Abstract
Epidermal growth factor (EGF) is involved in alveolar epithelial repair, lung fluid clearance, inflammation, and is regulated by sex hormones. An unmatched, nested case-control study was conducted to evaluate the associations of EGF variants with acute respiratory distress syndrome (ARDS) and the role of sex on the associations between EGF variants and ARDS.
Patients with ARDS risk factors upon ICU admission were enrolled. Cases were 416 Caucasians who developed ARDS and controls were 1052 Caucasians who did not develop ARDS. Cases were followed for clinical outcomes and 60-day mortality. One functional single nucleotide polymorphism (SNP) rs4444903 and six haplotype-tagging SNPs spanning entire EGF gene were genotyped with TaqMan assays.
No individual SNP or haplotype was associated with ARDS risk or outcomes in all subjects. Sex-stratified analyses showed opposite effects of EGF variants on ARDS in males versus in females. SNPs rs4444903, rs2298991, rs7692976, rs4698803, and haplotypes GGCGTC, ATCAAG were associated with ARDS risk in males. No associations were observed in females. Interaction analysis showed that rs4444903, rs2298991, rs7692976 and rs6533485 significantly interacted with sex for ARDS risk.
This study suggests that genetic associations of EGF with ARDS risk are modified by sex. Our findings should be replicated in other populations.
Keywords: Acute respiratory distress syndrome, epidermal growth factor, genetic susceptibility, haplotypes, lung injury, molecular epidemiology
INTRODUCTION
Among patients with sepsis, pneumonia, trauma and other triggering conditions, only a subset will develop acute respiratory distress syndrome (ARDS), and only approximately 60% of those developing ARDS will survive, suggesting that genetics may influence the susceptibility to and recovery from this syndrome. ARDS is characterized by diffuse damage to the alveolar barrier, which leads to increased permeability and influx of protein-rich edema into the interstitial and airspace. In addition to endothelial injury, epithelial damage also plays an important role in the development of and recovery from this disorder [1, 2]. The loss of epithelial integrity contributes to the formation of alveolar edema, while the repair of epithelial injury and the restoration of alveolar epithelial fluid transport function facilitate the resolution of pulmonary edema [2, 3]. Epidermal growth factor (EGF) is a key growth factor among the ligands of EGF receptors (EGFR). The family of epidermal growth factors and receptors is important in regulating cell growth, maturation, function and maintenance in epithelial tissues [4]. It is suggested that acute lung injury elicits growth factor responses that trigger repair mechanisms to restore lung integrity [5]. Studies show that EGF regulates bronchial and alveolar epithelial repair after lung injury [6, 7]. Moreover, EGF decreases alveolar epithelial junctional permeability, up-regulates alveolar epithelial Na+-K+-ATPase, and increases lung fluid clearance [8–10]. On the other hand, EGF has also been increasingly regarded as a pro-inflammatory mediator. In asthma, the EGFR pathway is involved not only in the bronchial epithelial repair [11] but also in lung inflammation [12]. EGF increases the production of interleukin-8 (IL-8), modulates the inflammatory effects of tumor necrosis factor-α (TNF-α), and enhances the neutrophil-mediated immunity [13–15]. Taken together, EGF might play a critical role in the pathogenesis of ARDS, and therefore, EGF gene polymorphisms could be potential risks for ARDS.
The EGF gene is located on chromosome 4 (4q25), spans about 99 kb and has 24 exons and 23 introns. Genetic variation within the EGF gene has been studied in relation to EGF phenotype and disease susceptibility [16]. However, most of these studies have focused on the single nucleotide polymorphism (SNP) rs4444903, the GG genotype of which is associated with a higher secretion of EGF protein than the AA genotype [17]. Recently, it has been recognized that haplotype-based tagging SNP approach can comprehensively scan the common variation of an entire gene and provide greater power than single-marker tests for genetic disease association [18]. In addition, it is evident that the EGFR signaling pathways are regulated by sex hormones [19–21]. Studies have shown sex difference of salivary and tear EGF levels [22, 23], as well as the EGF effects in ulcer healing [24]. Moreover, sex-specific associations of EGF polymorphisms with phenotypes have been found in schizophrenia [25, 26]. In this study, we hypothesized that common genetic variation of EGF is associated with the risk and outcomes of ARDS, and that such association is modified by sex. We conducted a hospital-based, unmatched, modified, nested case-control study of patients at risk for ARDS, and used a haplotype-tagging SNP approach to test our hypotheses.
