Abstract
Two crystal structures are determined for Δ-Rh(bpy)2(chrysi)3+ (chrysi = 5,6-chrysenequinone diimine) bound to the oligonucleotide duplex 5′-CGGAAATTACCG-3′ containing two adenosine-adenosine mismatches (italics) through metalloinsertion. Diffraction quality crystals with two different space groups (P3221 and P43212) were obtained under very similar crystallization conditions. In both structures, the bulky rhodium complex inserts into the two mismatched sites from the minor groove side, ejecting the mismatched bases into the major groove. The conformational changes are localized to the mismatched site; the metal complex replaces the mismatched base pair without an increase in base pair rise. The expansive metal complex is accommodated in the duplex by a slight opening in the phosphodiester backbone; all sugars retain a C2′-endo puckering, and flanking base pairs neither stretch nor shear. The structures differ, however, in that in one of the structures, an additional metal complex is bound by intercalation from the major groove at the central 5′-AT-3′ step. We conclude that this additional metal complex is intercalated into this central step because of crystal packing forces. The structures described here of Δ-Rh(bpy)2(chrysi)3+ bound to thermodynamically destabilized AA mismatches share critical features with binding by metalloinsertion in two other oligonucleotides containing different single base mismatches. These results underscore the generality of the metalloinsertion as a new mode of non-covalent binding by small molecules with a DNA duplex.
Almost fifty years ago, Lerman proposed four different non-covalent binding modes for small molecules with DNA: (1) electrostatic binding to the sugar phosphate backbone, (2) hydrophobic association with the minor groove, (3) intercalation into the helix by π-stacking between adjacent base pairs, and (4) insertion into the helix by separation and displacement of a base pair.1 The first three are frequently observed and have been extensively characterized both in solution and in the solid state.2–6 In contrast, the fourth binding mode, insertion, has eluded researchers almost completely.7 Recently, however, we have structurally characterized both by crystallography8 and NMR9 first examples of insertion into DNA by a small molecule, the mismatch-specific, octahedral metal complex Rh(bpy)2(chrysi)3+ (chrysi = 5,6-chrysenequinone diimine) (Figure 1).
Figure 1.
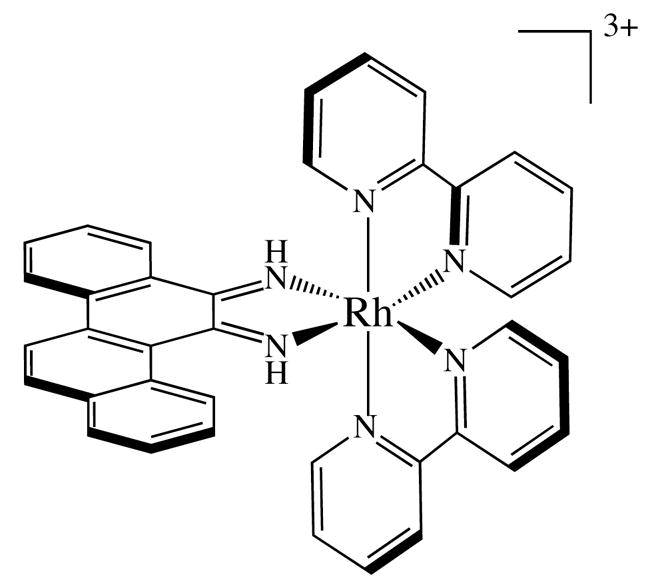
Δ-Rh(bpy)2(chrysi)3+
Because insertion requires the separation of a base pair and the ejection of the bases from the double helix, this binding mode occurs more readily at thermodynamically destabilized sites such as single-base mismatches. Indeed, to date, insertion has only been definitively observed with inert metal complexes bearing sterically expansive ligands, such as chrysi or phzi (benzo[a]phenanzine-5,6-quinone diimine); in both cases, the bulky ligands are 0.5 Å wider than the 10.85 Å span of a matched AT or GC base pair.10 This difference precludes the intercalation of the complex at matched sites and thus confers specificity for binding at thermodynamically destabilized mismatch sites.
