Abstract
Reagents for recognition and efficient cleavage of mismatched DNA without photoactivation were designed. They contain a combination of a mismatch-directing metalloinsertor, [Rh(bpy)2(chrysi)]3+ (chrysi = 5,6-chrysenequinone diimine), and an oxidative cleavage functionality, [Cu(phen)2]+ (Cu). Both unconjugated (Rh+Cu) and conjugated (Rh−Cu) frameworks of the Rh insertor and Cu were prepared. Compared to Cu, both constructs Rh+Cu and Rh−Cu exhibit efficient site-specific DNA scission only with mismatched DNA, confirmed by experiments with 32P-labeled oligonucleotides. Furthermore, these studies indicate that DNA cleavage occurs near the mismatch in the minor groove and on both strands. Interestingly, the order of reactivity of the three systems with a CC mismatch is Rh+Cu > Rh−Cu ≫ Cu. Rh binding appears to direct Cu reactivity with or without tethering. These results illustrate advantages and disadvantages in bifunctional conjugation.
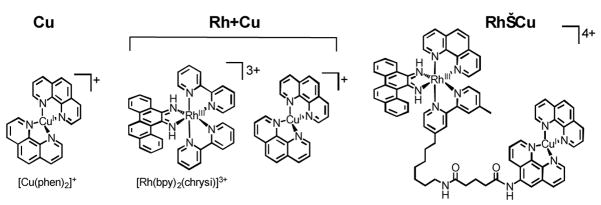
Base pair mismatches in the genome can arise from damage by environmental agents (i.e. UV light), a wide range of genotoxic chemicals as well as errors made by DNA polymerase during synthesis.1,2 These base mispairs are generally corrected by the mismatch repair machinery; if they are not repaired, however, mutations arise, which subsequently lead to increased cancer susceptibility.1–4 To recognize DNA mismatches, we have developed rhodium complexes containing an intercalating ligand that is too bulky to insert at stable matched sites and instead preferentially binds to thermodynamically destabilized mismatched sites.5–8 The complex [Rh(bpy)2(chrysi)]3+ (Figure 1), designed by our laboratory, targets more than 80% of mismatches with high selectivity and promotes single-stranded cleavage of the DNA backbone next to the mismatch site upon photoactivation.5–7 More interestingly, this Rh complex can detect and photocleave a single base mismatch in a linearized 2725 base pair plasmid.6 We have recently elucidated how the complex [Rh(bpy)2(chrysi)]3+ interacts with the mismatched sites in DNA by crystal structural determination and NMR analysis.9,10 The binding mode of this complex to the mismatch site is not classical intercalation but rather insertion. The expansive chrysi ligand is deeply inserted into the mismatch site in the minor groove resulting in the complete ejection of mismatched nucleotides from the base stack.
Figure 1.
Chemical structures of Cu, Rh+Cu and Rh−Cu.
Recently, this mismatch-specific metalloinsertor has shown promise in cell-selective strategies for chemotherapeutic design.11 The Rh complex selectively inhibits cellular proliferation in mismatch repair-deficient cells as compared to mismatch repair-proficient cells. In another possible chemotherapeutic approach, bifunctional conjugates that combine a metalloinsertor targeting DNA mismatches with a reactive species such as an aniline mustard or a cisplatin analog have been designed.12–14 Studies of these conjugates demonstrate that the Rh moiety tunes the reactivity of the agent linked to the metalloinsertor.
Following this conjugation strategy, a construct can be devised for targeting and cleaving mismatched DNA without light activation. To achieve this, we have investigated the reactivity of a chemical DNA cleavage agent in the presence of [Rh(bpy)2(chrysi)]3+. For efficient oxidative cleavage, we have pursued the use of [Cu(phen)2]2+ with the Rh insertor. In the presence of a reducing agent, the newly formed [Cu(phen)2]+ (Cu, Figure 1) promotes light-independent DNA backbone cleavage showing more localized cleavage sites than those generated by [Fe(EDTA)]2−.15–17 The complex Cu tethered to various DNA-binding moieties exhibits efficient DNA scission.17 The redox chemistry of [Cu(phen)2]2+ is feasible in the reducing environment of living cells, suggesting also a potential biological application of the copper species with the Rh insertor. Here we report the preparation of combined Rh and Cu systems that are composed of the metalloinsertor and the Cu complex as well as an examination of their DNA cleavage activity without photoactivation.