METHODS
Study design and subjects
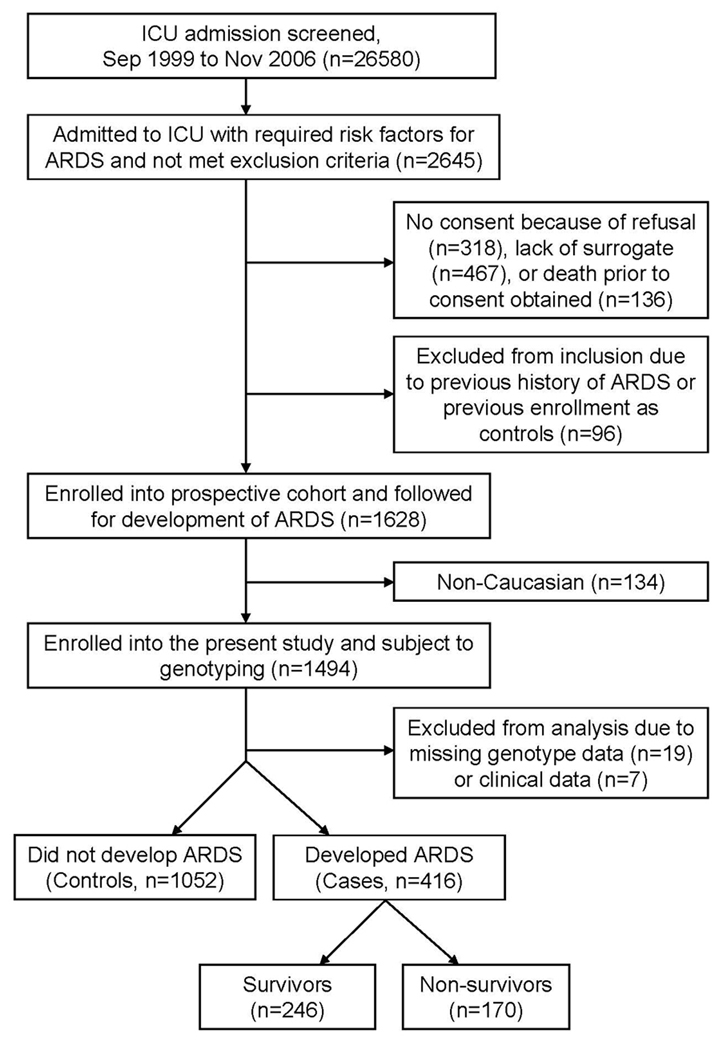
This study is part of an ongoing molecular epidemiology project investigating the influences of genetic factors on the development and outcomes of ARDS. Details of the study have been described previously [27]. Briefly, study subjects in the present study were selected from patients admitted to the intensive care units (ICU) at Massachusetts General Hospital (Boston, MA) from September 1999 to November 2006. Patients with clinical risk factors for ARDS such as sepsis, septic shock, trauma, pneumonia, aspiration, or multiple transfusions were eligible for inclusion (supplementary table 1). Exclusion criteria included age <18, diffuse alveolar hemorrhage, chronic lung diseases other than COPD or asthma, directive to withhold intubation, immunosuppression not secondary to corticosteroid, and treatment with granulocyte colony-stimulating factor. Baseline characteristics and Acute Physiology and Chronic Health Evaluation (APACHE) III scores were recorded on ICU admission. The enrolled patients who fulfilled the American-European Consensus Committee (AECC) criteria for ARDS upon ICU admission or during the daily follow-up were considered as ARDS cases, whereas at-risk patients who did not meet the criteria for ARDS were considered as controls. All enrolled patients with ARDS were then followed for clinical outcomes and all-cause 60-day mortality after development of ARDS. To reduce the potential confounding from ethnic backgrounds, only Caucasian (Non-Hispanic White) patients were analyzed. A schematic of study design and patient selection was illustrated in figure 1. The study was approved by the Human Subjects Committees of the Massachusetts General Hospital and the Harvard School of Public Health. Written informed consents were obtained from all subjects or surrogates.
FIGURE 1.
Schematic of study design and patient selection.
SNP selection
We selected the haplotype-tagging SNPs in the EGF gene based the HapMap data (release 22, on NCBI B36 assembly, dbSNP b126) for the CEU population with northern and western Europe ancestries. We used the multimarker tagging algorithm with criteria of r2 >0.8 and minor allele frequency (MAF) ≥0.1. The entire EGF gene was covered, including 5 kb on each side of the gene encompassing the promoter and 3’ untranslated region (UTR). In addition, we also included a functional SNP rs4444903 (+61 A>G) that has not been included in the HapMap data.
