′-AGGACATCCTGCGGCAGC-3′ (forward) and 5′-GCTTTAACCCCTCCTGTA-3′ (reverse). Primers for Oct-1 and β-Actin were purchased from Santa Cruz and Clontech Inc. (Mountain View, CA), respectively. The PCR products were run on 1.5 % agarose gel.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed with ChIP-IT kit (Active Motif, Carlsbad, CA) following the manual. A549 cells in 15-cm dish were divided into two groups. One group was untreated with CM as control and the other one was treated with CM for 4 h. The pre-cleared chromatin was added 4 μg of IgG for negative control, Oct-1 antibody (Santa Cruz), or NF-κB p50 antibody (Santa Cruz), respectively and incubated for overnight on a rotator at 4°C. Polymerase chain reaction (PCR) was performed with specific primers for 30 cycles. Amplified DNA fragments are analyzed on a 1.2% agarose gel by electrophoresis. The sequences of primers are listed as followed:
Forward primer: 5′-CACTAGTTAGATGCTGCCCTCTC-3′;
Reverse primer: 5′-GACAGGGAAAGCCAGGCAAGAG-3′.
Statistical analysis
Results are given as means ± SEM. Multiple comparisons were performed by ANOVA, followed by group comparisons using the Student's unpaired t test with correction by the Bonferroni method. Comparisons vs. control were performed using the Student's paired t test.
Acknowledgments
Supported in part by NIH grants GM52021, and DK62313
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kroncke KD, Fehsel K, Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clin Exp Immunol. 1998;113:147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suschek CV, Schnorr O, Kolb-Bachofen V. The role of iNOS in chronic inflammatory processes in vivo: is it damage-promoting, protective, or active at all? Curr Mol Med. 2004;4:763–775. doi: 10.2174/1566524043359908. [DOI] [PubMed] [Google Scholar]
- 3.Stuehr DJ, Marletta MA. Mammalian nitrate biosynthesis: mouse macrophages produce nitrite and nitrate in response to Escherichia coli lipopolysaccharide. Proc Natl Acad Sci U S A. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowenstein CJ, Glatt CS, Bredt DS, Snyder SH. Cloned and expressed macrophage nitric oxide synthase contrasts with the brain enzyme. Proc Natl Acad Sci U S A. 1992;89:6711–6715. doi: 10.1073/pnas.89.15.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons CR, Orloff GJ, Cunningham JM. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J Biol Chem. 1992;267:6370–6374. [PubMed] [Google Scholar]
- 6.Xie Q, Cho HJ, Calaycay J, Mumford RA, Swiderek KM, Lee TD, Ding A, Troso T, Nathan C. Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 7.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 8.Taylor BS, Geller DA. Molecular regulation of the human inducible nitric oxide synthase (iNOS) gene (review) Shock. 2000;13:413–424. doi: 10.1097/00024382-200006000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Nussler AK, DiSilvio M, Billiar TR, Hoffman RA, Geller DA, Selby R, Madariaga J, Simmons RL. Stimulation of the nitric oxide pathway in human hepatocytes by cytokines and endotoxin. J Exp Med. 1992;176:261–264. doi: 10.1084/jem.176.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vera ME, Shapiro RA, Nussler AK, Mudgett JS, Simmons RL, Morris SM, Billiar TR, Geller DA. Transcriptional regulation of human inducible nitric oxide synthase (iNOS) gene by cytokines: initial analysis of the human iNOS promoter. Proc Natl Acad Sci USA. 1996;93:1054–1059. doi: 10.1073/pnas.93.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geller DA, Lowenstein CJ, Shapiro RA, Nussler AK, Di Silvio M, Wang SC, Nakayama DK, Snyder SH, Simmons RL, Billiar TR. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc Natl Acad Sci USA. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, Forstermann U, Kleinert H. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J Biol Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 13.Fechir M, Linker K, Pautz A, Hubrich T, Förstermann U, Rodriguez-Pascual F, Kleinert H. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol Pharmacol. 2005;67:2148–2161. doi: 10.1124/mol.104.008763. [DOI] [PubMed] [Google Scholar]