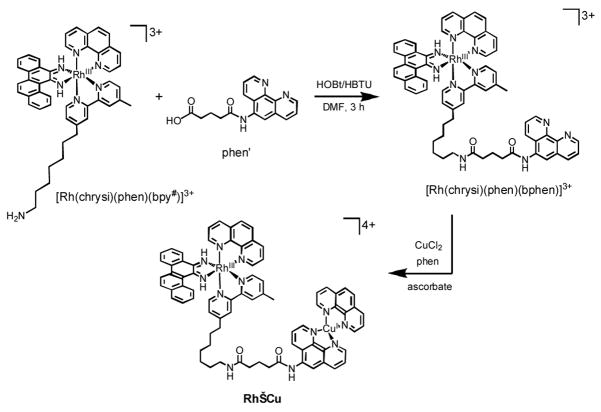
The reactivity of both nonconjugates (Rh+Cu) and conjugates (Rh−Cu) of [Rh(bpy)2(chrysi)]3+ and Cu in DNA scission was examined. The Rh complex [Rh(bpy)2(chrysi)]3+ in Rh+Cu (Figure 1) was synthesized as described.18 For the conjugate Rh−Cu, we prepared its precursor [Rh(chrysi)(phen)(bphen)]3+ (bphen = N1-[7-(4′-methyl-2,2′-bipyridin-4-yl)heptyl]-N5-(1,10-phenanthrolin-5-yl)pentanediamide) through the coupling reaction of [Rh(chrysi)(phen)(bpy#)]3+ (bpy# = 7-(4′-methyl-2,2′-bipyridin-4-yl)heptan-1-amine)12 with the modified phen ligand (phen’ = 5-(1,10-phenanthrolin-5-ylamino)-5-oxopentanoic acid),19 as described in Scheme 1 and Supporting Information. Copper coordination was accomplished in situ (vide infra), affording the conjugate Rh−Cu.
Scheme 1.
Preparation of Rh−Cu.
DNA cleavage experiments of Rh+Cu and Rh−Cu with 31mer oligonucleotides either lacking or containing a CC mismatch were performed and monitored by denaturing polyacrylamide gel electrophoresis (Figure 2). In the case of the reactions of Rh+Cu with DNA, duplex (1 μM) was incubated with 1 μM [Rh(bpy)2(chrysi)]3+ at 37 °C for 5 min, followed by addition of 1 μM CuCl2 and 3 μM phen; upon treatment with 5 mM ascorbate, the reagents were allowed to react with DNA. After 5 or 10 min, the reactions were quenched by 5 min treatment with 5 mM dmp (dmp = 2,9-dimethyl-1,10-phenanthroline) followed by freezing. For the conjugate Rh−Cu, the Rh complex [Rh(chrysi)(phen)(bphen)]3+ (1 μM) was incubated in the following order: duplex (1 μM, 5 min, 37 °C), CuCl2 (1μM, 5 min, 37 °C), phen (3 μM) and ascorbate (5 mM). The reaction was then quenched in the same manner as described above. To compare the reactivies of Cu, Rh+Cu and Rh−Cu, Cu was also generated in situ by reacting CuCl2 (1 μM) with phen (3 μM) followed by ascorbate (5 mM) and was similarly allowed to react with duplex. As shown in Figure 2, site-specific DNA cleavage occurs only in the presence of mismatched DNA, and the sites cleaved are near the mismatched site (two or three bases away). No DNA cleavage without light is evident for the parent conjugate without Cu or any of the individual reagents. More interestingly, the nonconjugate Rh+Cu exhibits more efficient DNA scission than the conjugate Rh−Cu (cleavage yield20 for 5 min = 35 and 19%; for 10 min = 65 and 41% by Rh+Cu and Rh−Cu, respectively, in Figure 2). Note that some cleavage is evident also with Cu alone.21 Binding of the Rh moiety of Rh+Cu and Rh−Cu to the mismatch site was visualized independently by performing photoactivated DNA cleavage with [Rh(bpy)2(chrysi)] 3+ and [Rh(chrysi)(phen)(bphen)]3+ without Cu present (Figure S1). These results together indicate that the recognition of mismatched DNA by the Rh moiety enhances the cleavage activity of Cu close to the mismatch.
Figure 2.