Rhodium metalloinsertors, most notably Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+, bind single base mismatches with very high selectivity and with binding affinities that correlate directly with the local destabilization associated with the single base pair mismatch.11–13 This correlation reflects the ease of separation and ejection of the mismatched bases from the double helix. Importantly, in both cases, mismatch binding is enantiospecific: the right-handed helix can only accommodate the right-handed (Δ) enantiomer. In the years since their discovery, these metal complexes have shown significant promise not only in the detection of single base mismatches14–16, abasic sites,17,18 and single nucleotide polymorphisms,19 but also as chemotherapeutic agents.14,20–23
The crystallographic structure of Δ-Rh(bpy)2(chrysi)3+ bound to a palindromic oligonucleotide containing two CA-mismatches has recently been determined.7 This structure first revealed that the mismatch-specific rhodium complex does not bind DNA through classical metallointercalation but rather by metalloinsertion: the complex approaches the DNA from the minor groove side and inserts the bulky chrysi ligand at the mismatch site, extruding the mismatched base pairs into the major groove and replacing them in the DNA π-stack. The sugar-phosphate backbone of the DNA opens slightly to accommodate the sterically expansive ligand at the mismatch site. Overall, the DNA is disturbed very little beyond the insertion site, for all sugars remain in the C2′-endo conformation and all bases retain an anti configuration. Somewhat surprising, however, was the presence of a third rhodium complex in the structure that is bound not through insertion at the mismatch sites but through intercalation at a central AT step. Given that no detectable binding to a matched site had been observed for these bulky complexes in solution, we considered that this intercalation was the result of crystal packing forces. Subsequent NMR studies of Δ-Rh(bpy)2(chrysi)3+ bound to a similar oligonucleotide containing a CC-mismatch confirmed the insertion binding mode in solution and, significantly, showed no evidence of an intercalated rhodium moiety.8
In order to explore more generally the characteristics of the metalloinsertion mode, here we describe two crystal structures of Δ-Rh(bpy)2(chrysi)3+ bound to an AA mismatch. Both structures provide examples of metalloinsertion at a new mismatch, but the two structures differ principally in the presence or absence of a third, intercalated rhodium. The comparison of these structures with studies of the metalloinsertor bound to a CA and a CC mismatch illuminates the general architecture of the metalloinsertion binding mode at destabilized sites in DNA.
EXPERIMENTAL
Synthesis and Purification
The metalloinsertor Δ–Rh(bpy)2(chrysi)3+ was co-crystallized with a self-complementary oligonucleotide containing two AA mismatches (5′-C1G2G3A4A5A6T7T8A9C10C11G12-3′). The enantiopure rhodium complexwas synthesized and isolated as described previously.24 Standard oligonucleotides were synthesized from phosphoramidites on an ABI 3400 DNA synthesizer (reagents from Glen Research) and purified both with and without the dimethoxytrityl protecting group via two rounds of reverse-phase HPLC (HP1100 HPLC system with Varian DynaMax™ C18 semi-preparative column, gradient of 5:95 to 45:55 MeCN:50 mM NH4OAc (aq) over 30 min for DMT-on purification and 2:98 to 17:83 MeCN:50 mM NH4OAc (aq) over 30 min for DMT-off purification).
Crystal Preparation and Data Collection
Annealed oligonucleotides were incubated with the rhodium complex before crystallization. Subsequent manipulations were performed with minimal exposure of the complex to light. Two different sets of bright orange crystals, henceforth referred to as 1 and 2, were obtained, each under a distinct set of conditions. In both cases, thirteen different sequences were screened before crystals were obtained with the sequence described above. Crystal set 1 was grown from a solution of 1 mM double-stranded duplex, 3 mM enantiomerically pure Δ–Rh(bpy)2(chrysi)3+, 20 mM sodium cacodylate (pH 7.0), 6 mM spermine·4HCl, 40 mM NaCl, and 5% 2-methyl-2,4-pentanediol (MPD) equilibrated in sitting drops versus a reservoir of 35% MPD at ambient temperature. The crystals grew in space group P3221 with unit cell dimension a = b = 48.34 Å; c = 69.50 Å, α = β = 90°, γ = 120°, with one biomolecule per asymmetric unit (Table 1).
Table 1.
Data collection and refinement statistics
| Structure 1 | Structure 2 | |
|---|---|---|
| Data Collection | ||
| Space group | P3121 | P43212 |
| Cell dimensions: | ||
| a, b, c | 48.3, 48.3, 69.5 | 39.0, 39.0, 57.4 |
| α, β, γ | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Wavelength | 1.0046 | 1.5418 |
| Resolution | 35.0-1.60 (1.69-1.60) | 28.71- 1.80 (1.90-1.80) |
| Rmerge | 0.035 (0.499) | 0.061 (0.782) |
| Rpim | 0.013 (0.288) | 0.031 (0.342) |
| I/σI | 26.7 (2.0) | 19.1 (2.3) |
| Completeness, % | 99.5 (98.9) | 98.7 ( 97.4) |
| Redundancy | 7.9 (4.2) | 6.5 ( 6.6) |
| Refinement | ||
| No. of Reflections | 22677 | 4469 |
| Rwork/Rfree | 0.184/0.227 | 0.183/0.213 |
| No. of atoms (DNA) | 524 | 262 |
| No. of atoms (RhL6) | 120 | 90 |
| No. of atoms (water) | 89 | 63 |
| B-factors (DNA) | 43.44 | 25.7 |
| B-factors (complex) | 43.44 | 22.1 |
| B-factors (water) | 48.86 | 41.4 |
| RMS dev. (lengths) | 0.013 | 0.032 |
| RMS dev. (angles) | 2.450 | 4.281 |
Crystal set 2 was grown from a solution of 1 mM double-stranded duplex, 2 mM enantiomerically pure Δ–Rh(bpy)2(chrysi)3+, 20 mM sodium cacodylate (pH 7.0), 6 mM spermine·4HCl, 40 mM KCl, and 5% MPD equilibrated in sitting drop versus a reservoir of 35% MPD at ambient temperature. The crystals grew in space group P43212 with unit cell dimensions a = b = 39.02 Å; c = 57.42 Å, α = β = γ = 90°, with half of a biomolecule per asymmetric unit (Table 1).
