Abstract
The luminescent characteristics of Ru(bpy)2dppz2+ (dppz = dipyrido[3,2-a:2′,3′-c]phenazine), a DNA light switch, were investigated in the presence of oligonucleotides containing single base mismatches or an abasic site. In water, the ruthenium luminescence is quenched, but, bound to well matched duplex DNA, the Ru complex luminesces. Here we show that with DNAs containing a defect, rac-, Δ- and Λ-Ru(bpy)2dppz2+ exhibit significant luminescent enhancements above that with well matched DNA. In the presence of a single base mismatch, large luminescent enhancements are evident for the Δ-Ru isomer; the Λ-isomer shows particularly high luminescence bound to an oligonucleotide containing an abasic site. Similar increases are not evident with two common DNA-binding organic fluorophores, ethidium bromide and TO-PRO-3. Titrations with hairpin oligonucleotides containing a variable mismatch site show correlation between the level of luminescent enhancement and the thermodynamic destabilization associated with the mismatch. This correlation is reminiscent of that found earlier for a bulky rhodium complex that binds mismatched DNA sites through metalloinsertion, where the complex binds the DNA from the minor groove side, ejecting the mismatched bases into the major groove. Differential quenching studies with minor and major groove quenchers and time resolved emission studies support this metalloinsertion mode for the dppz complex at the defect site. Certainly these data underscore the utility of Ru(bpy)2dppz2+ as a sensitive luminescent reporter of DNA and its defects.
INTRODUCTION
Cells have evolved intricate enzymatic pathways to screen for and repair DNA lesions such as base pair mismatches and abasic sites in order to maintain the integrity of the genome.1,2 Because unrepaired DNA defects can lead to cancerous transformation,3,4 a means of recognizing DNA lesions is a crucial step in the development of early cancer diagnostics. Many approaches have been developed to target DNA lesions and particularly single base mismatches in DNA.5–18 Our laboratory has focused on the design of bulky metalloinsertors as probes of DNA mismatches.17 Although these bulky complexes target mismatches with high specificity, they have not yet achieved sufficient sensitivity to serve as luminescent probes for mismatches in cellular assays.18 Here we explore the use of the simple luminescent probe for DNA, Ru(bpy)2dppz2+, in targeting single base mismatches and abasic sites.
Ru(bpy)2dppz2+, which has been dubbed a “molecular light switch for DNA,” displays a remarkable increase in luminescence upon intercalation into duplex DNA (Figure 1).19,20 In non-aqueous solvents, the complex luminesces brightly owing to the excitation to a metal to ligand charge transfer state, but in water the luminescence is quenched through hydrogen bonding with the phenazine nitrogen atoms.21 Studies have shown that the Ru complex binds well matched duplex DNA avidly through intercalation, where the planar dppz ligand intercalates into the helix, stacking with the DNA but not disturbing base pairing.19,22 Our results have indicated that the complex, like other metallointercalators,23 binds the duplex from the major groove side.22 Others have provided some evidence in support of minor groove association.24 In any case, through this intercalative stacking, the phenazine nitrogen atoms of the dppz become somewhat protected from water, and hence, bound to DNA in aqueous solution, Ru(bpy)2dppz2+ shows luminescence.
Figure 1.
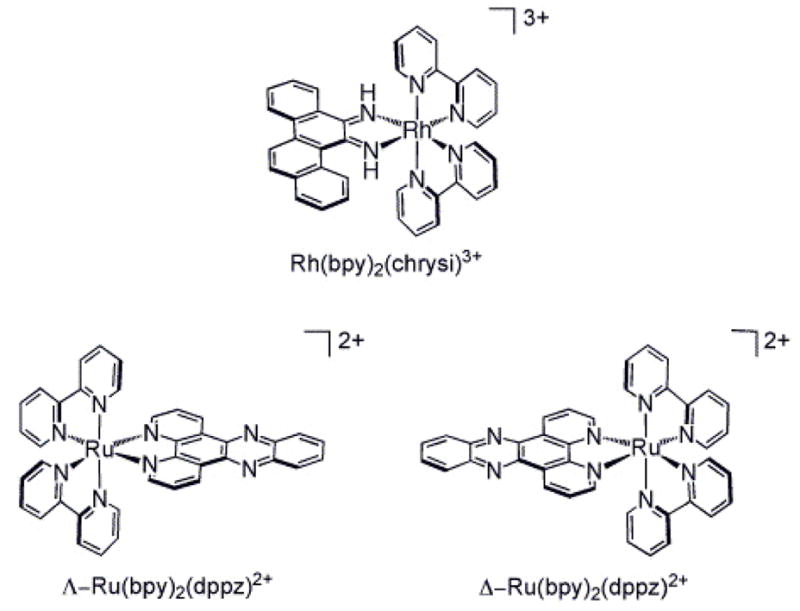
Chemical structures of Rh(bpy)2chrysi3+ and Ru(bpy)2dppz2+ isomers.