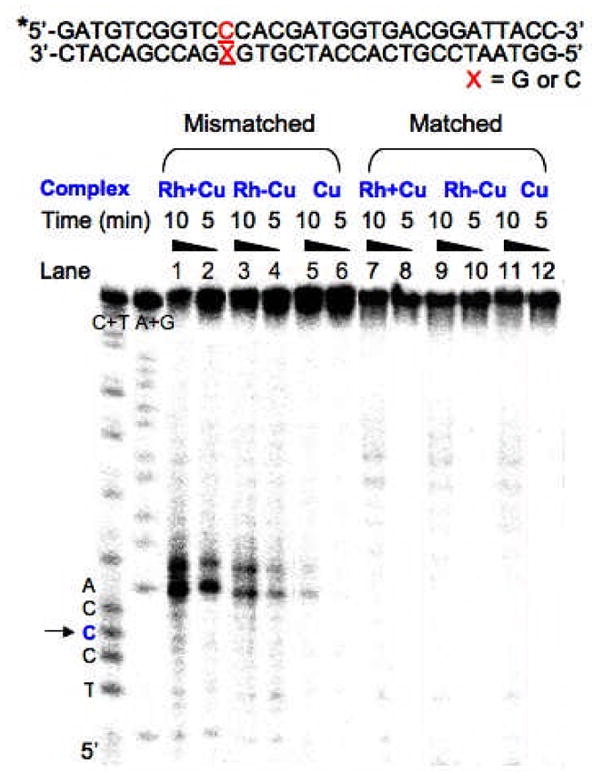
Sequences of 31mer oligonucleotides (top). Autoradiogram of a denaturing 20% polyacrylamide gel presenting DNA cleavage by Cu, Rh+Cu and Rh−Cu with matched and mismatched oligonucleotides (bottom). A+G and C+T, Maxam-Gilbert sequencing reactions. * indicates 5′-32P-end-label of the strand. Conditions: duplex (1 μM), Cu (1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), Rh+Cu (1 μM [Rh(bpy)2(chrysi)]3+, 1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), Rh−Cu (1 μM [Rh(chrysi)(phen)(bphen)]3+, 1 μM CuCl2, 3 μM phen, 5 mM sodium ascorbate), in 10 mM Tris, pH 7.5, 50 mM NaCl at 37 oC. Lanes 1–2 and 7–8, fragments with Rh+Cu; lanes 3–4 and 9–10, fragments with Rh−Cu; lanes 5–6 and 11–12, fragment with Cu. The time shown reflects incubation time after treatment with sodium ascorbate before quenching. The arrow marks the mismatched site.
To obtain further structural information regarding the DNA cleavage site, we performed the cleavage experiments using the 32P-end-labeled complementary strands (Figures S2). Site-specific DNA scission is again observed only with mismatched DNA.20 On both strands, the cleavage sites are mainly 2–3 bases from the mismatch toward the 3′-end (Figure 3). Furthermore, more efficient DNA cleavage is found with the unconjugated framework Rh+Cu than with the conjugate, Rh−Cu.
Figure 3.
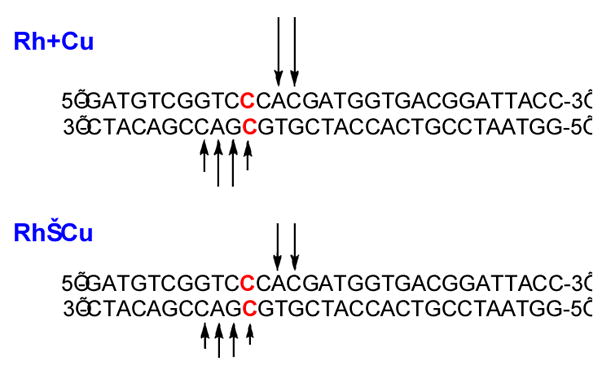
Representation of DNA cleavage sites by Rh+Cu (top) and Rh−Cu (bottom). The arrows indicate the cleavage sites on the 5′-32P-end-labeled strands. The length of each arrow at the given site reflects the relative percent cleavage on the strand. The mismatched site is highlighted in red.
The 3′ asymmetry in cleavage sites by Rh+Cu and Rh−Cu on both strands indicates that DNA cleavage occurs in the minor groove (Figure 3).22 This result is not surprising, since both the parent Rh complex and Cu(phen)2+ are known to bind DNA from the minor groove side.9,15–17 Thus both covalently and noncovalently, the Rh insertor directs reactivity of the Cu center in the minor groove.