The data for crystal 1 were collected on beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (Menlo Park, CA; λ= 1.00 Å, 100 K, Marrsearch 325 CCD detector). The data for crystal 2 was collected from a flash-cooled crystal at 100 K on an R-axis IV image plate using CuKα radiation produced by a Rigaku (Tokyo, Japan) RU-H3RHB rotating-anode generator with double-focusing mirrors and an Ni filter. Both sets of data were processed with MOSFLM and SCALA from the CCP4 suite of programs.25
Crystal Structure Determination and Refinement
Both structures were solved by single anomalous dispersion using the anomalous scattering of rhodium (f″ = 3.6 electrons for Rh at λ= 1.54 Å, and f″ = 1.7 electrons for Rh at λ= 1.00 Å) with the CCP4 suite of programs. For crystal 1, 2 heavy atoms were located per asymmetric unit; for crystal 2, 1.5 heavy atoms were located per asymmetric unit, with one on a special position. Structure 1 was refined with PHENIX v. 1.3 against 1.6 Å data taking into account the anomalous contribution of rhodium; for non-hydrogen atoms, anisotropic temperature factors were refined.26 The final Rcryst and Rfree were 0.18 and 0.23, respectively. Structure 2 was refined using REFMAC5 v. 5.5.0066 against 1.8 Å data to a final Rcryst = 0.18 and Rfree = 0.21.27, 28
In crystal 2, the rhodium complex located near the crystallographic twofold axis perpendicular to the helical axis of the DNA intercalates in two different orientations linked by symmetry. In crystal 1, residual density with anomalous contribution was also present near a crystallographic two-fold axis at the end of the duplex, most likely the result of disordered cacodylate or chloride ions. In the later stages of refinement for both crystals, riding hydrogens were included. Figures were drawn with Pymol.29
RESULTS AND DISCUSSION
Two types of crystals
The palindromic oligonucleotide 5′-C1G2G3A4A5A6T7T8A9C10C11G12-3′ contains two adenosine-adenosine mismatches, each situated three bases from the end of the strand and separated from one another by a central 5′-AATT-3′ step. Here, the duplex was co-crystallized with Δ-Rh(bpy)2(chrysi)3+ for high-resolution x-ray structure determination in order to improve our understanding of metalloinsertion at DNA single base mismatches. Interestingly, diffraction quality crystals with two different space groups (P3221 and P43212) were obtained under very similar crystallization conditions. Indeed, both crystals were grown with the same temperature, buffer, pH, type and concentration of precipitant, concentration of DNA, and concentration of spermine. The only differences are the concentration of metalloinsertor and the identity of salt employed: crystal 1 (P3221), containing 2 rhodiums per duplex, was obtained using 3 mM complex and 40 mM NaCl, and crystal 2 (P43212), containing 3 rhodiums per duplex, was obtained using 2 mM metalloinsertor and 40 mM KCl. Taken together, the structures of crystal 1 (1.6 Å) and 2 (1.8 Å) provide insights into the structure and generality of metalloinsertion.
Structure 1
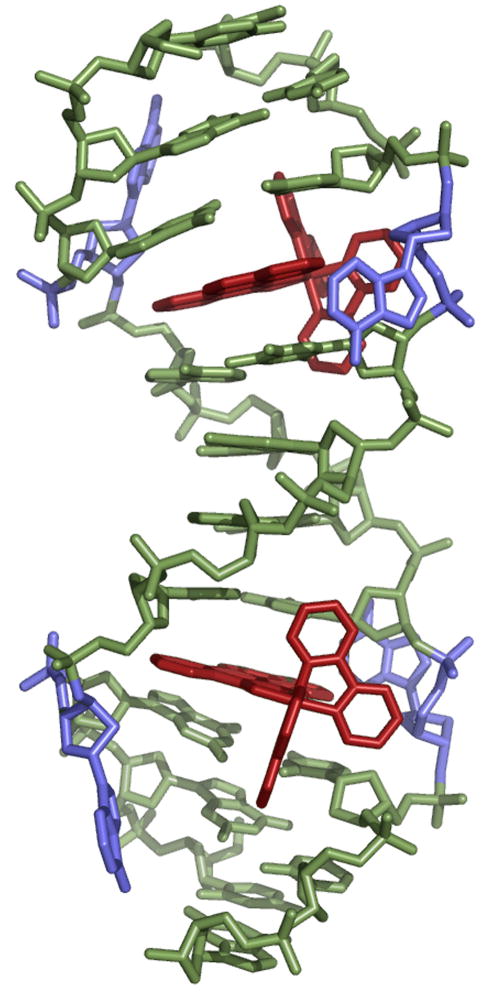
In crystal 1, the oligonucleotide co-crystallizes with the metalloinsertor in the space group P3221, with six asymmetric units per unit cell. The asymmetric unit contains one DNA duplex complexed with two metalloinsertors (Figure 2). Significantly, crystallization breaks the C2 symmetry of the DNA-metalloinsertor palindromic assembly, rendering the two mismatch sites inequivalent and providing two independent views of the mismatched site. Inspection of the unit cell reveals that the duplexes do not stack head-to-tail to form a longer double helix, as is frequently observed with DNA. Instead, it is the inter-duplex π-stacking of the ejected adenosines, either interwoven with the ancillary bpy ligand of a nearby rhodium moiety or stacked with adjacent, ejected adenosines, that determines the overall crystal packing and thus the space group.