Our laboratory has also carried out extensive studies to explore rhodium diimine complexes that contain a bulky ligand that is inhibited from binding duplex DNA by intercalation; owing to the expanse of the ligand, the complex instead targets single base mismatches in DNA through metalloinsertion.17 Rh(bpy)2chrysi3+ (chrysi = 5,6-chrysenequinone diimine), for example, binds both single base mismatches and abasic sites in DNA with high specificity through metalloinsertion (Figure 1).25,26 X-ray crystallography and NMR studies have shown that, in contrast to intercalation, in this metalloinsertion mode, the sterically expansive chrysi ligand binds deeply into the mismatch site from the minor groove with complete ejection of the mismatched base pair into the major groove.27 In short, the Rh complex behaves as a π-stacking replacement of the mismatched pair in the DNA base stack. Upon light activation, the Rh complexes promote direct strand cleavage adjacent to the DNA lesion. DNA photocleavage studies on the full range of single base mismatches in all sequence contexts have shown a strong correlation between mismatch binding affinity by the metal complex and the thermodynamic destabilization associated with the mismatch.25 Thus, the easier it is to eject the mismatched base pairs, the tighter the binding through metalloinsertion.
The intercalating dppz ligand of Ru(bpy)2dppz2+ is narrow and long compared with the chrysi ligand and therefore does not seem suitable for binding through metalloinsertion (Figure 1). Moreover extensive studies have shown the tight binding of dppz complexes to well matched DNA through classical intercalation.19,20,22,24 Here we explore the luminescent properties of Ru(bpy)2dppz2+ in the presence of DNA duplexes that contain a single base mismatch or an abasic site. We find significant luminescent enhancements associated with binding to these defects compared to binding to well matched duplex DNA. Our results suggest binding to these defects is through metalloinsertion. These data indicate a powerful new application of Ru(bpy)2dppz2+ in probing DNA defects.
EXPERIMENTAL
Materials
All reagents and solvents were purchased from commercial suppliers and used without further purification. The DNA-binding organic fluorophores, ethidium bromide (EB) and TO-PRO-3 were obtained from Sigma-Aldrich and Invitrogen, respectively. Ruthenium complexes were prepared and enantiomers separated by previously reported methods; all complexes were utilized as chloride salts.19,28 The oligonucleotides used for measurements of steady state luminescence and excited state lifetimes were synthesized on an ABI 392 DNA/RNA synthesizer (Applied Biosystems) and purified as previously described.29 The copper complex Cu(phen)22+ was generated in situ by reacting the phen ligand with CuCl2 in a ratio of 3:1.30
Methods
Luminescence spectra with emission intensities ranged from 560 to 800 nm were recorded on an ISS-K2 spectrophotometer at ambient temperature in aerated solutions and then integrated. For all titrations, the experiments were performed at least three times. UV-visible spectra were taken on a Beckman DU7400 spectrophotometer. Time-resolved emission measurements were carried out at the Beckman Institute Laser Resource Center where samples were excited using a Nd:YAG-pumped OPO (Spectra-Physics Quanta-Ray).31 Laser power at 470 nm ranged from 4.0–4.5 mJ per pulse at 10 Hz. Emitted light was collected and focused onto the entrance slit of an ISA double grating (100 mm) monochromator and detected by a PMT (Hamamatsu R928). Each measurement is the average of 500 shots. Emission decays were fit to biexponential functions using non-linear least squares minimization.31
RESULTS AND DISCUSSION
Steady state luminescence of rac–, Δ– and Λ–Ru(bpy)2dppz2+ bound to oligonucleotides
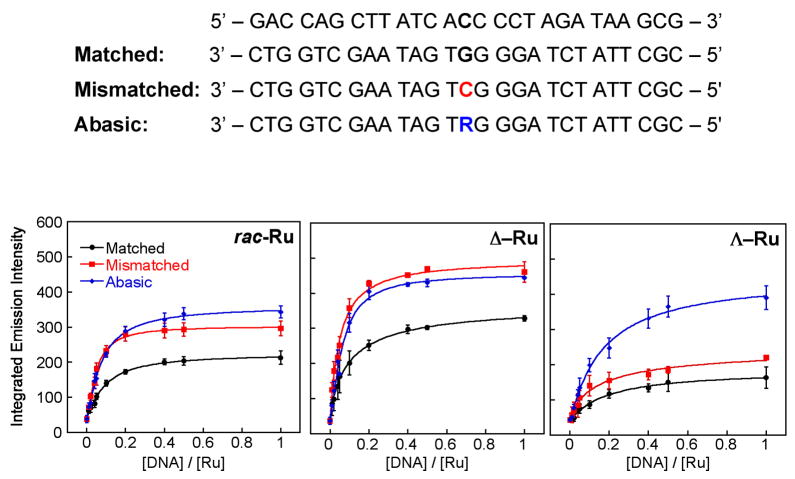
We first investigated the luminescent response of Ru(bpy)2dppz2+ in the presence of increasing concentrations of 27-mer oligonucleotide duplexes that were either fully matched, or contained a single base mismatch or an abasic site (Figure 2). After adding the Ru complex (100 nM, rac-, Δ-, or Λ-), to a solution containing various concentrations of DNA (0 – 100 nM oligomer) in 50 mM NaCl, 5 mM Tris, pH 7.5, the resulting steady state luminescence was measured. We find a marked increase in Ru luminescence as the concentration of DNA increases until saturating conditions are reached. Significantly, the luminescence increase is greater with those oligonucleotides containing the single site defect.
