Abstract
HLA disparity between hematopoietic stem cell donors and recipients is one of the most important factors influencing transplant outcomes, but there are no well accepted guidelines to aid in selecting the optimal donor amongst several HLA mismatched donors. In this report, HLA-A is used as a model to illustrate factors that are barriers to delineating the relationship between specific HLA mismatches and transplant outcomes in the United States. Patients in this investigation received transplants for hematological malignancies that were facilitated by the National Marrow Donor Program (NMDP) between 1990 and 2002 (n=4,226). High resolution HLA typing was performed for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1. HLA-A mismatches were observed in 745 donor-recipient pairs and 62% of these pairs also had disparities at HLA-B, -C and/or -DRB1. The HLA-A mismatches involved 190 different combinations of HLA-A alleles and 51% of these were observed in only one pair. Addition of a single HLA-A disparity when HLA-B, -C, and -DRB1 were matched (n=282) was associated with increased mortality (OR=1.32, CI 1.07-1.63). When HLA-B, -C, and DRB1 were matched, the most frequent HLA-A mismatches were HLAA*0201:0205 (n=28), HLA-A *0301:0302 (n=15), HLA-A *0201:0206 (n=15), HLAA *0201:6801 (n=12), HLA-A*0101:1101 (n=11) and HLA-A*0101:0201 (n=10). There were no statistically significant relationships between any of these disparities and transplant outcomes (engraftment, acute and chronic GVHD, relapse, transplant-related mortality or overall survival) when adjustments for multiple comparisons were considered. Achieving 80% power to detect an effect of any one of these six HLA-A disparities on survival is estimated to require a total transplant population of 11,000 to more than one million U.S. donor-recipient pairs depending upon the HLA disparity. Thus, alternative approaches are required to develop a clinically relevant ranking system for specific HLA disparities in the U.S.
Keywords: HLA, Histocompatibility, Bone Marrow Transplantation, Hematopoietic stem cell transplantation
INTRODUCTION
For blood and bone marrow transplantation (BMT), one of the few factors that the physician can modify to improve transplant outcomes is donor compatibility [1]. An HLA identical donor is optimal, but matched donors are not available for most patients. It is often necessary to prioritize several candidate donors who have different HLA disparities. For example, it may be necessary to choose between (1) a mismatch at the HLA-A or HLA-DRB1 locus, (2) an allele-level mismatch and an antigen-level mismatch, or (3) an allele with one amino acid difference and another with three amino acid differences. There is limited and sometimes contradictory evidence to support these decisions. Many factors including numerous confounding clinical variables, limitations in the ability to adjust for the effects of multiple HLA disparities, and statistical limitations imposed by the size of data sets are barriers to solving this problem [2, 3].
For studies comparing different HLA loci, HLA-A, -B, -C, and -DRB1 disparities have most consistently associated with adverse outcomes [1, 4-8]. In one of the largest of these studies, there was a 9 to 10% decrement in survival for each HLA-A, -B, -C, or -DRB1 mismatch [1]. Several studies have suggested differences between allele-level and antigen-level disparities [4, 8], but one of the largest studies using a rigorous statistical model did not confirm this relationship for a single allele versus a single antigen level mismatch [1]. It has been more difficult to detect associations between HLA-DQ and -DP disparities and transplant outcomes, but associations have been reported under certain conditions [1, 6, 9-11].
Some recent clinical investigations have attempted to investigate particular HLA mismatches or specific amino acid differences. A Japanese study reported 15 combinations of mismatched alleles and six specific amino acid differences that were associated with graft-versus-host disease (GvHD) [12]. Since large numbers of comparisons were required to detect this small number of statistically significant combinations and there were many confounding variables, independent confirmation or other supporting data are essential [13]. An international study focused on disparities involving HLA-A*02 alleles reported that mismatches between HLA-A*0201 and HLA-A*0206 were deleterious but mismatches between HLA-A*0201 and HLA-A*0205 were tolerated [14]. Interestingly, HLA-A*0201 and HLA-A*0206 are distinguished by a single amino acid difference at position 9. HLA-A*0201 and HLA-A*0205 have the same amino acid difference along with two additional mismatched amino acids. The HLA-A*0206 mismatches were predominantly observed in Japanese patients while the HLA-A*0205 mismatches were in non-Japanese patients.
Fundamental knowledge about the molecular mechanisms underlying allorecognition should be useful in prioritizing HLA mismatches. There is a large body of research using animal models as well as in vitro assays which show that the molecular characteristics of Major Histocompatibility Complex (MHC) disparities influence allorecognition. Alloreactive T lymphocytes play a key role in graft rejection, graft-versus-host disease, and graft-versus leukemia effects. These T cells respond to differences between HLA molecules and/or peptide antigens bound by HLA molecules [15, 16]. Elsner et al. used these concepts to rank HLA disparities based upon the number of amino acid differences between HLA molecules with adjustments for amino acid position and a similarity index (Risler score) [17]. However, in vitro and clinical observations have not supported this particular approach [18, 19]. Another algorithm, HLA Matchmaker, which uses amino acid triplets within the HLA molecule, showed some promise in solid organ transplantation, but was not useful for bone marrow transplantation [20].
The long-term goal of this investigation is to utilize a combination of fundamental knowledge about allorecognition along with clinical outcomes to develop a new system for evaluating candidate donors who are HLA mismatched. The purpose of the study described here is to delineate the HLA factors that need to be considered to investigate the impact of HLA disparities and to test the hypothesis that there are certain common HLA disparities that are deleterious. HLA-A was selected as the model for testing this hypothesis because HLA-A mismatches have been associated with adverse outcomes in every large clinical investigation and a recent NMDP report suggests that mismatches at HLA-A or -DRB1 may have greater effects on survival than HLA-B or -C mismatches [1]. The data reported here show the limitations posed by the diversity of HLA disparities in unrelated donor transplants performed in the U. S. and present transplant outcomes for the most frequent HLA-A disparities.