Figure 2.
Structure 1: two Δ-Rh(bpy)2(chrysi)3+ (red) are inserted, one in each AA mismatch of the oligonucleotide 5′-CGGAAATTACCG-3′ (green). The ejected adenosines are shown in blue.
The two mismatched sites, not related by symmetry, provide separate views of the metalloinsertion. In both cases, the metal complex inserts from the minor groove by separating and ejecting the mismatched bases. The sterically expansive chrysi ligand of the metalloinsertor replaces the destabilized bases in the helical π-stack. The two ejected purines are pushed outward into the major groove. One of them remains close and perpendicular to the base stack, while the other folds back to the minor groove in a position stabilized by crystal packing. In both cases, deep insertion in the double helix is not inhibited by the increased steric hindrance of the minor groove: the distance between the rhodium center and the helical axis is 4.8 Å, approximately half the radius of the duplex.
Upon binding, the rhodium complex inserts deeply to enable complete overlap and stacking with both the purines and pyrimidines of the flanking base pairs. Importantly, these flanking base pairs neither stretch nor shear despite the considerable width of the ligand. All sugars retain their original C2′-endo puckering, and all bases maintain their initial anti conformation. To accommodate the inserted rhodium complex, the minor groove at the binding site widens to 19 Å from phosphate to phosphate, between 1 and 1.5 Å wider than other points in the duplex. Aside from the opening of the phosphodiester junctions at the insertion site, however, very little distortion of the DNA is observed (Tables 2 and 3).
Table 2.
| Base-pair | Shift (Å) | Slide (Å) | Rise (Å) | Tilt (°) | Roll (°) | Twist (°) |
|---|---|---|---|---|---|---|
| CG/CG | 0.8 | 2.2 | 3.4 | 12.0 | −1.6 | 37.6 |
| GG/CC | −0.3 | 2.7 | 3.2 | −6.1 | 5.7 | 34.3 |
| GA/TC | - | - | - | - | - | - |
| AA/TT | −1.3 | 1.2 | 3.3 | −4.7 | 3.8 | 37.6 |
| AT/AT | 0.0 | 0.1 | 3.4 | 1.3 | −0.7 | 29.6 |
| TT/AA | 1.3 | 1.0 | 3.4 | 2.2 | 5.6 | 36.1 |
| TC/GA | - | - | - | - | - | - |
| CC/GG | 0.4 | 2.7 | 3.3 | 4.7 | 6.5 | 34.5 |
| CG/CG | −1.0 | 2.5 | 3.2 | −8.2 | 2.1 | 37.3 |
|
| ||||||
| B-DNA | −0.1 | −0.8 | 3.3 | −1.3 | −3.6 | 36 |
Geometrical relationships between consecutive base pairs: shift, translation into the groove; slide, translation toward the phosphodiester backbone; rise, translation along the helix axis; tilt, rotation about the pseudo-two-fold axis relating the DNA strands; roll, rotation about a vector between the C1′ atoms; and twist, rotation about the helix axis.
Data were calculated by using the program 3DNA.29
Table 3.