Genotyping
Genomic DNA was extracted from whole blood using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). The selected SNPs of EGF were genotyped using TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA). Primers and probes were ordered from Applied Biosystems. All PCR amplifications were performed in a 384-well format on GeneAmp® PCR Systems 9700 (Applied Biosystems). The fluorescence of PCR products was detected using the ABI Prism® 7900HT Sequence Detection System (Applied Biosystems). Genotyping was performed by laboratory personnel blinded to case-control status. A random 10% samples were inserted in different 384-well plates as duplicates for quality-control purposes. Two investigators reviewed all genotyping results independently. The concordance rate for the duplicate samples was >99% and the overall genotyping success rate was 98.8%. Samples not yielding the genotypes of all SNPs were excluded from analysis.
Statistical analyses
All statistical analyses were performed using the SAS software (version 9.1.3). The homogeneity of baseline characteristics between two groups was tested by Fisher’s exact test for categorical variables and by Student's t-test for continuous variables. The differences of genotype distributions between groups were compared by χ2 test. We used SAS/Genetics to calculate the allele frequencies, test the deviation from Hardy-Weinberg equilibrium (HWE), and estimate pairwise D’ and r2 values for linkage disequilibrium (LD).
Haplotype frequencies were estimated from the unphased genotype data in the combined population (cases and controls), using the expectation maximization (EM) algorithm as implemented in SAS/Genetics. The associations between EGF haplotypes and the risk and survival of ARDS were analyzed using the expectation-substitution approach as implemented in the SAS macro HAPPY (http://www.hsph.harvard.edu/faculty/kraft/soft.htm by Kraft, P. et al.) [28]. This approach treats subject-specific expected haplotype indicators, calculated by an additive mode, as observed covariates for regression models. We considered haplotypes with a frequency ≥5% in the total population to be “common” and used the most common haplotype as the referent in the regression model to assess the haplotype-specific risk for ARDS. All other haplotypes were pooled into a separate “rare haplotypes” category.
We used multivariate logistic regression to estimate the genotype- and haplotype-specific odds ratio (OR) and 95% confidence interval (CI) for ARDS risk. To evaluate the associations of individual genotypes and haplotypes with ARDS survival, we used the Cox proportional hazard model to estimate the hazard ratio (HR) and 95% CI. Covariates for the logistic regression and Cox models included age, sex (not included in sex-stratified analyses), APACHE III score and the potential risks for ARDS development and mortality based on univariate analysis. Global tests for the associations between haplotypes and ARDS risk and survival were carried out using the likelihood ratio test (LRT). For statistically significant associations, adjusted p-values were calculated to correct for multiple comparisons, using the False Discovery Rate (FDR) procedure of Benjamini and Hochberg [29]. The gene-sex interactions were examined by sex-stratification and their strength was evaluated in multivariate logistic regression models including an interaction term. A two-sided p-value <0.05 was considered to be statistically significant.
RESULTS
Study population
The flow diagram of patient selection in this study is illustrated in figure 1. The baseline characteristics of the study population, including 416 ARDS cases and 1052 at-risk controls, are shown in table 1. Patients who developed ARDS were younger and had higher APACHE III scores than those who did not develop ARDS. Among patients with ARDS, the survivors were younger and had lower APACHE III scores than the non-survivors. There were no differences of sex distributions between ARDS and controls, and between ARDS survivors and non-survivors. The comparisons of ARDS risk factors and comorbidities between ARDS and controls, and between ARDS survivors and non-survivors are also shown in table 1. The comparisons of clinical characteristics between males and females are provided in supplementary table 2.
TABLE 1.