Figure 2.
Titrations of Ru(bpy)2dppz2+ with DNAs containing defects. Top: DNA sequences of matched, mismatched and abasic 27-mer duplex DNA (R denotes a tetrahydrofuranyl abasic site). Bottom: plots of the integrated emission intensity (λ x = 440 nm) of rac– (left),Δ– (middle) and Λ–Ru(bpy)2dppz2+ (right) (100 nM) upon increasing the concentration of DNA in 50 mM NaCl, 5 mM Tris, pH 7.5. Error bars indicate standard deviations in the measurements.
Both Λ- and Δ-Ru enantiomers exhibit an increase in luminescence in the presence of DNA defects versus well matched DNA (Figure 2). Specifically, rac- and ΔRu show a 1.5-fold enhancement in luminescence with DNA containing a CC mismatch or an abasic site compared to well-matched DNA. It should be noted that Δ-Ru exhibits higher integrated luminescence intensity with all three duplexes, reflecting that the Δ-enantiomer binds more tightly than the Λ-enantiomer to right-handed B-form DNA.20,32 Nonetheless, for rac- and Δ-Ru, 90% saturation is reached at a Ru:DNA ratio of 3:1 with the matched duplex and 4:1 with the mismatched duplex. This stoichiometry reveals that an additional Ru is bound to the mismatch without affecting the loading of Ru at matched sites. Interestingly, Λ-Ru, unlike rac-Ru and Δ-Ru, presents a significant increase in luminescence with abasic DNA over well matched DNA (2.5-fold) or mismatched DNA (1.8-fold), as shown in Figure 2 (bottom right). This characteristic of Λ–Ru suggests that it may be useful in the detection of abasic sites in DNA.
Comparison with other DNA-binding fluorophores
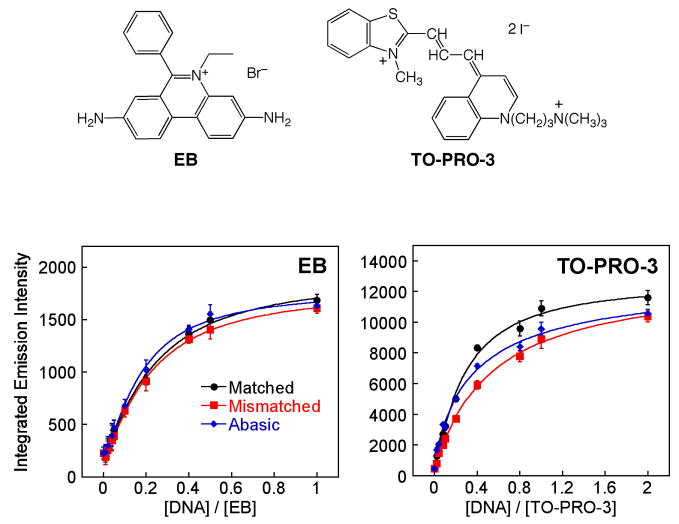
To compare the luminescent properties of the Ru complex to other commonly used DNA-binding fluorescent probes, we employed ethidium bromide (EB)33 and TO-PRO-334 (Figure 3). When the intercalator EB is incubated with each DNA duplex, no distinguishable difference in luminescence intensity is observed among the three oligonucleotides (Figure 3, bottom left). TO-PRO-3, a known minor groove binding agent, shows a small decrease in luminescence in the presence of DNA containing a mismatch or abasic site compared with well matched DNA. Neither of these two commonly used luminescent DNA-binding agents show any evidence of luminescence enhancement with DNAs containing a defect.
Figure 3.
Comparisons with common DNA-binding fluorophores. Structures of EB and TO-PRO-3 are shown along with plots of the integrated emission intensity of EB (100 nM) and TO-PRO-3 (100 nM) in the presence of 27-mer duplex DNAs (Figure 2;λex for EB = 512 nm, λex for TO-PRO-3 = 642 nm) in 50 mM NaCl, 5 mM Tris, pH 7.5. Error bars indicate standard deviations in the measurements.
The different luminescence behavior of Ru in the presence of mismatched and abasic DNA versus the well matched duplex reports on its structural characteristics at the defect binding site: the defect sites afford the complex a higher degree of protection from solvent water molecules versus a well matched duplex site. This result also suggests that Ru binding at mismatches and abasic sites is fundamentally different from intercalation between matched bases or groove binding, because if Ru(bpy)2dppz2+ were to bind to mismatches or abasic sites through classical intercalation or groove binding, we might expect the luminescence response to resemble that of EB or TO-PRO-3.