MATERIALS AND METHODS
Patients
This investigation was approved by the NMDP Institutional Review Board. Adults transplanted under the auspices of the NMDP between 1990-2002 for acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), myelodysplastic syndrome (MDS) or non-Hodgkin’s lymphoma (NHL) who had high resolution typing for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 were eligible for this investigation. The subject population was adjusted as described previously to correct for potential bias that may have been created by consent practices [1]. Early stage disease was defined as AML and ALL in first complete remission, CML in first chronic phase and MDS subtype refractory anemia. Intermediate stage disease was AML or ALL in second or subsequent complete remission or in first relapse, and CML in accelerated phase or second chronic phase. Advanced phase disease was AML in second or higher relapse or primary induction failure, CML in blast phase, MDS subtypes refractory anemia with excess blasts or in transformation, or MDS, not otherwise classified. NHL cases were not classified for disease stage.
Characteristics of the patients (n=4,226) and donors are provided in Supplemental Table 1. Demographic data were submitted by the transplant centers and subjected to a rigorous quality assurance process including on-site auditing against medical records, double data entry, on-line edit checks and review of the data for inconsistencies and omissions. Race was self reported. The majority of the recipients and donors were Caucasian (recipients: 87% and donors: 85%). For cases where race was reported for both donor and recipient, 87% of the pairs reported the same race/ethnicity. The majority of patients received myeloablative conditioning regimens (93%) and calcineurin inhibitor based GVHD prophylaxis (78%).
HLA Matching
Allele-level HLA typing was performed for HLA-A, -B, -C,-DRB1, -DQ, and -DP using methods that were previously described in detail [21]. Briefly, DNA-based HLA typing was based on exons 2 and 3 for HLA Class I and exon 2 for HLA Class II. Known null alleles were excluded using serological typing or extended DNA-based typing.
HLA mismatching was determined by comparing the peptide sequences encoded by the exons that were used for HLA typing (i.e., exons 2 and 3 for HLA Class I and exon 2 for HLA Class II) using a sequence database for the human MHC [22]. Low resolution HLA mismatches were defined as differences in the first two digits of the HLA types (e.g., HLA-A*0101 vs. HLA-A*0201) and high resolution mismatches were defined as differences in only the last two of four digit HLA types (e.g., HLA-A*0201 vs. HLA-A*0205). Particular combinations of mismatched alleles in this study are reported as HLA locus*first allele:second allele. For example, a mismatch involving HLA-A*0101 and HLA-A*0201 is denoted A*0101:0201. HLA-DQ and -DP disparities were not included in the assessment of overall HLA matching because mismatches at these loci did not individually have detectable effects on survival. If a donor had a homozygous HLA type and the recipient was heterozygous at the same locus, the mismatch was treated as a unidirectional mismatch for GvHD. Conversely, mismatches were treated as unidirectional for engraftment if a recipient had a homozygous HLA type and the donor was heterozygous.
For analysis of HLA-A mismatches, the orientation of amino acid side chains was based upon the position of the side chain in HLA-A crystal structures [23-25] as well as peptide contact residues defined by Chelvanayagam [26]. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Clinical Outcomes
The primary endpoint for this analysis was overall survival which was defined as the time from graft infusion (day 0) to death from any cause. Several secondary endpoints were also examined. Engraftment was defined as maintaining an absolute neutrophil count greater than 500 × 106/L for three consecutive measurements by day 28. Acute GvHD was graded using the Glucksberg scale [27] and chronic GvHD was graded using the method of Shulman et al. [28]. Disease relapse was defined using criteria determined by the Center for International Blood and Marrow Transplant Research (CIBMTR). Transplant-related mortality (TRM) was defined as death while in continuous complete remission from the primary disease.
Statistics
Continuous variables are presented as the medians and ranges. Categorical variables are reported as percentages. The Kaplan-Meier estimator was used for determining probabilities for survival endpoints with variance estimated by Greenwood’s formula. Comparisons of survival curves were done with the logrank test. Multivariate analyses were performed using the proportional hazards model to compare the HLA matched and mismatched groups. Models were fit to determine which risk factors may be related to a given outcome. All models were tested for significant clinical covariates including disease, disease stage, Karnofsky performance status, donor-patient cytomegalovirus (CMV) serology, patient race, patient age, T cell depletion, use of total body irradiation (TBI), graft source (peripheral blood or bone marrow), donor age, patient-donor sex match and year of transplant. Models included any clinical factors that were related to a given outcome at p less than 0.05. All variables were tested for the affirmation of the proportional hazards assumption. Factors violating the proportional hazards assumption were adjusted first before the stepwise model building approach was used in developing models for the primary and secondary outcomes. Due to multiple comparisons a threshold of p less than 0.01 for the main effect was used for significance.
RESULTS
Characteristics of HLA-A Disparities
HLA-A mismatches were observed in 745 donor-recipient pairs and 62% of these pairs also had disparities at HLA-B, -C and/or -DRB1. One of the factors that many transplant centers use to classify HLA disparities is the level of typing resolution required to detect the mismatch: high resolution types (allele-level) or low resolution types (antigen level). Figure 1 shows that these classifications are generally indicative of the number of amino acid differences between two HLA-A molecules with low resolution mismatches having 3-28 amino acid differences and high resolution mismatches having 1-6 amino acid differences. For low resolution disparities, there are a mean 13.5 amino acid differences (median 12) and most of the disparities have more than 6 amino acid differences. In this study there are four “low resolution” mismatch combinations that have only 3 amino acid differences: A*0101:3601 (n=2), A*2301:2402 (n=11), A*2601:6601 (n=8), and A*3101:3303 (n=3). The HLA-A mismatches with the largest number of amino acid differences (n=28) are A*2301:3601 (n=1) and A*2301:6601 (n=1).
Figure 1.
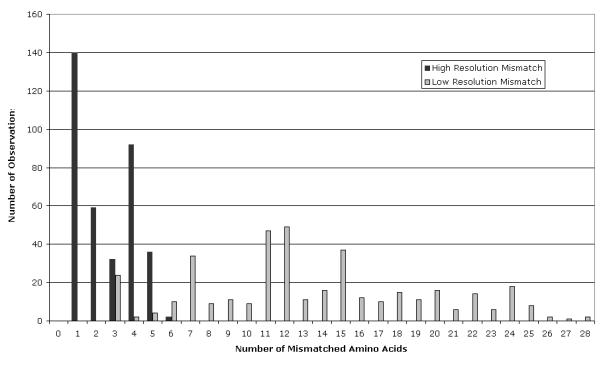
Number of mismatched amino acid residues in each HLA-A disparity in 4,226 donor-recipient pairs.