Baseline characteristics of study population
| ARDS Development |
ARDS 60-Day Mortality |
|||||
|---|---|---|---|---|---|---|
| ARDS (n=416) |
Controls (n=1052) |
p-value | Non-survivors (n=170) |
Survivors (n=246) |
p-value | |
| Age-yr, mean ± SD | 59 ± 18 | 63 ± 17 | <0.001 | 67 ± 15 | 53 ± 18 | <0.001 |
| Male, n (%) | 246 (59.1%) | 641 (60.9%) | 0.554 | 100 (58.8%) | 146 (59.4%) | 0.920 |
| APACHE III score, mean ± SD | 68 ± 23 | 61 ± 21 | <0.001 | 79 ± 21 | 60 ± 20 | <0.001 |
| ARDS risk factor, n (%) | ||||||
| Sepsis | 112 (26.9%) | 383 (36.4%) | <0.001 | 43 (25.3%) | 69 (28.1%) | 0.575 |
| Septic shock | 249 (59.8%) | 461(43.8%) | <0.001 | 111 (65.3%) | 138 (56.1%) | 0.067 |
| Pneumonia | 284 (68.3%) | 455 (43.3%) | <0.001 | 123 (72.4%) | 161 (65.5%) | 0.164 |
| Aspiration | 41 (9.9%) | 88 (8.4%) | 0.359 | 19 (11.2%) | 22 (8.9%) | 0.505 |
| Multiple transfusion | 37 (8.9%) | 126 (12.0%) | 0.097 | 17 (10.0%) | 20 (8.1%) | 0.600 |
| Trauma | 31 (7.5%) | 81 (7.7%) | 0.914 | 2 (1.2%) | 29 (11.8%) | <0.001 |
| Comorbidity, n (%) | ||||||
| Post-operative | 23 (5.5%) | 77 (7.3%) | 0.251 | 11 (6.5%) | 12 (4.9%) | 0.518 |
| Diabetes | 73 (17.6%) | 285 (27.1%) | <0.001 | 32 (18.8%) | 41 (16.7%) | 0.601 |
| End-stage renal disease | 25 (6.0%) | 55 (5.2%) | 0.610 | 15 (8.8%) | 10 (4.1%) | 0.058 |
| Liver cirrhosis / failure | 29 (7.0%) | 41 (3.9%) | 0.020 | 21 (12.4%) | 8 (3.3%) | <0.001 |
| Metastatic cancer | 12 (2.9%) | 51 (4.9%) | 0.115 | 9 (5.3%) | 3 (1.2%) | 0.018 |
| History of steroid use, n (%) | 44 (10.6%) | 94 (8.9%) | 0.323 | 27 (15.9%) | 17 (6.9%) | 0.005 |
| History of alcohol abuse, n (%) | 60 (14.4%) | 105 (10.0%) | 0.017 | 27 (15.9%) | 33 (13.4%) | 0.482 |
ARDS: acute respiratory distress syndrome; APACHE: Acute Physiology and Chronic Health Evaluation.
SNP selection and genotype frequencies
Seven SNPs of the EGF gene were selected for this study (in the order of 5’ to 3’): the functional SNP rs4444903, and six tagging SNPs rs2298991, rs11568993, rs6850557, rs7692976, rs4698803 and rs6533485 (table 2). All SNPs conformed to HWE, with the exception of rs6850557 (FDR adjusted p<0.05), which was then excluded from further analyses. Pairwise LD analysis revealed that all alleles among selected SNPs are in high LD (supplementary figure 1). The alleles, locations, chromosome positions, and minor allele frequencies of these seven SNPs are presented in table 2. The minor allele frequencies between ARDS and controls were not different in all subjects for all SNPs, but were significantly different in males for rs7692976 and rs4698803 (p=0.030, 0.024, respectively). The genotype distributions are shown in supplementary table 3. We found no differences of genotype frequencies between ARDS and controls in all subjects, between males and females, and between ARDS survivors and non-survivors. However, the sex-stratified analyses showed that the genotype frequencies of rs4444903, rs7692976 and rs4698803 were significantly different between ARDS and controls in males (p=0.029, 0.037, 0.032, respectively) (supplementary table 4).
TABLE 2.
Summary of six single nucleotide polymorphisms evaluated in this study
| Minor Allele Frequency (ARDS / Controls) |
HWE | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Allele | Location | Position# | All Subjects | Male | Female | p-value+ |
| rs4444903 | A>G | 5’ UTR | 111053559 | 0.436 / 0.416 | 0.441 / 0.393 | 0.429 / 0.453 | 0.140 |
| rs2298991 | T>G | Intron 11 | 111111461 | 0.433 / 0.421 | 0.439 / 0.400 | 0.424 / 0.453 | 0.169 |
| rs11568993 | C>T | Exon 13 | 111116764 | 0.096 / 0.095 | 0.083 / 0.094 | 0.115 / 0.097 | 0.592 |
| rs6850557 | G>A | Intron 18 | 111130464 | 0.359 / 0.347 | 0.364 / 0.333 | 0.353 / 0.367 | <0.001§ |
| rs7692976 | A>G | Intron 18 | 111131016 | 0.440 / 0.417 | 0.455 / 0.399¶ | 0.418 / 0.447 | 0.134 |
| rs4698803 | T>A | Exon 19 | 111133876 | 0.168 / 0.192 | 0.152 / 0.199¶ | 0.191 / 0.181 | 0.026 |
| rs6533485 | G>C | Intron 22 | 111147012 | 0.499 / 0.491 | 0.504 / 0.481 | 0.509 / 0.494 | 0.630 |
ARDS: acute respiratory distress syndrome; HWE: Hardy-Weinberg equilibrium; SNP: single nucleotide polymorphism; UTR: untranslated region.
chromosome positions are based on National Center for Biotechnology Information Build 36.
p-value by χ2 test <0.05.
p-value is the probability of the χ2 test for deviation from Hardy-Weinberg equilibrium in all controls
FDR adjusted p-value <0.05.