- 14.Pautz A, Linker K, Hubrich T, Korhonen R, Altenhöfer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 15.Korhonen R, Linker K, Pautz A, Förstermann U, Moilanen E, Kleinert H. Post-transcriptional regulation of human inducible nitric-oxide synthase expression by the Jun N-terminal kinase. Mol Pharmacol. 2007;71:1427–1434. doi: 10.1124/mol.106.033449. [DOI] [PubMed] [Google Scholar]
- 16.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Billiar TR, Geller DA. A novel NF-κB enhancer element regulates cytokine induction of the human inducible nitric oxide synthase gene promoter. J Biol Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 17.Lowenstein CJ, Alley EW, Raval P, Snowman AM, Snyder SH, Russell SW, Murphy WJ. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon gamma and lipopolysaccharide. Proc Natl Acad Sci USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie QW, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon- and bacterial LPS. J Exp Med. 1993;177:1779–1785. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganster RW, Taylor BS, Shao L, Geller DA. Complex regulation of human iNOS gene transcription by Stat1 and NF-κB. Proc Natl Acad Sci USA. 2001;98:8638–43. doi: 10.1073/pnas.151239498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganster RW, Guo Z, Shao L, Geller DA. Differential effects of TNF-α and IFN-γ on gene transcription mediated by NF-κB-Stat1 interactions. J Interferon Cyctokine Res. 2005;25:707–719. doi: 10.1089/jir.2005.25.707. [DOI] [PubMed] [Google Scholar]
- 21.Marks-Konczalik J, Chu SC, Moss J. Cytokine-mediated transcriptional induction of the human inducible nitric oxide synthase gene requires both activator protein 1 and nuclear factor κB-binding sites. J Biol Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Guo Z, Nourbakhsh M, Hauser H, Ganster R, Shao L, Geller DA. Identification of a negative response element in the human inducible nitric oxide synthase (hiNOS) promoter: the role of NF-κB repressing factor (NRF) in basal repression of the hiNOS gene. Proc Natl Acad Sci U S A. 2002;99:14212–14217. doi: 10.1073/pnas.212306199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z, Shao L, Feng X, Reid K, Mardersten E, Geller DA. A critical role for C/EBPβ binding to the AABS promoter response element in the human iNOS gene. FASEB J. 2003;17:1718–1720. doi: 10.1096/fj.02-1172fje. [DOI] [PubMed] [Google Scholar]
- 24.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175:3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 25.Du Q, Park KS, Guo Z, He P, Nagashima M, Shao L, Sahai R, Geller DA, Hussain SP. Regulation of human NOS2 expression by wnt β-catenin signaling. Cancer Res. 2006;66:7024–7031. doi: 10.1158/0008-5472.CAN-05-4110. [DOI] [PubMed] [Google Scholar]
- 26.Herr W, Sturm RA, Clerc RC, Corcoran LM, Baltimore D, Sharp PA, Ingraham HA, Rosenfeld MG, Finney M, Ruvkun G, et al. The POU Domain: a large conserved region in the mammalian Pit-1, Oct-1, Oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 27.Verrijzer CP, Van der Vliet PC. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 28.Sturm RA, Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- 29.Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 30.Clerc RG, Corcoran LH, LeBowitz JH, Baltimore D, Sharp PA. The B-cell specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 1988;2:1570–1581. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- 31.LeBowitz JH, Kobayashi T, Staudt L, Baltimore D, Sharp PA. Octamer-binding proteins from B or HeLa cells stimulate transcription of the immunoglobulin heavy-chain promoter in vitro. Genes Dev. 1988;2:1227–1237. doi: 10.1101/gad.2.10.1227. [DOI] [PubMed] [Google Scholar]
- 32.Schaffner W. How do different transcription factors binding the same DNA sequence sort out their jobs. Trends Genet. 1989;5:37–39. doi: 10.1016/0168-9525(89)90017-6. [DOI] [PubMed] [Google Scholar]