Luminescence behavior of Ru with different base mismatches
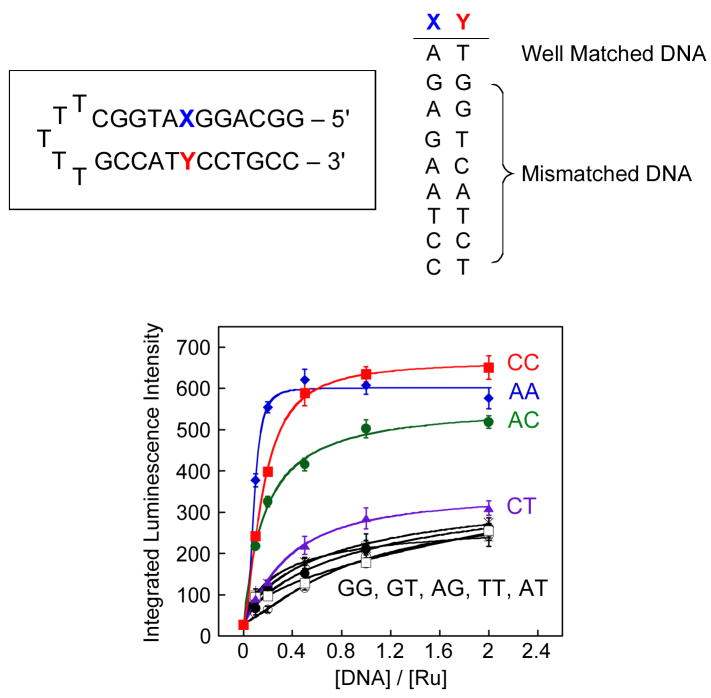
We investigated also the ability of Ru to report on other types of DNA base mismatches by using a short hairpin oligonucleotide containing a mismatch near the center of the duplex. As evident in Figure 4, we detect enhanced luminescence intensities with AA, AC, and CC mismatches, which are relatively destabilized compared to Watson-Crick base pairs.35 With the thermodynamically stable G-containing mismatches (GG, GA, GT), the Ru complex acts in a manner similar to that with well matched DNA (AT). Indeed, the luminescence intensity of Ru is correlated with the relative thermodynamic stability of each mismatch.
Figure 4.
Titrations of Ru(bpy)2dppz2+ with hairpin DNAs containing different mismatches. Top: Hairpin DNA sequences. Bottom: plots of the integrated emission intensity (λex = 440 nm) of Δ–Ru (100 nM) with with increasing concentrations of hairpin DNA containing mismatches: GG (○), GT (□), AT (●), AG (×), TT (+), CT ( ), AC (
), AC ( ), AA (
), AA ( ), and CC (
), and CC ( ) in 50 mM NaCl, 5 mM Tris, pH 7.5. Error bars indicate standard deviations in the measurements.
) in 50 mM NaCl, 5 mM Tris, pH 7.5. Error bars indicate standard deviations in the measurements.
An exception to this thermodynamic correlation that must be noted is that there is no significant increase in the luminescence of Ru with CT and TT mismatches, even though the dppz ligand may insert into pyrimidine-pyrimidine mismatches more easily than purine-purine ones (Figure 4). The absence of an increase in luminescence with CT and TT may be related to an intermolecular hydrogen bonding interaction between thymine and the phenazine moiety of dppz, which, like hydrogen bonding with water, yields quenching.
This dependence on mismatch thermodynamic stability strikingly resembles that observed in DNA binding studies of the metalloinsertor Rh(bpy)2chrysi3+.25 Binding of the chrysi complex to a mismatched site occurs via insertion with the ejection of the mismatched base and this binding is correlated with the thermodynamic instability of the mismatch; the easier it is to eject the mismatched base pairs from the stack, the tighter the binding of the metalloinsertor. Thus, these luminescence data, showing a similar correlation with mismatch instability, suggest that Ru(bpy)2dppz2+ complexes may bind similarly to thermodynamically destabilized sites via insertion of the dppz ligand from the minor groove, possibly causing the ejection of one or both bases into the major groove. Accordingly, because insertion into the minor groove allows for deeper binding, one could explain the greater luminescence at these mismatched sites.
It should also be noted that a higher differential luminescence intensity of Ru between mismatched and matched DNA is observed with the hairpin oligonucleotides (3-fold increase) compared with the longer 27-mer duplex (1.5-fold increase) for the same mismatch (CC). This observation is expected; only one Ru complex can bind to the single mismatch on both duplexes, while the longer duplex can accommodate more Ru in matched duplex sites. Thus, shortening the DNA increases the probability of binding to the destabilized site which in turn enhances the differential in luminescence intensity.