Since binding of the Rh insertor into the mismatch site results in ejection of the mismatched bases, we also explored whether the Cu complex might react with the ejected bases. To evaluate this, alkaline and piperidine treatment were performed after the reaction of the mismatched DNA with Rh+Cu (Figure S3). No new scission products are observed, suggesting that the Cu complex does not react with the bases or the ejected bases upon insertion of the Rh moiety in mismatched DNA to form alkaline- or piperidine-sensitive lesions. Thus, the observed DNA cleavage, induced by reaction of Cu, appears primarily with the sugar ring.15–17
Why do we find preferential reaction with Rh+Cu rather than the tethered conjugate, Rh−Cu? One possible explanation is that Rh binding may locally distort the helix to facilitate Cu(phen)2+ reaction. Reaction by Cu(phen)2+ is localized, directed by an intermediate high-valent Cu(O) species.17, 23 Tethering to the Rh center may also somewhat confine the orientation of the damaging agent. Additionally, tethering affects binding of the Rh complex within the narrow minor groove. Measurements of the binding of [Rh(bpy)2(chrysi)]3+ and [Rh(chrysi)(phen)-(bphen)]3+ by photocleavage assay (Figure S1) show a decrease in binding affinity to the mismatched site associated with functionalizing the bpy ligand. Thus the relative reactivities of Rh−Cu and Rh+Cu are likely a consequence of confinement of both the Rh and Cu moieties in the particularly narrow minor groove of the DNA duplex.
This work establishes a new approach for developing an efficient cleavage agent without light that is specific for mismatched DNA. In order to exploit the individual contributions of the Rh and Cu complexes in recognition of mismatched DNA and strand cleavage, both species were combined. This remains a useful strategy to couple recognition and reaction. Conjugation of these two species, however, results in a reduction in both binding and reactivity of the complexes with mismatched DNA. Indeed, for the CC mismatch, the parent Rh complex can direct Cu reactivity without cumbersome synthesis. Although unexpected, these results illustrate both the advantages and disadvantages of preparing bifunctional conjugates.
Supplementary Material
Acknowledgments
We are grateful to the NIH (GM33309) and Applied Biosystems for their financial support. We also thank the tobacco-related disease research program (TRDRP) for a postdoctoral fellowship to M.H.L.
Footnotes
Supporting Information Available: Preparation of the conjugate Rh–Cu and Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hoeijmakers JH. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.(a) Jiricny J, Marra G. Curr Opin Genet Dev. 2003;13:61–69. doi: 10.1016/s0959-437x(03)00004-2. [DOI] [PubMed] [Google Scholar]; (b) Jiricny J. Nat Rev Mol Cell Bio. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 3.Schärer OD. Angew Chem, Int Ed. 2003;42:2946–2974. doi: 10.1002/anie.200200523. [DOI] [PubMed] [Google Scholar]
- 4.Duval A, Hamelin R. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 5.Jackson BA, Barton JK. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 6.Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 7.Jackson BA, Barton JK. Biochemistry. 2000;10:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 8.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. Proc Natl Acad Sci USA. 2003;100:3337–3342. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierre VC, Kaiser JT, Barton JK. Proc Natl Acad Sci USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordier C, Pierre VC, Barton JK. J Am Chem Soc. 2007 doi: 10.1021/ja0739436. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart JR, Glebov O, Ernst RJ, Barton JK. Proc Natl Acad Sci USA. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schatzschneider U, Barton JK. J Am Chem Soc. 2004;126:8630–8631. doi: 10.1021/ja048543m. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A, Barton JK. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 14.A bifunctional conjugate has also been prepared with an unreactive fluorophore. See Zeglis BM, Barton JK. J Am Chem Soc. 2006;128:5654–5655. doi: 10.1021/ja061409c.
- 15.(a) Sigman DS, Mazumder A, Perrin DM. Chem Rev. 1993;93:2295–2316. and references therein. [Google Scholar]; (b) Meijler MM, Zelenko O, Sigman DS. J Am Chem Soc. 1997;119:1135–1136. [Google Scholar]
- 16.Stubbe J, Kozarich JW. Chem Rev. 1987;87:1107–1136. and references therein. [Google Scholar]
- 17.(a) Sigman DS, Chen CH. Annu Rev Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]; (b) Bruice TW, Wise JG, Rosser DSE, Sigman DS. J Am Chem Soc. 1991;113:5446–5447. [Google Scholar]
- 18.Zeglis BM, Barton JK. Nat Protocols. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardesai NY, Lin SC, Zimmermann K, Barton JK. Bioconjugate Chem. 1995;6:302–312. doi: 10.1021/bc00033a011. [DOI] [PubMed] [Google Scholar]
- 20.Cleavage yield (%) = Icp/(Icp + Ip) × 100 (Icp = intensity of the cleavage product bands; Ip = intensity of the parent strand band).
- 21.Cleavage with Cu(phen)2+ alone on the mismatched oligonucleotide may reflect dynamic fraying associated with the destabilized mismatch.
- 22.Dervan PB. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 23.Thyagarajan S, Murthy NN, Narducci-Sarjeant AA, Karlin KD, Rokita SE. J Am Chem Soc. 2006;128:7003–7004. doi: 10.1021/ja061014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.