High resolution HLA-A mismatches have only 1-6 mismatched amino acids (mean 2, median 2). Six amino acid differences are observed in only one relatively uncommon disparity, A*3001:3004 (n=2). In this dataset there are 26 different HLA-A high resolution mismatches that have a single amino acid difference. These single amino acid differences are located at 19 different positions within the HLA-A molecule (3, 9, 19, 36, 54, 66, 70, 74, 87, 97, 99, 102, 116, 127, 131, 144, 156, 163, and 171). Ten of these are predicted to have side chains that contact the peptide bound to the HLA molecule [26]. These data show that a HLA disparities defined by a single amino acid difference constitute a diverse group because the functional role of the mismatched amino acid is extremely variable.
There has been interest in determining if HLA mismatches involving particular amino acid positions are associated with unfavorable outcomes. In general, the amino acid positions that are observed in the largest number of different mismatch combinations (Table 1) influence the peptide binding groove of the HLA molecule (Figure 2). However, there are also 54 different amino acid positions that are involved in only one mismatch combination. These are distributed throughout the HLA molecule and each of these combinations is infrequent (1-5 observations/combination). Since the impact of a particular amino acid position is influenced by the molecular context of the amino acid disparity, position alone is unlikely to be sufficient for predicting effects. Position 9 exemplifies the potential for numerous interactions that are likely to confound comparisons which are focused on particular positions within the HLA molecule. In this study position 9 is mismatched in 389 of the HLA-A disparities and these disparities involve 105 different combinations of HLA-A molecules. Three of these combinations involve a single amino acid mismatch (n=64) with the majority being A*0201:0206 (n=60). The remaining mismatch combinations that involve a difference in position 9 have up to 27 additional amino acid differences. The most frequent numbers of additional amino acid differences are 4 (n=67, 3 combinations), 12 (n=47, 7 combinations), and 11 (n=41, 9 combinations). Considering this extensive variability, a study grouping any amino acid difference at position 9 is questionable from the functional perspective.
Table 1.
Amino Acids that Are Different in the Largest Number of HLA-A Mismatch Combinations
| Amino Acid Position* |
HLA-A Mismatch Combinations (n) |
|---|---|
| 9 | 105 |
| 62 | 99 |
| 63 | 72 |
| 70 | 74 |
| 76 | 84 |
| 77 | 76 |
| 97 | 91 |
| 114 | 97 |
| 116 | 71 |
| 156 | 90 |
| 152 | 85 |
Based upon numbering of amino acids in the mature protein.
Figure 2.
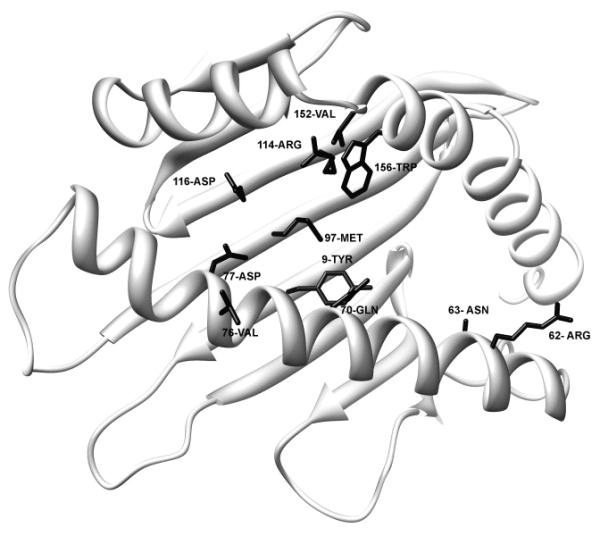
Structure of HLA*6801. Amino acids that are observed in the largest number of different mismatch combinations are distributed throughout the peptide binding domain. The side chains of the amino acids that are observed in the largest number of different mismatch combinations are shown on an HLAA*6801 structure (2HLA in the Protein Data Bank). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Many of the highly variable amino acid positions are predicted to influence the peptides that can bind to the HLA molecule and/or their precise position within the peptide binding groove. This can be examined using nine peptide environments which have been shown to alter the amino acid preference for each of the positions in a nonamer peptide [26]. In the donor-recipient pairs in this study, more than half of the 54 amino acid positions that were disparate influence these peptide binding environments. If the frequency of each mismatch combination is considered, the amino acids that are most often mismatched are all located in positions that alter these nine peptide binding environments: positions 9 (n=389), 76 (n=255), 97 (n=251),114 (n=251), 156 (n=362), 152 (n=248), 77 (n=241), 95 (n=239), 62 (n=229), 70 (n=225), and 116 (n=204), where n is the number of donor-recipient pairs with a mismatch at the position. With the exception of position 95, these frequently mismatched amino acids influence several of the nine peptide binding environments. The effects on peptide binding are expected to be extremely variable.
Multiple HLA Disparities
The clinical impact of a particular HLA-A disparity may be confounded by the effects of additional mismatches at other HLA loci. The high frequency of multiple HLA locus mismatches is demonstrated in Figure 3 which shows each amino acid position that was mismatched at HLA-A in this subject population along with the proportion of mismatches that involve additional mismatches at HLA-B, -C, and -DRB1. Overall, 62% of the HLA-A mismatches are accompanied by additional disparities at HLA-B, -C, and/or -DRB1.
Figure 3.
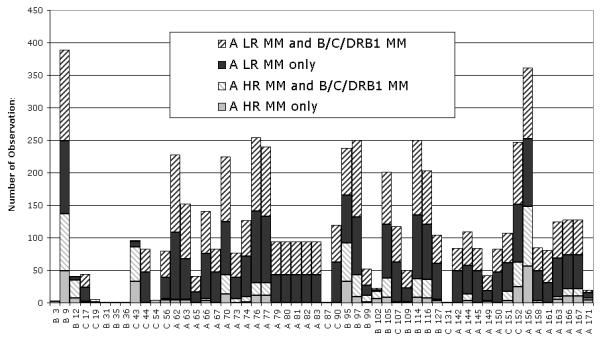
Amino acid differences that are involved in HLA-A disparities are distributed throughout the peptide binding domain, present in both low resolution (LR) and high resolution (HR) mismatches, and often accompanied by mismatches (MM) at HLA-B, -C, and DRB1 loci. The position and side-chain orientation of each mismatched amino acid are indicated below the x-axis (A=alpha helix, B= beta sheet, and C=connecting loop). The amino acids that are most frequently mismatched line the peptide binding groove of the HLA molecule. Most of the mismatched amino acids are involved in both low and high resolution mismatches.