Excited state lifetimes of Ru
To elucidate further the luminescent characteristics of the Ru complexes bound to DNA defects, we examined their excited state lifetimes in the presence of the 27-mer oligonucleotides. The excited state decay profiles for the Ru complexes have been fit to biexponential luminescent decay (Table 1). We have previously seen biexponential decays for the Ru complexes with B-form DNAs and, through quenching studies and NMR experiments, have characterized this biexponential decay structurally in terms of side-on and perpendicular components.19,22 In the perpendicular binding mode, the dppz ligand intercalates such that the Ru dppz axis lies along the DNA dyad axis; in contrast, when Ru intercalates via a side-on approach, the Ru dppz axis lies along the long axis of the base pairs. As a result of these intercalative differences, the perpendicular binding mode places the phenazine moiety between the base pairs in a way that is more protected from quenching by water compared to the side-on bound phenazine moiety, yielding a longer fluorescent lifetime for the perpendicular versus side-on mode.
Table 1.
Luminescence Decay Parameters for rac–, Δ– and Λ–Ru(bpy)2dppz2+ with DNAa
| Complex | DNA | τ1 (ns)b | τ2 (ns)b | τ1:τ2b |
|---|---|---|---|---|
| rac-Ru | Matched | 72 | 212 | 83:17 |
| Mismatched | 74 | 213 | 77:23 | |
| Abasic | 86 | 192 | 69:31 | |
| Δ-Ru | Matched | 83 | 245 | 88:12 |
| Mismatched | 91 | 296 | 86:14 | |
| Abasic | 90 | 204 | 79:21 | |
| Λ-Ru | Matched | 41 | 199 | 89:11 |
| Mismatched | 37 | 156 | 78:22 | |
| Abasic | 69 | 167 | 41:59 |
Samples containing 10 μM Ru and 20 μM DNA (5 mM Tris, pH 7.5, 50 mM NaCl) were used for the excited state lifetime measurements (λex = 470 nm, λem = 610 nm). The 27-mer oligonucleotides (matched, mismatched and abasic DNA, Figure 2) were used.
Uncertainties in excited state lifetimes are ≤ 10%.
Relative contributions of each lifetime to the overall decay.
In the presence of a DNA mismatch, the Δ-Ru complex shows an increase in the long-lived excited state lifetime (Table 1). This observation is consistent with binding through metalloinsertion, where the complex is expected to be more deeply held and certainly more protected from water in the small minor groove. Rh(bpy)2chrysi3+ appears to bind a mismatched site in a strictly perpendicular orientation,27 but the longer dppz complex enjoys a higher degree of rotational freedom than the short chrysi complex, likely allowing both orientations of the inserting dppz ligand (Figure 1). For the Λ-Ru complex, little significant difference is evident with the mismatch; this result is not surprising based upon the steady state titration. It is noteworthy here that Δ-Rh(bpy)2chrysi3+ binds enantiospecifically through metalloinsertion at the mismatched site because of the very small size of the right-handed minor groove at the mismatched binding site.25,27
We have previously found for Rh(bpy)2chrysi3+ that binding to an abasic site resembles closely binding to a mismatched site.26 We see similar results for Δ-Ru(bpy)2dppz2+. It appears that the luminescent enhancement associated with binding to the abasic site is reflected in an increase in the excited state lifetime of the side-on component but a higher population of the perpendicular component. For the Λ-isomer these effects are still more substantial. These enhancements likely reflect a looser binding of the Λ-isomer within the abasic site pocket.
Luminescence response of Ru to DNA in the presence of Cu(phen)22+
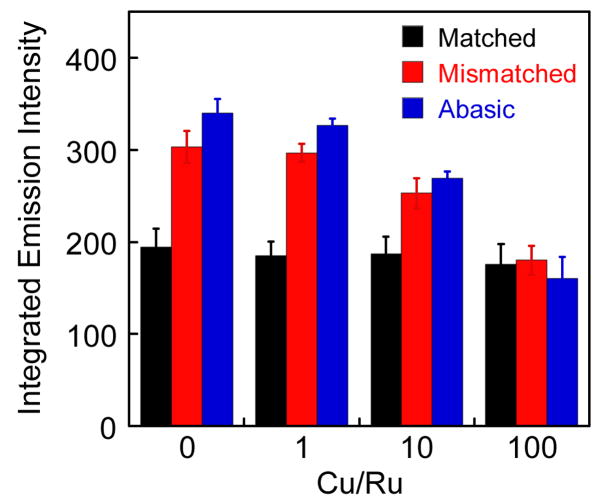
To explore further whether the Ru complex interacts with DNA defects from the minor groove, we measured the steady state luminescence of Ru with DNA in the presence of the DNA-binding quencher, Cu(phen)22+. Upon increasing the concentration of Cu(phen)22+,30 the luminescence intensity of Ru with matched DNA is unchanged, while that with mismatched and abasic DNA decreases to the same level as that with matched DNA (Figure 5). Thus Cu binding selectively quenches the luminescence from Ru bound at the defect sites but not of Ru bound at well-matched sites. Since the Cu complex binds in the minor groove,30 these data further support binding by Ru(bpy)2dppz2+ to well matched DNA in the major groove. Moreover, since the quenching is selectively at the defect sites, these data also indicate that binding of the Ru complex to the defect sites appears to occur from the minor groove side. We expect that Cu quenching at these sites is the result of paramagnetic quenching rather than displacement of the Ru complex, since titrations indicate binding to the mismatched DNA is comparable in affinity to binding to well matched DNA and thus far tighter than weak minor groove association by the copper complex. Nonetheless, irrespective of the quenching mechanism, this loss of the enhanced luminescence associated with Ru binding to defects by the minor groove-binding Cu(phen)22+ strongly implies that Ru binds to the DNA defects via the minor groove. Like the metalloinsertor Rh(bpy)2chrysi3+,25 the binding mode of Ru into the destabilized site thus is likely to be insertion from the minor groove.27
Figure 5.