DNA helical parameters for the base pairs of structure 1.a
| Base-pair | Shear (Å) | Stretch (Å) | Stagger (Å) | Buckle (°) | Propeller (°) | Opening (°) | Sugar pucker |
|---|---|---|---|---|---|---|---|
| C-G | 0.1 | −0.1 | 0.9 | −10.0 | −2.5 | −2.8 | C2′-endo |
| G-C | −0.2 | −0.1 | −0.0 | −0.2 | 1.1 | −3.8 | C2′-endo |
| G-C | −0.4 | −0.1 | 0.6 | 15.0 | −7.2 | −1.3 | C2′-endo |
| A-A | - | - | - | - | - | - | C2′-endo |
| A-T | −0.1 | −0.1 | 0.0 | −8.6 | 7.2 | 1.5 | C2′-endo |
| A-T | 0.1 | −0.1 | 0.1 | −0.0 | −8.3 | 3.2 | C2′-endo |
| T-A | −0.0 | −0.0 | 0.0 | −0.8 | −8.0 | 1.1 | C2′-endo |
| T-A | −0.1 | −0.2 | 0.1 | 5.8 | 9.4 | 1.7 | C2′-endo |
| A-A | - | - | - | - | - | - | C2′-endo |
| C-G | 0.3 | −0.1 | 0.6 | −18.8 | −5.8 | −0.5 | C2′-endo |
| C-G | 0.2 | −0.1 | 0.1 | −3.9 | 2.0 | −0.5 | C2′-endo |
| G-C | −0.3 | −0.1 | 0.4 | 6.2 | −5.3 | 0.7 | C2′-endo |
|
| |||||||
| B-DNA | 0 | 0.1 | 0.1 | 0.1 | 4.1 | −4.1 | C2′-endo |
Data were calculated by using the program 3DNA.29
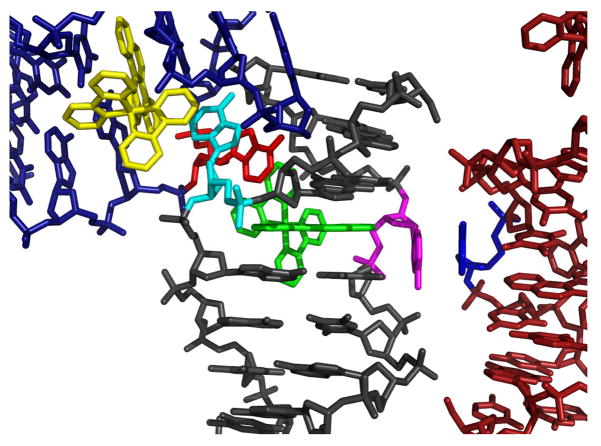
The difference between the two insertion sites lies only in the crystal packing of the ejected adenosines. In one of the two insertion sites, one of the ejected adenosines is stacked tightly within the major groove, where it lies perpendicular to the DNA base stack and is not involved in any interduplex interactions or hydrogen-bonding (Supporting Information). In contrast, the other adenosine at this site is interwoven with and π-stacks between the ejected adenosine from an adjacent duplex and the ancillary bipyridine ligand of the rhodium complex inserted in that nearby oligonucleotide. At the second insertion site, one of the ejected adenosines again π-stacks between the ejected adenosine from a second adjacent oligonucleotide and the ancillary bipyridine of the rhodium complex intercalated in that nearbyDNA. Unlike at the first insertion site, however, the other ejected adenosine here does partake in π-stacking, in this case with an extruded adenosine of yet another nearby duplex (Figure 3).
Figure 3.
Crystal packing by the ejected adenosines at one of the metalloinsertion sites in structure 1. At both insertion sites of the duplex, one ejected adenosine (cyan) π-stacks in an interwoven fashion with the bipyridine ligand of a rhodium complex (yellow) inserted in a nearby crystallographically related oligonucleotide and its corresponding ejected adenosine (red). The bipyridine ligand of the rhodium complex in the original duplex (green) completes the four-component stacking. In only one of the two insertion sites, as shown here, the second mismatched adenosine ejected in the major groove (magenta) π-stacks with a crystallographically equivalent ejected major groove adenosine (blue).
Structure 2
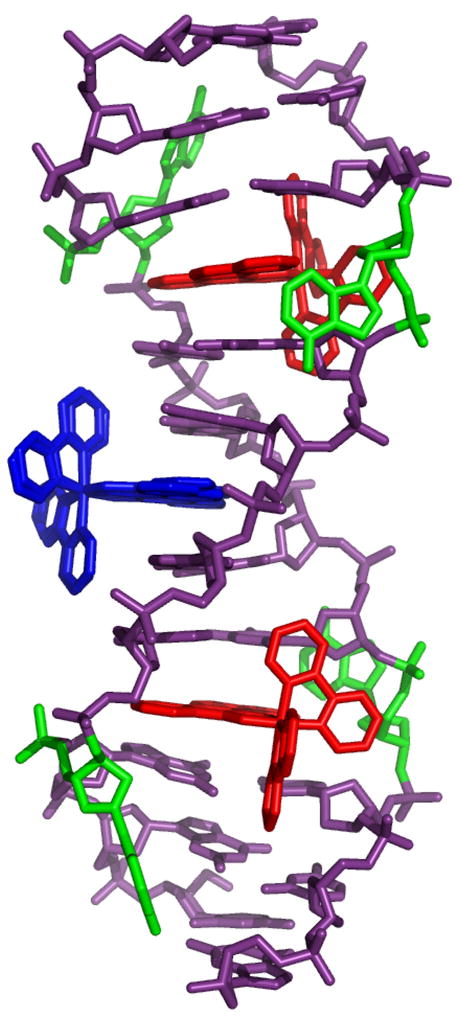
In crystal 2, the oligonucleotide co-crystallizes with the metalloinsertor in the space group P43212. In this case, the asymmetric unit is a single DNA strand with 1.5 metalloinsertors. Each duplex thus contains three rhodium complexes, one inserted at each of the mismatched sites and a third intercalated between the adenosine and thymine of the central 5′-AT-3′ step (Figure 4). Due to its position on a crystallographic two-fold axis, the central rhodium intercalates in two different orientations. The rhodium complexes at the two mismatched sites are also related by C2 symmetry, providing a single, independent view of the insertion site. Interestingly, in all respects other than the identity of the mismatch, this structure is virtually identical to that previously published for Δ-Rh(bpy)2(chrysi)3+ bound to a CA mismatch.8
Figure 4.