Most Frequent HLA-A Disparities
Table 2 lists the 16 HLA-A disparities that were observed at least 10 times in the entire data set (including those with additional mismatches at HLA-B, -C, and/or DRB1). Investigating the effects of particular HLA mismatches or mismatched positions is simplified by focusing on single HLA mismatches because strategies to compensate for additional disparities are currently suboptimal. If HLA-A disparities are considered only when HLA-B, -C, and -DRB1 are matched, there are only six mismatch combinations that are observed at least 10 times in a population of 4,226 transplants. These HLA-A mismatch combinations have very different properties including differences in the number of mismatched amino acids (1-24 amino acids, Table 4). One common feature is that all of them have at least one amino acid difference located in a position that is predicted to influence the peptides bound to the HLA molecules.
Table 2.
HLA-A Disparities Observed in at Least 10 Donor-Recipient Pairs
| HLA MM | Number of Observations in 4,226 Pairs |
Number of Observations in HLA-B, -C, -DRB1 Matched Pairs (%) |
Number of Mismatched Amino Acids |
|---|---|---|---|
| 0201-0205 | 65 | 28 (43) | 4 |
| 0201-0206 | 60 | 15 (25) | 1 |
| 6801-6802 | 34 | 7 (21) | 5 |
| 0201-6801 | 30 | 12 (40) | 12 |
| 0301-0302 | 27 | 15 (56) | 2 |
| 3001-3002 | 23 | 7 (30) | 4 |
| 0101-1101 | 23 | 11 (48) | 11 |
| 0301-1101 | 19 | 8 (42) | 7 |
| 0101-0301 | 19 | 9 (47) | 15 |
| 2901-2902 | 17 | 6 (35) | 1 |
| 2402-2403 | 17 | 9 (53) | 2 |
| 0201-0202 | 16 | 4 (25) | 3 |
| 0101-0201 | 13 | 10 (77) | 24 |
| 2501-2601 | 12 | 7 (58) | 7 |
| 3101-3201 | 12 | 5 (42) | 12 |
| 2301-2402 | 11 | 3 (27) | 3 |
Table 4.
Transplant Outcomes for the Six Most Frequent HLA-A Disparities in HLA-B, -C, and -DRB1 Matched Pairs
| HLA Mismatch | A*0201: 0205 |
A*0301: 0302 |
A*0201: 0206 |
A*0201: 6801 |
A*0101: 1101 |
A*0101: 0201 |
|---|---|---|---|---|---|---|
|
Number of Amino Acid Differences |
4 | 2 | 1 | 12 | 11 | 24 |
|
Position and Single Letter Code for Amino Acid Differences |
9FY 43QR 95VL 156LW |
152EV 156LQ |
9FY | 9FY 62GR 63EN 66KN 70HQ 74HD 95VI 97RM 107WG 114HR 116YD 156LW |
9FY 44KR 67MV 70HQ 76AV 77ND 150VA 156RQ 158VA 166DE 167GW |
44KR 62QG 66NK 67MV 74DH 76AV 77ND 90DA 95IV 97IR 107GW 114RH 116 DY 127NK 142IT 145RH 150VA 152AV 156RL 158VA 163RT 105PS 166DE 167GW |
| N | 27* | 15 | 15 | 11* | 11 | 9* |
|
OR for Survival 95% CI |
2.08 0.86-4.80 (p=0.07) |
1.10 0.25-3.73 |
2.64 0.81-8.38 |
2.52 0.60-9.94 |
1.13 0.19-4.74 |
0.86 0.09-4.55 |
|
OR for Acute GvHD 95% CI |
1.41 0.55-3.33 |
1.03 0.24-3.48 |
3.22 1.02-10.50 (p=0.03) |
1.06 0.18-4.43 |
2.35 0.56-9.29 |
0.0 0.0-1.12 |
Three cases (1 A*0201:0205, 1 A*0201:6801 and 1 A*0101:0201) were not included in the multivariate analysis due to missing data.
Clinical Outcomes for the Most Frequent HLA-A Disparities
Since knowledge regarding the interactions caused by multiple HLA disparities is extremely limited, outcomes analysis was limited to pairs having a single HLA-A mismatch with matching at HLA-B, -C, and -DRB1. To select the reference population, donor-recipient pairs with 8/8 matches (HLA-A, -B, -C, -DRB1), 12/12 matches (HLA-A, -B, -C, -DRB1, -DQA1, -DQB1) and 16/16 matches (HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1) were compared. For overall survival, there were no differences between the 16/16 matched group and those with HLA-DP mismatches (12/12 matches, OR=1.00, 95% confidence interval 0.84-1.18) or those with HLA-DP and/or -DQ mismatches (8/8 matches, OR=1.01, 95% confidence interval 0.80-1.29). Therefore, the 8/8 matched group was used as the reference group for pairs with HLA-A mismatches. A single disparity at the HLA-A locus was associated with increased mortality (OR=1.32, 95% confidence interval 1.07-1.63).
To determine if it is possible to detect detrimental effects of any of the most common HLA-A mismatches, transplant outcomes were examined for HLA-A mismatches that were observed in at least 10 pairs when HLA-B, -C, and -DRB1 were matched. There are only six HLA-A mismatches that met these criteria: A*0201:0205, A*0301:0302, A*0201:0206, A*0201:6801, A*0101:1101, and A*0101:0201 (Table 3). The patient and donor characteristics of these pairs which are provided in Table 3 show that there were no major differences amongst these groups. For overall survival, with relatively small numbers of patients there are no statistically significant differences between pairs with these HLA disparities and the 8/8 matched pairs (data not shown). The most common single HLA-A mismatch is A*0201:0205 (OR=2.08, 95% confidence interval 0.86-4.80, p=0.07), but this did not reach statistical significance. Multivariate results for survival of pairs with the six HLA mismatches compared with the 8/8 matched pairs has an overall p-value of 0.075.
Table 3.