Associations between EGF variants and ARDS risk
Considering all subjects as a whole, none of the individual SNPs was significantly associated with ARDS risk (table 3). Upon the stratification by sex, we found that the variant genotypes of rs4444903 (OR 1.64, 95% CI 1.17–2.31, p=0.005), rs2298991 (OR 1.50, 95% CI 1.07–2.11, p=0.019) and rs7692976 (OR 1.64, 95% CI 1.17–2.31, p=0.005) were associated with increased risks, while the variant genotypes of rs4698803 (OR 0.67, 95% CI 0.48–0.96, p=0.025) were associated with reduced risks of developing ARDS in males. In females, none of the associations between EGF variants and ARDS risk were significant. However, we observed that the effects of variant genotypes on ARDS risk were opposite from that in males for five of the six SNPs.
TABLE 3.
Associations between Epidermal Growth Factor (EGF) genetic variants and acute respiratory distress syndrome (ARDS) risk
| All Subjects (n=1468) |
Male (n=887) |
Female (n=581) |
||||
|---|---|---|---|---|---|---|
| OR (95% CI)+ | p-value | OR (95% CI)+ | p-value | OR (95% CI)+ | p-value | |
| Genotype# | ||||||
| rs4444903 | 1.22 (0.94–1.59) | 0.134 | 1.64 (1.17–2.31) | 0.005§ | 0.74 (0.48–1.13) | 0.165 |
| rs2298991 | 1.13 (0.87–1.46) | 0.367 | 1.50 (1.07–2.11) | 0.019§ | 0.68 (0.45–1.04) | 0.077 |
| rs11568893 | 0.88 (0.64–1.21) | 0.435 | 0.86 (0.56–1.32) | 0.488 | 0.90 (0.55–1.48) | 0.678 |
| rs7692976 | 1.21 (0.94–1.58) | 0.145 | 1.64 (1.17–2.31) | 0.005§ | 0.73 (0.48–1.11) | 0.140 |
| rs4698803 | 0.83 (0.64–1.09) | 0.178 | 0.67 (0.48–0.95) | 0.025§ | 1.10 (0.72–1.69) | 0.650 |
| rs6533485 | 1.06 (0.80–1.39) | 0.707 | 1.39 (0.96–2.02) | 0.080 | 0.66 (0.42–1.03) | 0.068 |
| Haplotype | ||||||
| Global test¶ | χ2 = 5.78 | 0.328 | χ2 = 16.99 | 0.005 | χ2 = 2.75 | 0.738 |
| Hap1 (ATCATG) | (reference) | - | (reference) | - | (reference) | - |
| Hap2 (GGCGTC) | 1.10 (0.88–1.38) | 0.403 | 1.35 (1.00–1.81) | 0.048 | 0.76 (0.53–1.10) | 0.147 |
| Hap3 (ATCAAG) | 0.80 (0.60–1.06) | 0.118 | 0.64 (0.44–0.94) | 0.022 | 1.02 (0.65–1.60) | 0.941 |
| Hap4 (GGTGTC) | 0.91 (0.65–1.26) | 0.550 | 0.87 (0.55–1.36) | 0.538 | 0.91 (0.55–1.49) | 0.705 |
| Hap5 (ATCATC) | 0.93 (0.64–1.34) | 0.681 | 0.88 (0.55–1.42) | 0.605 | 1.00 (0.54–1.87) | 0.993 |
| All others (Freq < 5%) | 1.11 (0.81–1.52) | 0.528 | 1.22 (0.81–1.84) | 0.332 | 0.85 (0.50–1.43) | 0.532 |
OR: odds ratio; CI: confidence interval.
wildtype homozygote of each SNP was considered as reference
Likelihood-ratio test (LRT), with 5 degrees of freedom
adjusted for age, sex, APACHE III score, all risk factors for ARDS (listed in table 1), diabetes, liver cirrhosis/failure and alcohol abuse. APACHE III score was revised to remove age and PaO2/FiO2 to avoid colinearity. Sex was removed from the model during sex-stratified analyses
FDR adjusted p-value <0.05.