- 33.Mittal V, Cleary MA, Herr W, Hernandez N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol Cell Biol. 1996;16:1955–1965. doi: 10.1128/mcb.16.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturm RA, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 35.Delhase M, Castrillo JL, de la Hoya M, Rajas F, Hooghe-Peters EL. AP-1 and Oct-1 transcription factors down-regulate the expression of the human PIT1/GHF1 gene. J Biol Chem. 1996;271:32349–32358. doi: 10.1074/jbc.271.50.32349. [DOI] [PubMed] [Google Scholar]
- 36.Pankratova EV, Polanovasky OL. Oct-1 promoter region contains octamer sites and TAAT motifs recognized by Oct proteins. FEBS Letters. 1998;426:81–85. doi: 10.1016/s0014-5793(98)00316-0. [DOI] [PubMed] [Google Scholar]
- 37.Hitomi T, Matsuzaki Y, Yasuda S, Kawanake M, Yogosawa S, Koyama M, Tantin D, Sakai T. Oct-1 is involved in the transcriptional repression of the p15INK4b gene. FEBS Letters. 2007;581:1087–1092. doi: 10.1016/j.febslet.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 38.Scheidereit C, Cromlish JA, Gerster T, Kawakami K, Balmaceda CG, Currie RA, Roeder RG. A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature. 1988;336:551–557. doi: 10.1038/336551a0. [DOI] [PubMed] [Google Scholar]
- 39.Gstaiger M, Knoepfel L, Geogiev O, Schaffner W, Hovens CM. A B-cell coactivator-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 40.Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–24. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strubin M, Newell JW, Matthias P. OBF-1, a novel B-cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 42.Xie QW. A novel lipopolysaccharide-response element contributes to induction of nitric oxide synthase. J Biol Chem. 1997;272:14867–14872. doi: 10.1074/jbc.272.23.14867. [DOI] [PubMed] [Google Scholar]
- 43.Sawada T, Falk LA, Rao P, Murphy WJ, Pluznik DH. IL-6 induction of protein-DNA complexes via a novel regulatory region of the inducible nitric oxide synthase gene promoter. J Immunol. 1997;158:5267–5276. [PubMed] [Google Scholar]
- 44.Kim YM, Ko CB, Park YP, Kim YJ, Paik SG. Octamer motif is required for the NF-κB-mediated induction of the inducible nitric oxide synthase gene expression in RAW 264.7 macrophages. Mol Cells. 1999;9:99–109. [PubMed] [Google Scholar]
- 45.Gay RD, Dawson SJ, Murphy WJ, Russell SW, Latchman DS. Activation of the iNOS gene promoter by Brn-3 POU family transcription factor is dependent upon the octamer motif in the promoter. Biochim Biophys Acta. 1998;1443:315–322. doi: 10.1016/s0167-4781(98)00234-6. [DOI] [PubMed] [Google Scholar]
- 46.Darville MI, Terryn S, Eizirik DL. An octamer motif is required for activation of the inducible nitric oxide synthase promoter in pancreatic β-cells. Endocrinology. 2004;145:1130–1136. doi: 10.1210/en.2003-1200. [DOI] [PubMed] [Google Scholar]
- 47.Lee BS, Kim YM, Kang HS, Kim HM, Pyun KH, Choi I. Octamer binding protein-1 is involved in inhibition of inducible nitric oxide synthase expression by exogenous nitric oxide in murine liver cells. J Biochem. 2001;129:77–86. doi: 10.1093/oxfordjournals.jbchem.a002839. [DOI] [PubMed] [Google Scholar]
- 48.Wang B, Ma L, Tao X, Lipsky PE. Triptolide, an active component of the Chinese herbal remedy Tripterygium wilfordii Hook F, inhibits production of nitric oxide by decreasing inducible nitric oxide synthase gene transcription. Arthritis & Rheumatism. 2004;50:2995–3003. doi: 10.1002/art.20459. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez AC, Davis RL, Syapin PJ. The Oct DNA motif participates in the alcohol inhibition of the inducible nitric oxide synthase gene promoter in rat C6 glioma cells. Brain Res. 2007;1179:16–27. doi: 10.1016/j.brainres.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellott J, Nick HS, Waters MF, Billiar TR, Geller DA, Chesrown SE. Cytokine-induced changes in chromatin structure and in vivo footprints in the inducible NOS promoter. Am J Physiol. 2001;280:L390–L399. doi: 10.1152/ajplung.2001.280.3.L390. [DOI] [PubMed] [Google Scholar]
- 51.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 52.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]