Demographics of Patients and Donors for the Six Most Frequent HLA-A Disparities in HLA-B, -C, and -DRB1 Matched Pairs
| Variable | A*0201- A*0205 N (%) |
A*0301- A*0302 N (%) |
A*0201- A*0206 N (%) |
A*0201- A*6801 N (%) |
A*0101- A*1101 N (%) |
A*0101- A*0201 N (%) |
|---|---|---|---|---|---|---|
| Patients (n) | 28 | 15 | 15 | 12 | 11 | 10 |
| Centers (n) | 22 | 12 | 13 | 9 | 7 | 8 |
| Patient Age | ||||||
| Median (y) | 27 | 37 | 23 | 32 | 42 | 29 |
| Range (y) | 1-51 | 9-58 | 3-51 | 1-54 | 2-50 | 13-45 |
| < 20 y | 7 (25) | 1 (6) | 7 (46) | 4 (34) | 2 (18) | 4 (40) |
| 20-39 y | 14 (50) | 7 (47) | 4 (27) | 4 (33) | 3 (27) | 5 (50) |
| 40+ y | 7 (25) | 7 (47) | 4 (27) | 4 (33) | 6 (55) | 1 (10) |
| Male sex | 14 (50) | 7 (47) | 10 (67) | 7 (58) | 4 (36) | 7 (70) |
| Karnofsky ≥ 90 | 21 (78) | 12 (80) | 12 (80) | 8 (73) | 7 (64) | 9 (90) |
| Disease | ||||||
| AML | 7 (25) | 1 (7) | 5 (33) | 5 (42) | 3 (27) | 4 (40) |
| ALL | 10 (36) | 1 (7) | 3 (20) | 1 (8) | 0 | 0 |
| CML | 6 (21) | 11 (73) | 7 (47) | 3 (25) | 6 (55) | 4 (40) |
| MDS | 4 (14) | 2 (13) | 0 | 2 (17) | 2 (18) | 2 (20) |
| NHL | 1 (4) | 0 | 0 | 1 (8) | 0 | 0 |
| Disease status | ||||||
| Early | 7 (25) | 9 (60) | 6 (40) | 4 (33) | 4 (36) | 6 (60) |
| Intermediate | 11 (39) | 4 (27) | 9 (60) | 5 (42) | 4 (36) | 2 (20) |
| Advanced | 8 (29) | 2 (13) | 0 | 1 (8) | 1 (9) | 1 (10) |
| Other | 2 (7) | 0 | 0 | 2 (17) | 2 (18) | 1 (10) |
| Marrow | 28 (100) | 15 (100) | 15 (100) | 11 (92) | 10 (91) | 9 (90) |
| Conditioning | ||||||
| Ablative | 27 (96) | 14 (93) | 15 (100) | 12 (100) | 11 (100) | 10 (100) |
| Other | 1 (4) | 1 (7) | 0 | 0 | 0 | 0 |
| GvHD Prophylaxis | ||||||
| FK506 | 5 (18) | 2 (13) | 2 (13) | 4 (33) | 0 | 1 (10) |
| CsA | 17 (61) | 10 (67) | 13 (87) | 6 (51) | 10 (91) | 6 (60) |
| T-cell depletion | 6 (21) | 3 (20) | 0 | 1 ( 8) | 1 (9) | 3 (30) |
| Other | 0 | 0 | 0 | 1 ( 8) | 0 | 0 |
| Donor/recipient sex match |
||||||
| M/M | 10 (36) | 6 (40) | 7 (47) | 4 (33) | 2 (18) | 3 (30) |
| M/F | 10 (36) | 3 (20) | 3 (20) | 2 (17) | 5 (46) | 2 (20) |
| F/M | 4 (14) | 1 ( 7) | 3 (20) | 3 (25) | 2 (18) | 4 (40) |
| F/F | 4 (14) | 5 (33) | 2 (13) | 3 (25) | 2 (18) | 1 (10) |
| Donor/recipient CMV match |
||||||
| N/N | 14 (50) | 7 (47) | 2 (14) | 3 (25) | 5 (46) | 4 (40) |
| N/P | 6 (22) | 6 (40) | 3 (20) | 2 (17) | 3 (27) | 2 (20) |
| P/N | 4 (14) | 0 | 5 (33) | 1 ( 8) | 2 (18) | 3 (30) |
| P/P | 4 (14) | 2 (13) | 5 (33) | 5 (42) | 1 ( 9) | 0 |
| Unknown | 0 | 0 | 0 | 1 ( 8) | 0 | 1 (10) |
| Donor age (y) Median (range) |
40 (20-57) |
39 (21-50) |
36 (20-53) |
36 (29-47) |
39 (22-53) |
38 (22-49) |
| Year of transplant | ||||||
| 1990-1994 | 6 (21) | 3(20) | 3(20) | 1 ( 8) | 3(27) | 5(50) |
| 1995-2002 | 22(79) | 12(80) | 12(80) | 11(92) | 8(73) | 5(50) |
| Follow-up median mo (range) |
60 (48-101) |
111 (36-144) |
61 (22-99) |
77 (35-97) |
90 (60-157) |
55 (34-154) |
For severe acute GvHD (grades 3-4), the overall p-value for comparison of these mismatches with 8/8 matches is 0.014 and data for the individual HLA-A mismatches is shown in Table 4. Interestingly, A*0201:0206 which was found to have an association with acute GvHD in another study [12] is approaching significance in this study (OR=3.22, 95% CI 1.02-10.50, p=0.03). There were no significant differences detected for engraftment (overall p-value 0.36), chronic GvHD (overall p-value = 0.28), relapse (overall p-value = 0.19), transplant-related mortality (overall p-value = 0.14), or disease-free survival (overall p-value = 0.18) (data not shown).
DISCUSSION
One of the major challenges in blood and marrow transplantation is selection of the most compatible donor when an HLA matched donor is not available. Although several investigations have attempted to determine which HLA disparities are poorly tolerated, the results have often been contradictory. The investigation reported here uses one of the largest study populations to date (n=4,226) to illustrate that extensive HLA diversity is a major barrier to establishing the association between HLA disparity and transplant outcomes.
HLA-A was selected for this investigation because mismatches at this locus have been most consistently associated with adverse transplant outcomes [1, 4, 5, 12]. In the 4,226 pairs examined in this investigation, there were 190 different HLA-A mismatch combinations and 51% of these were observed in only one pair. Very few HLA-A disparities were observed frequently enough to study. Only four HLA-A disparities were observed in 30 or more donor-recipient pairs (A*0201:0205, A*0201:0206, A*6801:6802, A*0201:6801). Sixteen HLA disparities were observed in 10 or more donor-recipient pairs (Table 2). If pairs with additional HLA-B, -C, and -DRB1 disparity are excluded to diminish confounding HLA variables, only 6 mismatch combinations occur in 10 or more pairs (Table 4). Since the most frequent HLA-A mismatches in HLA-B, -C, and -DRB1 matched pairs occur only 2-6 times/1,000 U.S. patients, it is not feasible to directly determine associations between a particular HLA-A disparity and transplant outcomes for patients transplanted in the U.S.