Structure 2: two Δ-Rh(bpy)2(chrysi)3+ (red) are inserted, one in each AA mismatch of the oligonucleotide 5′-CGGAAATTACCG-3′ (purple). A third rhodium complex (blue) is intercalated at the central 5′-AT-3′ step. The ejected adenosines are shown in green.
At the AA mismatch site, the metalloinsertor approaches the DNA from the minor groove, ejects the mispaired adenines from the helix, and replaces them in the DNA base stack with its own sterically expansive chrysi ligand. Indeed, the metalloinsertor π-stacks with the flanking AT and CG base pairs and penetrates so deeply from the minor groove that it is solvent accessible from the major groove. One of the ejected adenosines sits in the major groove, positioned perpendicular to the DNA base stack. The other adenosine bends back into the minor groove, where it π-stacks between the ejected adenosine of an adjacent duplex and a bipyridine ligand of a metalloinsertor bound to that oligonucleotide. Insertion of the rhodium complex into the site is facilitated by a slight widening of the phosphate backbone, from an average of 17.5 Å for well-matched sites to 19 Å for the metalloinsertion sites. Indeed, beyond this conformational change, metalloinsertion again distorts the DNA very little. Some buckling of the external flanking CG basepair is observed, but all riboses exhibit C2′-endo puckering, and all bases retain an anti configuration (Supporting Information).
As in the CA-mismatch structure,8 a third Δ-Rh(bpy)2(chrysi)3+ is also found intercalated at the central 5′-AT-3′ step. At this site, the rhodium complex approaches the duplex from the major groove and intercalates the chrysi ligand between adjacent AT and TA base pairs, doubling the rise at the intercalation site to 7.1 Å and slightly unwinding the duplex (Supporting Information). This binding interaction resembles closely that previously observed in the crystal structure of the sequence-specific metallointercalator Δ-α-Rh[(R,R)-Me2trien](phi)3+ bound by classical intercalation to its target site.6 The intercalative binding, like insertion, is accommodated by a slight widening of the phosphate backbone at the intercalation site and is accompanied by some buckling of the adjacent base pairs. Given the exquisite mismatch selectivity of the metalloinsertors in solution, such intercalative binding is a surprise and is almost certainly the result of crystal packing forces. The bipyridines of the intercalated metal complex π-stack with the terminal CG base pairs of two crystallographically related duplexes, in essence making the intercalated rhodium complex a linchpin for the crystal packing (Supporting Information).
Differences between the two structures
Certainly the telling difference between the two structures is the presence or absence of a Δ-Rh(bpy)2(chrysi)3+ intercalated at the central 5′-AT-3′ step. Given the similarity in crystallization conditions for crystals 1 and 2, the rhodium complex likely has comparable affinity for this central matched site in both cases. That the intercalated rhodium complex is not observed in structure 1 therefore strongly substantiates our conclusion that Δ-Rh(bpy)2(chrysi)3+ has negligible affinity for matched DNA and only binds to such sites when intercalation is stabilized by crystal packing-driven π-stacking. In structure 2 and the previously reported CA-mismatch structure,8 intercalation at the matched site is supported by π-stacking between the ancillary bipyridines of the intercalated rhodium complex and the terminal CG base pairs of two adjacent helices. Moreover, interwoven stacking between rhodium moieties in these latter duplexes and ejected purines further serves to lock the helices in an orientation that favors intercalative binding. These interactions, taken together, promote the binding of the metalloinsertor in a mode that is not detectable in solution. In fact, the interactions are insufficient to enforce complete intercalation into the double helix (the Rh-helical axis distance in the CA-structure, for example, is 1.24 Å longer than in the case of the DNA-bound metallointercalator Δ-α-Rh[(R,R)-Me2trien](phi)3+). These structures, taken together, provide a cautionary example of how crystal packing forces may alter the binding of small molecules with DNA.
The intercalated Δ–Rh(bpy)2(chrysi)3+ in structure 2 is likely also responsible for a second major difference between the structures. Upon superposition of the two structures, it becomes evident that the duplex in structure 1 is slightly bent relative to that in structure 2 (Supporting Information). Examination of the two mismatch-bound chrysi ligands in each structure is particularly instructive in this regard: in structure 2, the two ligands are nearly coplanar, whereas in structure 1, they are clearly skewed relative to one another. Because few perturbations to the duplex are observed beyond the mismatched base pair itself in either structure, it is improbable that the metalloinsertors are responsible for this bend in the duplex. Rather, the slight bending is most likely a result of the flexibility associated with the base step. It follows that in structure 2, the centrally intercalated and well-stacked rhodium complex rigidifies and straightens the helix.
A third major difference between the two structures lies in the stacking of the extrahelical adenosines. The interduplex, four component π-stacking interactions of one of the ejected adenosines at each mismatch site is common to both structures reported here, as well as the previously published CA-mismatch structure. It is with the second ejected base at each mismatch site that differences arise. At each AA-mismatch site in structure 2 and in the CA-mismatch structure, the second ejected adenine or ejected cytosine, respectively, sits tightly within the major groove, perpendicular to the DNA base stack and uninvolved in any π-stacking or hydrogen bonding. The same is true for the second ejected adenine at one of the two AA-mismatch sites in structure 1. At the other AA-site in structure 1, however, the second ejected adenine lies near the major groove, remains close to the phosphate backbone, and π-stacks with the ejected adenine of a nearby duplex (Figure 3).