Plot of the integrated emission intensity of rac–Ru(bpy)2dppz2+ (100 nM) with increasing concentration of Cu(phen)22+ in the presence of 27-mer duplex DNA (Figure 2, λex = 440 nm). The Ru complex was incubated with DNA previously treated with Cu(phen)22+ for 30 min (5 mM Tris, pH 7.5, 50 mM NaCl). Error bars indicate standard deviations in the measurements.
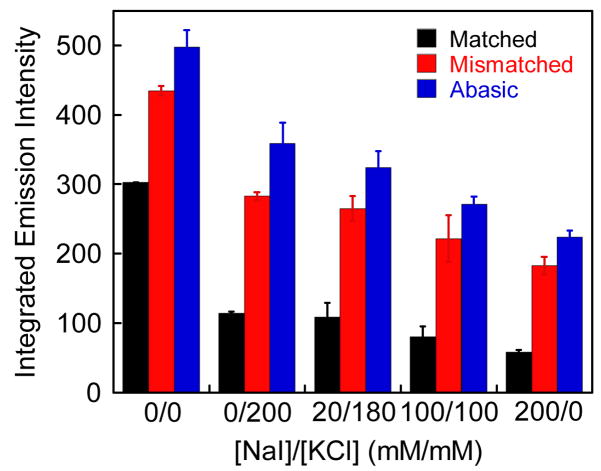
Selective quenching of Ru luminescence with matched DNA by NaI
To improve the luminescence differential of bound Ru between matched and mismatched or abasic DNA, we used NaI to quench preferentially the Ru luminescence associated with matched DNA (Figure 6). Iodide is an anionic luminescent quencher, and its efficiency in quenching a small molecule bound to DNA depends upon how closely the small molecule is protected from the quencher by the DNA polyanion.36 Based upon the electrostatic profile of B-DNA, along with the greater exposure of a Ru complex bound to the major groove versus the minor groove, we might expect greater quenching by I- of Ru bound in the major versus minor groove. Appropriate amounts of non-quenching KCl were added to maintain a constant ionic strength in all samples. Upon initial addition of KCl alone, we observe a marked decrease in luminescence for all three duplexes; and interestingly, simply increasing this ionic strength leads to some increase in the ratio of luminescence for mismatched:matched DNAs. The increased counter-ion concentration of the solution must inhibit Ru binding electrostatically and more so for the matched versus mismatched binding. Moreover, as the concentration of the quencher NaI increases, Ru luminescence with all three duplexes decreases further. As a result, in comparing the luminescence ratios without 200 mM salt versus with 200 mM NaI, the relative differential luminescence improves from 1.5-fold to 4-fold. At constant ionic strength, comparing ratios for 200 mM KCl versus 200 mM NaI, we see the ratio for mismatched to matched luminescence change from 2.3 to 3.8. Thus we see some preferential quenching of matched DNA with iodide. This result too suggests that Ru binds to DNA defects from the minor groove. DNA defect-bound Ru is expected to be less accessible to an anionic quencher and thus show less decrease in luminescence upon addition of the quencher.
Figure 6.
Plot of the integrated emission intensity of rac–Ru(bpy)2dppz2+ (200 nM) with increasing NaI in the presence of 27-mer duplex DNA (Figure 2, λex = 440 nm). Appropriate amounts of KCl were added to keep the ionic strength constant. Error bars indicate standard deviations in the measurements.
Changes in the excited state lifetimes for Δ-Ru in the presence of KCl and/or NaI support our conclusions from the steady state measurements (Table 2). Again, all data were fit to biexponential decays. Compared with the lifetimes before the addition of salt (Table 1), in the presence of 1 M KCl, the longer-lived component shows a substantial increase in excited state lifetime for matched, mismatched and abasic DNA. This increase may reflect deeper perpendicular stacking of the dppz moiety between DNA bases. As the NaI concentration increases, the relative proportion of the longer-lived component increases as well, indicating that the shorter-lived species is more accessible to the quencher. The lifetime of both components decreases with higher quencher concentrations, revealing that the excited state is dynamically quenched by iodide. However, both of the components continue to show a longer lifetime with mismatched DNA even at the highest quencher concentration, thus maintaining a high luminescence differential between matched and mismatched DNA. It should be noted that in all cases the instantaneous emission intensity decreases as iodide concentration increases, suggesting that high concentrations of the quencher also result in sphere of action static quenching. Importantly, this decrease is most substantial in the case of matched DNA for both binding modes. These observations are consistent with the notion that minor groove-bound Ru is buried deeper in the duplex and thus is less likely to be in the proximity of a quencher. Consequently, Ru bound in the major groove is preferentially quenched, thereby accentuating the differential luminescence between defective and matched DNA.