One of the goals of this investigation was to identify frequent HLA disparities that are deleterious. However, there were only 6 mismatch combinations observed more than 10 times in HLA-B, -C, and -DRB1 matched pairs and when survival of each of these small groups was compared with that of the HLA-matched control group, no statistically significant associations were detected. A*0201:0205, the most frequent HLA-A disparity observed when HLA-B, -C, and -DRB1 were matched, had a trend toward increased mortality at one year (p=0.07, OR=2.08, 95% CI 0.86-4.80). A study from the 14th International Histocompatibility Workshop examined 51 pairs with A*0201:0205 mismatches using additional HLA mismatches as a co-variable and was unable to detect any association between this disparity and survival [14]. Additional donor recipient pairs are required to evaluate the impact of this disparity.
A trend for increased severe acute GvHD was detected for one of the most frequent mismatches, A*0201:0206 (OR=3.22, 95% CI 1.02-10.50, p=0.03). This is provocative because two recent publications reported associations between A*0201:0206 and adverse outcomes. Kawase et al., who studied transplants facilitated by the Japan Marrow Donor Program (JMDP), reported that a graft from an A*0206 donor to an A*0201 recipient was significantly associated with GvHD (OR=1.78, p<0.001) [12]. An analysis of data from the 14th International Histocompatibility Workshop suggested that A*0201:0206 mismatches were associated with mortality [14]. One limitation in these two studies is that it is likely that there is some overlap in the subjects because many of the A*0201:0206 mismatched pairs examined in the Workshop were obtained from the JMDP. Nevertheless, the A*0201:0206 mismatch combination has now been identified as a potential risk factor in several studies, suggesting that this particular mismatch may be associated with acute GvHD and perhaps mortality.
A high incidence of certain HLA-A disparities in the JMDP data set allowed Kawase et al. to examine directional differences in HLA disparities [12]. For example, in their analysis an A*0201 donor:A*0206 recipient was not detrimental (OR=1.23, p=.22) while A*0201 recipient:A*0206 donor (OR=1.78, p<0.001) was significantly associated with GvHD. In their study, there were three HLA-A disparities that were associated with acute GvHD and all of these associations were directional. The possibility of directional effects has also been suggested by laboratory studies involving B*4402:4403 where (1) in vitro assays, (2) characteristics of the peptides bound by the two HLA molecules, and (3) crystal structures of the molecules have shown that the same peptide in the HLA binding groove can have different orientations in HLA-B*4402 and *4403. These differences can be recognized by T lymphocytes [15, 16, 29, 30] and may be clinically important [31, 32]. For studies involving heterogeneous patient populations such as those from the NMDP, there are too few subjects with particular HLA disparities to further subdivide the mismatches according to their presence in the donor or recipient and this limitation may obscure significant effects.
One of the striking differences between reports from the JMDP and NMDP data sets is the incidence of particular HLA disparities. A study of 5,210 subjects whose transplants were facilitated by the JMDP reported very large frequencies for certain HLA-A mismatch combinations: A*0201:0206 (n=269), A*2402:2420 (n=90), and A*2601:2603 (n=69). The allelic lineages HLA-A2, A26 and A24 present rich diversity in the Japanese population [33]. In contrast the European populations present only a limited number of alleles in these groups because there is one predominant allele in each lineage. The NMDP data set is largely composed of patients and donors with European ancestry where most of the pairs are HLA matched for common alleles and the mismatches often involve low frequency alleles.
Using the frequencies of the six most common HLA-A disparities observed in this study when HLA-B, -C, and -DRB1 are matched and the 100 day mortality difference observed between each disparity and an 8/8 match, the number of U.S. donor recipient pairs needed to achieve 80% power to detect an association with survival ranges is 11,000 to more than 1.3 million, depending upon the HLA mismatch (Table 5). For example, it is estimated that 72 pairs with a single A*0201:0205 would be required to achieve 80% power to detect an association with survival versus an 8/8 match. Using the frequency of 27 pairs with this HLA disparity in 4,226 subjects, it is estimated that 11,269 pairs would be required to detect an association with survival. Attempts to overcome this limitation by including pairs with multiple HLA mismatches creates another problem--adjusting for the effects of particular HLA disparities are not yet well understood.
Table 5.
Number of Subjects Needed to Achieve 80% Power for Detecting an Association with 100 day Mortality for the Six Most Frequent HLA-A Disparities in HLA-B, -C, and -DRB1 Matched Pairs*
| HLA-A Mismatch | Number of Mismatches in 4,226 Subjects |
Number Required for 80% Power |
Total Number of Subjects Required to Achieve 80% Power |
|---|---|---|---|
| A*0201:0205 | 27 | 72 | 11,269 |
| A*0301:0302 | 15 | 4,687 | 1,320,484 |
| A*0201:0206 | 15 | 39 | 10,988 |
| A*0201:6801 | 11 | 43 | 16,520 |
| A*0101:1101 | 11 | 2,653 | 1,019,234 |
| A*0101:0201 | 9 | 2,014 | 945,685 |
| A*2402:2403 | 8 | 30 | 15,848 |
Using the difference observed between each group and an 8/8 match.
One alternative which has been explored is to investigate the impact of mismatching particular amino acids within the HLA molecule. Kawase et al. used bootstrap analysis to examine each amino acid involved in an HLA mismatch and concluded that two specific amino acid substitutions in HLA-A were associated with acute GvHD: position 9 Tyr vs Phe and position 116 Asn vs Asp [12]. This approach however has some limitations and the conclusions about some specific residues may be biased since context of the amino acid differences is ignored. For example, in the NMDP data set residue 9 can be mismatched along with 0-28 additional amino acids; a large body of laboratory investigation suggests that context of an amino acid difference is crucial for allorecognition. The analysis conducted by Kawase et al. may also be confounded by the occurrence of additional HLA mismatches in each donor-recipient pair. In this scenario specific amino acid mismatches may occur in one or a few mismatch combinations that may be accompanied by an additional mismatch in another locus as the result of linkage disequilibrium. Therefore, the effect attributed to a particular amino acid mismatch at given locus may in fact result from the occurrence of multiple mismatches. Another concern is that multiple comparisons were made in the study by Kawase et al. Multiple comparisons without support from functional data can detect relationships that are statistically significant but may not be clinically or biologically relevant [13]. In vitro assays may help to clarify the situation for certain HLA disparities [34], but this approach is not practical for studying the large number of combinations of HLA mismatches that are observed in clinical transplantation.