There were five common haplotypes (frequencies: 30.6%, 29.0%, 14.0%, 8.8%, 7.5%, respectively) inferred from the six polymorphisms analyzed. Similar with the results from genotypes analyses, we observed significant associations between haplotypes and ARDS risk only in the male subgroup, in which global test for association was significant (LRT, p=0.005) (table 3). In addition, Hap2 (GGCGTC) was associated with an increased ARDS risk (OR 1.35, 95% CI 1.00–1.81; p=0.048), whereas Hap3 (ATCAAG) was associated with a reduced ARDS risk (OR 0.64; 95% CI 0.44–0.94, p=0.022). The haplotype-specific associations were assessed in one regression model, thus correcting for multiple comparisons is not necessary.
Associations between EGF variants and clinical outcomes of ARDS
The clinical outcomes of ARDS patients among different genotype groups are shown in supplementary table 5. In 28 days since ARDS diagnosis, no significant differences of ICU-free days, ventilator-free days, successful extubation rates and mortality rates were observed between genotypes of all SNPs. The 60-day mortality rates were also not different between genotypes. In analyses stratified by sex (data not shown), male ARDS patients with variant genotypes of rs4698803 had a higher successful extubation rate (68.7% vs. 54.2%, p=0.043) and longer ventilator-free days (11.9±9.8 vs. 8.5±9.7 days, p=0.014) than those with wildtype homozygote. There were no differences of clinical outcomes between genotypes in females.
None of the EGF genotypes or haplotypes was significantly associated with ARDS 60-day survival in all subjects, or in male and female subgroups (table 4). Only Hap3 was marginally associated with decreased mortality in male ARDS patients (HR 0.57, 95% CI 0.31–1.05,p=0.069).
TABLE 4.
Cox proportional hazard analysis of associations between genetic variants of Epidermal Growth Factor (EGF) and 60-day survival in patients with acute respiratory distress syndrome (ARDS)
| All ARDS (n=416) |
Male (n=246) |
Female (n=170) |
||||
|---|---|---|---|---|---|---|
| HR (95% CI)+ | p-value | HR (95% CI)+ | p-value | HR (95% CI)+ | p-value | |
| Genotype# | ||||||
| rs4444903 | 0.93 (0.67–1.29) | 0.662 | 0.84 (0.54–1.32) | 0.452 | 1.09 (0.66–1.83) | 0.731 |
| rs2298991 | 0.86 (0.62–1.20) | 0.378 | 0.76 (0.48–1.19) | 0.229 | 1.06 (0.63–1.77) | 0.829 |
| rs11568893 | 0.76 (0.49–1.17) | 0.213 | 0.95 (0.53–1.68) | 0.849 | 0.58 (0.28–1.20) | 0.142 |
| rs7692976 | 0.88 (0.63–1.23) | 0.460 | 0.73 (0.46–1.16) | 0.179 | 1.12 (0.67–1.86) | 0.664 |
| rs4698803 | 0.88 (0.62–1.24) | 0.470 | 0.70 (0.43–1.15) | 0.137 | 1.11 (0.66–1.89) | 0.689 |
| rs6533485 | 0.75 (0.53–1.06) | 0.102 | 0.68 (0.42–1.10) | 0.112 | 0.86 (0.51–1.46) | 0.577 |
| Haplotype | ||||||
| Global test¶ | χ2 = 2.60 | 0.761 | χ2 = 5.64 | 0.343 | χ2 = 6.93 | 0.226 |
| Hap1 (ATCATG) | (reference) | - | (reference) | - | (reference) | - |
| Hap2 (GGCGTC) | 1.03 (0.77–1.38) | 0.840 | 0.79 (0.52–1.20) | 0.267 | 1.32 (0.85–2.05) | 0.222 |
| Hap3 (ATCAAG) | 0.93 (0.63–1.38) | 0.734 | 0.57 (0.31–1.05) | 0.069 | 1.37 (0.79–2.39) | 0.268 |
| Hap4 (GGTGTC) | 0.79 (0.51–1.22) | 0.287 | 0.91 (0.48–1.70) | 0.759 | 0.62 (0.31–1.22) | 0.166 |
| Hap5 (ATCATC) | 0.84 (0.52–1.34) | 0.462 | 0.83 (0.45-1.53) | 0.545 | 0.89 (0.42–1.92) | 0.774 |
| All others (Freq < 5%) | 1.13 (0.76–1.67) | 0.548 | 1.28 (0.78–2.11) | 0.336 | 0.95 (0.47–1.89) | 0.872 |
HR: hazard ratio; CI: confidence interval.