Table 2.
Luminescence Decay Parameters for Δ–Ru(bpy)2dppz2+ with DNA in the Presence of NaI.a
| DNA | [NaI]/[KCl] (mM/mM) | τ1 (ns)b | τ2 (ns)b | τ1:τ2c |
|---|---|---|---|---|
| Matched | 0/1000 | 83 | 305 | 74:26 |
| 500/500 | 81 | 295 | 66:34 | |
| 1000/0 | 76 | 275 | 63:37 | |
| Mismatched | 0/1000 | 111 | 448 | 80:20 |
| 500/500 | 103 | 357 | 74:26 | |
| 1000/0 | 99 | 325 | 75:25 | |
| Abasic | 0/1000 | 97 | 300 | 85:15 |
| 500/500 | 90 | 200 | 75:25 | |
| 1000/0 | 85 | 177 | 73:27 |
Samples containing 2 μM Ru and 4 μM DNA (5 mM Tris, pH 7.5, 50 mM NaCl) were used for the excited state lifetime measurements (λex = 470 nm, λem = 610 nm). The 27-mer oligonucleotides (matched, mismatched and abasic DNA, Figure 2) were used.
Uncertainties in excited state lifetimes are ≤ 10%.
Relative contributions of each lifetime to the overall decay.
Implications
While small organic DNA binding molecules EB and TO-PRO-3 show either a small decrease or no change in luminescence when combined with defective DNA, Ru(bpy)2dppz2+ displays an increase in luminescence in the presence of mismatches and abasic sites. Several observations suggest that binding of Ru into the mismatch or abasic sites occurs in a manner analogous to that of the metalloinsertor Rh(bpy)2chrysi3+ (that is, insertion from the minor groove): (i) the correlation of the luminescent enhancement with the thermodynamic instability of mismatched sites, (ii) the preferential quenching of the enhanced luminescence at defects with Cu(phen)22+ and (iii) the increase in differential luminescence at defects upon iodide quenching. Remarkably, even though the intercalating dppz ligand is structurally very different from the chrysi ligand, its similar manner of binding at mismatches and abasic sites suggests that metalloinsertion may be the general binding mode of octahedral metal complexes into the destabilized mismatched and abasic sites.
In this work, we thus extend the utility of Ru(bpy)2dppz2+ in probing small local distortions in the structure of DNA by showing the ability of Ru(bpy)2dppz2+ to act as a light-activated signal for DNA defects. Using NaI, we have magnified the luminescence differential with Ru(bpy)2dppz2+ between matched and defective DNA by selectively quenching the luminescence from Ru bound to matched DNA. This selective quenching strategy may prove useful for the direct visualization of biological samples containing DNA defects. Certainly these data underscore the utility of Ru(bpy)2dppz2+ as a sensitive luminescent reporter of DNA and its defects.
Acknowledgments
We are grateful to the NIH (GM33309) for their financial support and the Tobacco-Related Disease Research Program (TRDRP) for a postdoctoral fellowship to M.H.L. We thank the Beckman Institute Laser Resource Center at Caltech for facilities.
References
- 1.Marnett LJ, Pastaras JP. Trends Genet. 2001;17:214. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 2.Iyer RR, Pluciennik A, Burdett V, Modrich PK. Chem Rev. 2006;106:302. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. Nature. 2001;411:366. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.(a) Jiricny J, Marra G. Curr Opin Genet Dev. 2003;13:61. doi: 10.1016/s0959-437x(03)00004-2. [DOI] [PubMed] [Google Scholar]; (b) Jiricny J. Nat Rev Mol Cell Biol. 2006;7:335. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 5.Broude NE. Trends Biotechnol. 2002;20:249. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]
- 6.Kohler O, Jarikote DV, Seitz O. ChemBioChem. 2005;6:69. doi: 10.1002/cbic.200400260. [DOI] [PubMed] [Google Scholar]
- 7.Chenoweth DM, Viger A, Dervan PB. J Am Chem Soc. 2007;129:2216. doi: 10.1021/ja0682576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruyama T, Takata T, Ichinose H, Park LC, Kamaiya N, Goto M. Biotech Letters. 2003;25:1637. doi: 10.1023/a:1025661730518. [DOI] [PubMed] [Google Scholar]
- 9.(a) Nakatani K, Sando S, Saito I. Nat Biotech. 2001;19:51. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]; (b) Kobori A, Horie S, Suda H, Saito I, Nakatani K. J Am Chem Soc. 2004;126:557. doi: 10.1021/ja037947w. [DOI] [PubMed] [Google Scholar]
- 10.Nakatani K, Hagihara S, Goto Y, Kobori A, Hagihara M, Hayashi G, Kyo M, Nomura M, Mishima M, Kojima C. Nat Chem Biol. 2005;1:39. doi: 10.1038/nchembio708. [DOI] [PubMed] [Google Scholar]