General architecture of the insertion binding mode
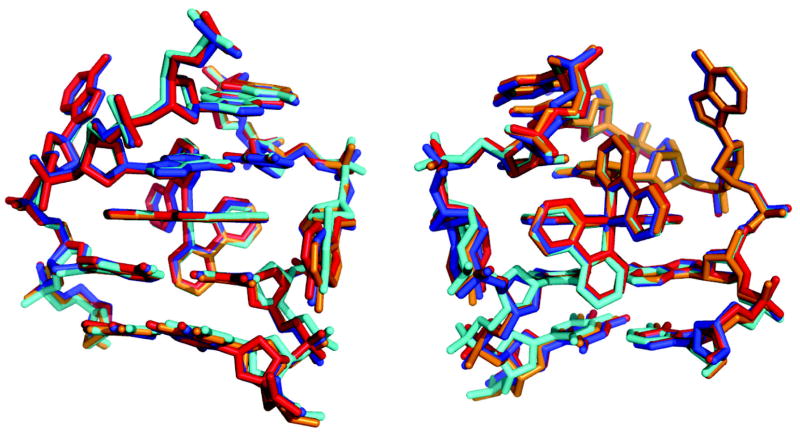
What is perhaps most remarkable about these crystal structures is not their differences but their similarity, not only to one another but also to the earlier structure we obtained.8 The superposition of the four independent views of Δ-Rh(bpy)2(chrysi)3+ bound to a mismatch site (3 AA-mismatch sites, 1 CA-mismatch site) reveals how every detail of the insertion binding mode is maintained regardless of the type of mismatch (Figure 5). In all cases, the DNA conformational changes are localized to the binding site. The metal complex essentially replaces the mismatched base pair; there is no increase in rise, no change in stacking, and no change in sugar puckering. In every case, Δ Rh(bpy)2(chrysi)3+ is well stacked with the matched DNA bases and penetrates the DNA so deeply that it protrudes from the opposite major groove. Furthermore, in each study, this binding is accommodated by a slight opening in the phosphodiester backbone, and the DNA is only minimally perturbed beyond the insertion site: all bases maintain their original anti conformation, all sugars retain a C2′-endo puckering, and flanking base pairs neither stretch nor shear. Perhaps most remarkable is that even the positions of the ejected bases, irrespective of their identities, assume nearly identical positions. The ejected bases are not splayed out in random positions, at least not in the structures in the solid state. Instead, the positions of the ejected bases seem to be defined, at least in part, by the sugar torsions. In fact, it may be more facile for the bases to be ejected from the minor groove side and accommodated in the major groove; this ejection into the major groove may then be a general characteristic of base pair displacement.31 Certainly, as evident in Figure 5, the distinct overlap of these different insertion sites, independent of the mismatch identity and crystal packing, must reflect the ease of adopting this conformation. These results, all taken together, indicate clearly that insertion into the double helix from the minor groove side with ejection of a base pair towards the major groove is a motif that is characteristic of binding of metal complexes bearing extended ligands to thermodynamically destabilized sites in DNA.
Figure 5.
Superposition of the three crystal structures showing insertion of Δ-Rh(bpy)2(chrysi)3+ into a single base mismatch viewed looking into the major groove (left) or minor groove (right). The red, blue, and orange structures represent insertion into an AA mismatch as reported in this work (red and blue are the two sites from structure 1, and orange is from structure 2). The cyan structure represents insertion into a CA mismatch as previously reported.7
CONCLUSIONS
The metalloinsertion of bulky metal complexes at DNA mismatches represents a new paradigm for how small molecules may bind non-covalently to the DNA duplex. The structures described here of Δ-Rh(bpy)2(chrysi)3+ bound to thermodynamically destabilized AA-mismatches illustrate the generality of this binding mode. Combined with previous crystallographic8 and NMR9 studies on different mismatched oligonucleotides, these structures reveal the architectural characteristics of metalloinsertion: in every case, without regard to the type of mismatch, the metal complex approaches the DNA from the minor groove, ejects the mismatched bases from the helix towards the major groove, replaces the extruded pair in the base stack with its own bulky ligand, and perturbs the DNA only minimally beyond the local binding site. The similarity in structures described here along with their clear differences serves furthermore to underscore metalloinsertion as a unique binding interaction, one distinct from intercalation. The presence of an intercalative rhodium in one of the structures also highlights how crystal packing forces can contribute to the solid state structures of small molecules bound non-covalently to DNA. While the information obtained from these structures yields critical and detailed insights, these data must also be considered in context with other data obtained in solution. In future work, it is hoped that these structures will not only prove useful as an illustration of a binding archetype but also in driving the design, synthesis, and application of new generations of small molecules that bind DNA through the insertion mode.
Supplementary Material
Acknowledgments
We thank Dr. Leonard Thomas for valuable discussions.