Wildtype homozygote of each SNP was considered as reference
Likelihood-ratio test (LRT), with 5 degrees of freedom
adjusted for age, sex, APACHE III score, septic shock, pneumonia, trauma, end-stage renal disease, liver cirrhosis/failure, metastatic cancer, recent use of steroid, and treatment with activated protein C. APACHE III score was revised to remove age from the score to avoid colinearity. Sex was removed from the model during sex-stratified analyses.
Gene-sex interaction for ARDS risk
The interaction analyses showed that the variant genotypes of rs4444903, rs2298991, rs7692976 and rs6533485 significantly interacted with sex for ARDS risk (p for interaction=0.010, 0.010, 0.007, 0.021, respectively) (table 5). Because sex hormone activities are decreasing with age, we further stratified the interaction analysis by age. In order to have the largest statistical power, we used the median age for stratification. We found the gene-sex interactions were even stronger in the younger group (age <65), but were not significant in the elder group (age ≥65), whose sex hormone effects were expected to be less active.
TABLE 5.
Genotype-sex interactions for acute respiratory distress syndrome (ARDS) risk.
| All Subjects (n=1468) |
Age <65 (n=736) |
Age ≥65 (n=732) |
||||
|---|---|---|---|---|---|---|
| Genotype-Sex Interaction | OR (95% CI)# | p-value ¶ | OR (95% CI)# | p-value ¶ | OR (95% CI)# | p-value ¶ |
| rs4444903*sex | 2.01 (1.18–3.43) | 0.010+ | 2.50 (1.18–5.33) | 0.017+ | 1.55 (0.72–3.33) | 0.261 |
| rs2298991*sex | 2.02 (1.18–3.43) | 0.010+ | 2.67 (1.26–5.69) | 0.011+ | 1.44 (0.67–3.07) | 0.350 |
| rs11568893*sex | 0.99 (0.52–1.89) | 0.973 | 1.20 (0.49–2.96) | 0.688 | 0.69 (0.26–1.85) | 0.464 |
| rs7692976*sex | 2.07 (1.22–3.54) | 0.007+ | 3.08 (1.45–6.57) | 0.004+ | 1.32 (0.62–2.82) | 0.473 |
| rs4698803*sex | 0.64 (0.37–1.09) | 0.100 | 0.68 (0.32–1.48) | 0.336 | 0.61 (0.28–1.30) | 0.200 |
| rs6533485*sex | 1.96 (1.11–3.46) | 0.021+ | 2.46 (1.09–5.56) | 0.031+ | 1.46 (0.66–3.26) | 0.353 |
OR: odds ratio; CI: confidence interval. Females with wildtype homozygote of each SNP were used as reference.
OR and 95% CI for interaction in multivariate logistic regression models, adjusted for age, sex, APACHE III score, all risk factors for ARDS (listed in table 1), diabetes, liver cirrhosis/failure and alcohol abuse. APACHE III score was revised to remove age from the score to avoid colinearity
for interaction
FDR adjusted p-value <0.05.
DISCUSSION
In the present study, we comprehensively evaluated the associations of EGF variants with ARDS risk and outcomes, and the role of sex in the association between EGF variants and ARDS. We found that the common EGF variants were associated with ARDS risk in a sex-specific manner. Variant genotypes of rs4444903, rs2298991, rs7692976, rs4698803, and haplotypes GGCGTC, ATCAAG were associated with ARDS risk in males. Although no significant associations was found in females, we observed that the association of EGF variants on ARDS development in females were mostly opposite from that observed in males. Such gene-sex interaction was further supported by the results from interaction analyses. On the other hand, EGF variants did not significantly influence the ARDS outcomes and survival in our study.
EGF was first discovered in submaxillary glands of adult male mice, while human EGF (beta-urogastrone) was first isolated from urine. The 6-kDa human EGF consists of 53 amino acids. It is initially synthesized as a transmembrane glycoprotein precursor (prepro-EGF) of 1207 amino acids, and is then processed through a pro-EGF stage to mature EGF protein [30, 31]. The large prepro-EGF polypeptide contains the EGF subunit and eight additional EGF-like subunits, the biological significance of which is unknown [30]. The heparin-binding 160-kDa pro-EGF, isolated from human urine, has been shown to be biologically active [31]. However, there seem to be increasing reports of molecular heterogeneity in mature EGF. The biological properties and physiological significance of the EGF precursors and the heterogeneity of mature EGF remain to be elucidated [32].