This study shows that extraordinary diversity of HLA creates complexity related to multiple HLA disparities between donor and recipient along with differences in the context in which each amino acid difference occurs within HLA molecules. Decades of research in transplant immunology have taught us that these are important factors influencing the ability to stimulate a clinically significant response against an alloantigen. Unfortunately, the relatively large study population examined here (n=4,226) provides only a few examples for investigation and none of these have sufficient subjects to apply robust multivariate statistical approaches. Given extensive HLA polymorphism and complex biology, it is unlikely that the approaches described above will be successful in ranking the relative risks of most of the HLA disparities that are encountered in clinical practice in the United States. Although it may be possible to study certain HLA disparities in specific populations such as the Japanese, these observations may not extend to other racial and ethnic groups because there may be other genetic differences that influence immune responses.
There have been attempts to identify permissive and detrimental HLA disparities using characteristics of HLA mismatches. Several studies have classified HLA disparities based upon typing resolution: low resolution (antigen level) and high resolution (allele level) mismatches. This approach assumes that HLA mismatches detected by serological HLA typing reagents (i.e., low resolution) have different clinical effects that those which are not. The reported studies have been conflicting with the largest detecting no statistically significant difference for a single allele vs a single antigen mismatch [1]. One explanation is that this comparison does not adequately classify HLA mismatches according to their risk. The number and function of amino acid differences in each of these groups is diverse and those with 3-6 amino acid differences can have either high or low resolution HLA disparities (Figure 1).
Scoring systems which group HLA disparities based upon their similarity have also been explored. One approach which assigned scores to HLA disparities based upon the number, position and properties of the amino acid differences has not correlated well with clinical outcomes [18, 19]. This may in part be caused by observations showing that the number of mismatched amino acids may not correlate with T lymphocyte responses [34]. Duquesnoy has created an algorithm which has shown some promise in solid organ transplantation, but ranking by this approach has not correlated with outcomes in a NMDP data set [20].
Although these attempts have not been successful, approaches that combine HLA disparities using similar functional characteristics have the highest likelihood for success in studying populations which have a large number of low frequency HLA disparities. One possibility would be to establish a group in which the amino acid differences are predicted to only alter the peptide binding grove (PBG group) versus all others or perhaps subsets thereof defined by differences in epitopes for T, B, and NK lymphocytes. In this scenario, many high resolution mismatches which have only one amino acid difference in the peptide binding groove (e.g., A*0201:0206) would be classified in the PBG group. Classification of HLA mismatches that have amino acid differences outside the peptide binding groove is more complicated. There is evidence suggesting that there are six amino acids that are frequently involved in docking of T cell receptors [35]. If these positions are the same between two HLA molecules and there are differences in the peptides bound by the HLA molecule, positive selection of T lymphocytes on a similar HLA surface may increase the frequency of alloreactive T lypmphocytes. In this case, A:0201:0206, which several studies suggest may be associated with adverse transplant outcomes, may be detrimental because T cells selected by similar docking residues respond to subtle differences in peptides bound by the HLA molecule. HLA disparities with differences in T cell contact residues may be less problematic [34]. Others such as A*0202:0224 would be classified in another group because some of the amino acid differences are located in loops that are predicted to create epitopes for alloantibodies.
Extreme HLA diversity, genetic variation involving other factors influencing immune responses, and numerous factors influencing transplant outcomes (e.g., disease and stage of disease, recipient age) create a formidable challenge for achieving compelling data to develop well accepted guidelines for ranking the HLA mismatches in potential donors. Until this problem can be solved, it is tempting to use available reports with statistically significant comparisons to guide donor selection. The observations reported here illustrate some of the limitations of published investigations and the need for caution in using the available data to guide donor selection. Ultimately, sophisticated models will likely be required to address this important question.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank David Edelshtein for his assistance with molecular graphics images. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
The CIBMTR is supported by Public Health Service Grant U24-CA76518 from the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National Heart, Lung and Blood Institute; Office of Naval Research (grant to the NMDP N00014-06-1-0704); Health Resources and Services Administration (DHHS); and grants from AABB; Aetna; American Society for Blood and Marrow Transplantation; Amgen, Inc.; Anonymous donation to the Medical College of Wisconsin; Association of Medical Microbiology and Infectious Disease Canada; Astellas Pharma US, Inc.; Baxter International, Inc.; Bayer HealthCare Pharmaceuticals; BloodCenter of Wisconsin; Blue Cross and Blue Shield Association; Bone Marrow Foundation; Canadian Blood and Marrow Transplant Group; Celgene Corporation; CellGenix, GmbH; Centers for Disease Control and Prevention; ClinImmune Labs; CTI Clinical Trial and Consulting Services; Cubist Pharmaceuticals; Cylex Inc.; CytoTherm; DOR BioPharma, Inc.; Dynal Biotech, an Invitrogen Company; Enzon Pharmaceuticals, Inc.; European Group for Blood and Marrow Transplantation; Gambro BCT, Inc.; Gamida Cell, Ltd.; Genzyme Corporation; Histogenetics, Inc.; HKS Medical Information Systems; Hospira, Inc.; Infectious Diseases Society of America; Kiadis Pharma; Kirin Brewery Co., Ltd.; Merck & Company; The Medical College of Wisconsin; MGI Pharma, Inc.; Michigan Community Blood Centers; Millennium Pharmaceuticals, Inc.; Miller Pharmacal Group; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Nature Publishing Group; New York Blood Center; Novartis Oncology; Oncology Nursing Society; Osiris Therapeutics, Inc.; Otsuka Pharmaceutical Development & Commercialization, Inc.; Pall Life Sciences; PDL BioPharma, Inc; Pfizer Inc; Pharmion Corporation; Saladax Biomedical, Inc.; Schering Plough Corporation; Society for Healthcare Epidemiology of America; StemCyte, Inc.; StemSoft Software, Inc.; Sysmex; Teva Pharmaceutical Industries; The Marrow Foundation; THERAKOS, Inc.; Vidacare Corporation; Vion Pharmaceuticals, Inc.; ViraCor Laboratories; ViroPharma, Inc.; and Wellpoint, Inc. Any opinions, findings and conclusions or recommendations expressed in this article are those of the author(s) and do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, National Marrow Donor Program, or any other agency of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lee SJ, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–83. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 2.Hurley CK, et al. National Marrow Donor Program HLA-matching guidelines for unrelated marrow transplants. Biol Blood Marrow Transplant. 2003;9(10):610–5. doi: 10.1016/j.bbmt.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Bray RA, et al. National marrow donor program HLA matching guidelines for unrelated adult donor hematopoietic cell transplants. Biol Blood Marrow Transplant. 2008;14(9 Suppl):45–53. doi: 10.1016/j.bbmt.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Flomenberg N, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–30. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 5.Morishima Y, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99(11):4200–6. doi: 10.1182/blood.v99.11.4200. [DOI] [PubMed] [Google Scholar]
- 6.Loiseau P, et al. HLA Association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13(8):965–74. doi: 10.1016/j.bbmt.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Petersdorf EW, et al. Limits of HLA mismatching in unrelated hematopoietic cell transplantation. Blood. 2004;104(9):2976–80. doi: 10.1182/blood-2004-04-1674. [DOI] [PubMed] [Google Scholar]
- 8.Petersdorf EW, et al. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med. 2001;345(25):1794–800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 9.Petersdorf EW, et al. Definition of HLA-DQ as a transplantation antigen. Proc Natl Acad Sci U S A. 1996;93(26):15358–63. doi: 10.1073/pnas.93.26.15358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw BE, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110(13):4560–6. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 11.Gallardo D, et al. Hla-DPB1 mismatch in HLA-A-B-DRB1 identical sibling donor stem cell transplantation and acute graft-versus-host disease. Transplantation. 2004;77(7):1107–10. doi: 10.1097/01.tp.0000122225.10296.10. [DOI] [PubMed] [Google Scholar]
- 12.Kawase T, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110(7):2235–41. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 13.Halloran PF, Reeve J, Kaplan B. Lies, damn lies, and statistics: the perils of the P value. Am J Transplant. 2006;6(1):10–1. doi: 10.1111/j.1600-6143.2005.01182.x. [DOI] [PubMed] [Google Scholar]
- 14.Morishima Y, et al. Effect of HLA-A2 allele disparity on clinical outcome in hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007;69(Suppl 1):31–5. doi: 10.1111/j.1399-0039.2006.759_3.x. [DOI] [PubMed] [Google Scholar]
- 15.Archbold JK, et al. T cell allorecognition and MHC restriction--A case of Jekyll and Hyde? Mol Immunol. 2008;45(3):583–98. doi: 10.1016/j.molimm.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat Rev Immunol. 2007;7(12):942–53. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- 17.Elsner HA, et al. HistoCheck: rating of HLA class I and II mismatches by an internet-based software tool. Bone Marrow Transplant. 2004;33(2):165–9. doi: 10.1038/sj.bmt.1704301. [DOI] [PubMed] [Google Scholar]
- 18.Heemskerk MB, et al. The HistoCheck algorithm does not predict T-cell alloreactivity in vitro. Bone Marrow Transplant. 2005;36(10):927–8. doi: 10.1038/sj.bmt.1705154. [DOI] [PubMed] [Google Scholar]
- 19.Shaw BE, et al. Scoring for HLA matching? A clinical test of HistoCheck. Bone Marrow Transplant. 2004;34(4):367–8. doi: 10.1038/sj.bmt.1704586. author reply 369. [DOI] [PubMed] [Google Scholar]
- 20.Duquesnoy R, et al. HLAMatchmaker-defined triplet matching is not associated with better survival rates of patients with class I HLA allele mismatched hematopoietic cell transplants from unrelated donors. Biol Blood Marrow Transplant. 2008;14(9):1064–71. doi: 10.1016/j.bbmt.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley CK, et al. A high degree of HLA disparity arises from limited allelic diversity: analysis of 1775 unrelated bone marrow transplant donor-recipient pairs. Hum Immunol. 2007;68(1):30–40. doi: 10.1016/j.humimm.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Robinson J, et al. IMGT/HLA Database--a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29(1):210–3. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–12. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 24.Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219(2):277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 25.Garrett TP, et al. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature. 1989;342(6250):692–6. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- 26.Chelvanayagam G. A roadmap for HLA-A, HLA-B, and HLA-C peptide binding specificities. Immunogenetics. 1996;45(1):15–26. doi: 10.1007/s002510050162. [DOI] [PubMed] [Google Scholar]
- 27.Glucksberg H, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Shulman HM, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69(2):204–17. doi: 10.1016/0002-9343(80)90380-0. [DOI] [PubMed] [Google Scholar]
- 29.DiBrino M, et al. Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry. 1995;34(32):10130–8. doi: 10.1021/bi00032a005. [DOI] [PubMed] [Google Scholar]
- 30.Burrows SR, et al. Human leukocyte antigen phenotype imposes complex constraints on the antigen-specific cytotoxic T lymphocyte repertoire. Eur J Immunol. 1997;27(1):178–82. doi: 10.1002/eji.1830270126. [DOI] [PubMed] [Google Scholar]
- 31.Keever CA, et al. HLA-B44-directed cytotoxic T cells associated with acute graft-versus-host disease following unrelated bone marrow transplantation. Bone Marrow Transplant. 1994;14(1):137–45. [PubMed] [Google Scholar]
- 32.Fleischhauer K, et al. Bone marrow-allograft rejection by T lymphocytes recognizing a single amino acid difference in HLA-B44. N Engl J Med. 1990;323(26):1818–22. doi: 10.1056/NEJM199012273232607. [DOI] [PubMed] [Google Scholar]
- 33.Imanishi T, et al. HLA 1991 Proceedings of the Eleventh Histocompatibility Workshop and Conference. Eleventh Histocompatibility Workshop and Conference; Yokahama Japan: Oxford University Press; 1991. [Google Scholar]
- 34.Heemskerk MB, et al. Allogeneic MHC class I molecules with numerous sequence differences do not elicit a CTL response. Hum Immunol. 2005;66(9):969–76. doi: 10.1016/j.humimm.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