Abbreviations
- chrysi
chrysene-5,6-quinone diimine
- phzi
benzo[a]phenazine-5,6-quinone diimine
- DMT
dimethoxyltrityl
- MPD
2-methyl-2,4-pentanediol
Footnotes
Financial support for this work from the National Institutes of Health (GM33309) is gratefully acknowledged. We also thank the Gordon and Betty Moore Foundation (Caltech Molecular Observatory). The coordinates described in this paper have been deposited in the RCSB Protein Data Bank at the Brookhaven National Laboratory (Structure 1, RCSB ID Code: RCSB052256, PDB ID: 3GSK; Structure 2, RCSB ID Code RCSB052255, PDB ID: 3GSJ).
Supporting Information. Figures showing crystal packing, DNA bending, and interduplex stacking along with Tables containing helical parameters for structure 2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lerman LS. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- 2.Neidle S. Nucleic Acid Structure and Recognition. Oxford University Press; USA: 2002. [Google Scholar]
- 3.Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. A structural basis for recognition of A•T and T•A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 4.Bennett M, Krah A, Wien F, Garman E, McKenna R, Sanderson M, Neidle S. A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling. Proc Natl Acad Sci USA. 2000;97:9476–9481. doi: 10.1073/pnas.160271897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coll M, Frederick CA, Wang AH, Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci USA. 1987;84:8385 –8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Structure of a photoactive rhodium complex intercalated into DNA. Nat Struct Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 7.Lipscomb LA, Zhou FX, Presnell SR, Woo RJ, Peek MR, Plaskon RR, Williams LD. Structure of a DNA-porphyrin complex. Biochemistry. 1996;35:2818–2823. doi: 10.1021/bi952443z. [DOI] [PubMed] [Google Scholar]
- 8.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc Natl Acad Sci USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordier C, Pierre VC, Barton JK. Insertion of a Bulky Rhodium Complex into a DNA Cytosine Cytosine Mismatch: An NMR Solution Study. J Am Chem Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeglis BM, Pierre VP, Barton JK. Metallo-intercalators and metallo-insertors. Chem Comm. 2007;44:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 12.Jackson BA, Alekseyev VY, Barton JK. A versatile mismatch recognition agent: Specific cleavage of a plasmid DNA at a single base mispair. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 13.Jackson BA, Barton JK. Recognition of base mismatches in DNA by 5,6-chrysenequinone diimine complexes of rhodium(III): A proposed mechanism for preferential binding in destabilized regions of the double helix. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 14.Hart JR, Glebov O, Ernst R, Kirsch IR, Barton JK. Mismatch specific targeting: Hypersensitivity of mismatch repair deficient cells to bulky rhodium(III) intercalators. Proc Natl Acad Sci U S A. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. A rhodium(III) complex for high-affinity DNA base-pair mismatch recognition. Proc Natl Acad Sci U S A. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeglis BM, Barton JK. A Mismatch-Selective Bifunctional Rhodium-Oregon Green Conjugate: A Fluorescent Probe for Mismatched DNA. J Am Chem Soc. 2006;128:5654–5655. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]
- 17.Zeglis BM, Boland JA, Barton JK. Targeting Abasic Sites and Single Base Bulges in DNA with Metalloinsertors. J Am Chem Soc. 2008;130:7530–7531. doi: 10.1021/ja801479y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeglis BM, Boland JA, Barton JK. Recognition of Abasic Sites and Single Base Bulges in DNA with a Metalloinsertor. Biochemistry ASAP. 2009 doi: 10.1021/bi801885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart JR, Johnson MD, Barton JK. Single-nucleotide polymorphism discovery by targeted DNA photocleavage. Proc Nat Acad Sci USA. 2004;101:14040–14044. doi: 10.1073/pnas.0406169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schatzschneider U, Barton JK. Bifunctional Rhodium Intercalator Conjugates as Mismatch-Directing DNA Alkylating Agents. J Am Chem Soc. 2004;126:8630–8631. doi: 10.1021/ja048543m. [DOI] [PubMed] [Google Scholar]
- 21.Petitjean A, Barton JK. Tuning the DNA Reactivity of cis-Platinum: Conjugation to a Mismatch-Specific Metallointercalator. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 22.Brunner J, Barton JK. Targeting DNA Mismatches with Rhodium Intercalators Functionalized with a Cell-Penetrating Peptide. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 23.Ernst RJ, Song H, Barton JK. DNA Mismatch Binding and Anti-proliferation Activity of Rhodium Metalloinsertors. J Am Chem Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeglis BM, Barton JK. DNA Base Mismatch Detection with Bulky Rhodium Intercalators: Synthesis and Applications. Nat Prot. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger RC. PHENIX: Building New Software for Automated Cystallographic Structure Determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.It should be noted that two different programs were used to solve the two structures, which needs to be considered in making comparisons between them.
- 29.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 30.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng T, Dohno C, Nakatani K. Mismatch-binding ligands function as a molecular glue for DNA. Agnew Chem Int Ed. 2006;45:5623–5626. doi: 10.1002/anie.200601190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.