- 11.Valis L, Amann N, Wagenknecht HA. Org Biomol Chem. 2005;3:36. doi: 10.1039/b414672g. [DOI] [PubMed] [Google Scholar]
- 12.Makrigiorgos GM, Chakrabarti S, Mahood A. Int J Radiat Biol. 1998;74:99. doi: 10.1080/095530098141762. [DOI] [PubMed] [Google Scholar]
- 13.Adamczyk M, Mattingly PG, Moore JA. Bioorg Med Chem Lett. 1998;8:3599. doi: 10.1016/s0960-894x(98)00658-1. [DOI] [PubMed] [Google Scholar]
- 14.Moran N, Bassani DM, Desvergne J-P, Keiper S, Lowden PAS, Vyle, Tucker JHR. Chem Commun. 2006;48:5003. doi: 10.1039/b611650g. [DOI] [PubMed] [Google Scholar]
- 15.Greco NJ, Tor Y. J Am Chem Soc. 2005;127:10784. doi: 10.1021/ja052000a. [DOI] [PubMed] [Google Scholar]
- 16.Dhar S, Kodama T, Greenberg MM. J Am Chem Soc. 2007;129:8702. doi: 10.1021/ja073014e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeglis BM, Pierre VC, Barton JK. Chem Commun. 2007;44:4565. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Ruba E, Hart JR, Barton JK. Inorg Chem. 2004;43:4570. doi: 10.1021/ic0499291. [DOI] [PubMed] [Google Scholar]; (b) Zeglis BM, Barton JK. J Am Chem Soc. 2006;128:5654. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]
- 19.(a) Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960. [Google Scholar]; (b) Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochemistry. 1992;31:10809. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]; (c) Hartshorn RM, Barton JK. J Am Chem Soc. 1992;114:5919. [Google Scholar]
- 20.(a) Hiort C, Lincoln P, Norden B. J Am Chem Soc. 1993;115:3448. [Google Scholar]; (b) Haq I, Lincoln P, Suh D, Norden B, Chowdhry B, Chaires J. J Am Chem Soc. 1995;117:4788. [Google Scholar]
- 21.(a) Olson EJC, Hu D, Hormann A, Jonkman AM, Arkin MR, Stemp EDA, Barton JK, Barbara PF. J Am Chem Soc. 1997;119:11458. [Google Scholar]; (b) Onfelt B, Lincoln P, Norden B, Baskin JS, Zewail AH. Proc Natl Acad Sci USA. 2000;97:5708. doi: 10.1073/pnas.100127397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupureur CM, Barton JK. Inorg Chem. 1997;36:33. [Google Scholar]
- 23.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Nature Struct Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 24.(a) Eriksson M, Leijon M, Hiort C, Norden B, Graslund A. Biochemistry. 1994;33:5031. doi: 10.1021/bi00183a005. [DOI] [PubMed] [Google Scholar]; (b) Lincoln P, Broo A, Norden B. J Am Chem Soc. 1996;118:2644. [Google Scholar]; Greguric A, Greguric ID, Hambley TW, Aldrich-Wright JR, Collins JG. J Chem Soc Dalton Trans. 2002:849–855. [Google Scholar]
- 25.(a) Jackson BA, Barton JK. J Am Chem Soc. 1997;119:12986. [Google Scholar]; (b) Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]; (c) Jackson BA, Barton JK. Biochemistry. 2000;39:6176. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 26.(a) Zeglis BM, Boland JA, Barton JK. J Am Chem Soc. 2008;130:7530. doi: 10.1021/ja801479y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zeglis BM, Boland JA, Barton JK. Biochemistry. 2009;48:839. doi: 10.1021/bi801885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Pierre VC, Kaiser JT, Barton JK. Proc Natl Acad Sci USA. 2007;104:429. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cordier C, Pierre VC, Barton JK. J Am Chem Soc. 2007;129:12287. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickeson JE, Summers LA. Aust J Chem. 1970;23:1023. [Google Scholar]
- 29.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 30.(a) Sigman DS, Mazumder A, Perrin DM. Chem Rev. 1993;93:2295. [Google Scholar]; (b) Lim MH, Lau IH, Barton JK. Inorg Chem. 2007;46:9528. doi: 10.1021/ic701598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Holmlin RE, Barton JK. Inorg Chem. 1995;34:7. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]; (b) Holmlin RE, Stemp EDA, Barton JK. J Am Chem Soc. 1996;118:5236. [Google Scholar]; (c) Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, O’Shea VL, David SS, Barton JK. Proc Natl Acad Sci USA. 2005;102:3546. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barton JK. Science. 1986;233:727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 33.Dervan PB. Biorg Med Chem Lett. 2001;9:2215. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 34.Armitage BA. Top Curr Chem. 2005;253:55. [Google Scholar]
- 35.SantaLucia J, Jr, Hicks D. Annu Rev Biophys Biomol Struct. 2004;33:415. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 36.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]