The EGF gene in humans is located on chromosome 4. As for functional polymorphisms, the variant G allele of rs4444903 has been associated with higher secretion of EGF protein than the A allele. The mechanism by which EGF levels are modulated has been recently proposed in a study of hepatocellular carcinoma risk [33]. Transcripts from the G allele exhibit longer half-life than those from the A allele, thus the EGF mRNA and EGF protein are increased in cell lines with more copies of G in the genotype. In our study, the variant genotypes (AG/GG) of this polymorphism significantly predisposed at-risk male patients to develop ARDS but were not associated with better clinical outcomes, suggesting that higher EGF levels in the early phase might contribute to the pathogenesis of ARDS, possibly through its pro-inflammatory properties. On the other hand, the proposed function of EGF to facilitate epithelial repair and alveolar fluid reabsorption might not be decisive in the recovery from ARDS. Hap2 (GGCGTC), carrying the variant alleles of rs4444903, rs2298991, rs7692976, and rs6533485, was identified as a common haplotype associated with increased ARDS risk in males. The variant genotypes of these four SNPs were all associated with increased ARDS risk (p <0.05 for the first three SNPs; p=0.08 for rs6533485). Based on the established functional significance of rs4444903, the positions of SNPs (rs4444903 is in 5’ UTR; the other three are in introns), and the high degree of pairwise LD within these SNPs (D’ >0.85), we inferred that the associations of rs2298991 and rs7692976 with ARDS observed in genotype analysis were mediated by their LD with rs4444903.
SNP rs4698803 is a missense T>A polymorphism located in the exon 19 of EGF gene, causing an amino acid substitution (V920E). We identified the A allele of rs4698803 as a potentially protective allele against ARDS. Based on our tagging algorithm, this SNP did not tag any other SNPs in the EGF gene. Hap3 (ATCAAG), carrying the variant allele of rs4698803 and the wildtype alleles of all other SNPs, was also identified as a potentially protective haplotype against ARDS. Our results from genotype and haplotype analyses suggest that rs4698803 is independently associated with ARDS. Future studies are needed to understand whether and how this single amino acid mutation V920E might alter the biochemical function of EGF precursor and mature EGF protein.
A similar sex difference as we found in this study has also been observed in the association studies of EGF with schizophrenia. The G allele at SNP rs4444903 is associated with the age of onset in male patients with schizophrenia, but not in females [25, 26]. The sex difference in the genetic influence of EGF on ARDS might be explained by the sex- and tissue-specific regulation of EGFR signaling pathways by sex hormones. In our study, the gene-sex interaction was not significant in the elder group, whose sex hormones effects are expected to be less active. It is evident that there is crosstalk between sex hormone receptors and EGF receptor pathways. In lung development, the expression and activity of EGFR appears to be sex-specific and cell-specific [21]. Androgen treatment has been shown to decrease EGFR density and EGF induced autophosphorylation of EGFR in fetal rabbit lung [19]. On the other hand, estrogen up-regulates the expression of EGFR, whereas progesterone up-regulates the expression of 133- and 71-kDa immunoreactive EGF (the prepro-EGF-like proteins) in uterine leiomyoma cells [20]. Indeed, the impacts of sex and sex hormones on acute lung injury have been studied in animal models. Androgens appear to be detrimental while estrogens tend to be protective in the pathogenesis of acute lung injury [34].
To our knowledge, this is the first study using the haplotype-tagging SNP approach to investigate genetic susceptibility to ARDS. A major advantage of this approach is that it allows a cost-effective identification of common susceptibility alleles across the entire gene region. In addition, this is so far the largest study for genetic epidemiology of ARDS and hence provided more statistical power to detect genetic associations with ARDS, particularly in sex-stratified analyses. Another major strength of this study is its study design. We used the AECC definition for ARDS diagnosis to clearly define the phenotype prospectively. We selected the at-risk critically ill patients as controls to reduce the possible confounding from associations between candidate polymorphisms and predisposing conditions for ARDS. Furthermore, we restricted the analysis to a single ethnic group, thus minimized false results due to population stratification.
One of the limitations in this study is that neither EGF nor sex hormones levels were determined, thus the functional significance of the EGF genotypes and haplotypes on ARDS and their interactions with sex hormones remain to be further defined. Although this is so far the largest population available for ARDS association study, we might not have adequate power to detect the association of EGF variants on ARDS survival with 416 ARDS cases, particularly when it is expected to be small because the resolution of lung edema and injury contributes only partially to surviving ARDS. Since this study included only a single cohort, our findings need to be validated in other independent populations. Finally, our results are based on the Caucasians. Additional studies in other ethnic groups will be needed.
In conclusion, this study demonstrates that genetic associations of EGF with ARDS risk are modified by sex. The variant genotypes of rs4444903, rs2298991, rs7692976, rs4698803 and haplotypes GGCGTC, ATCAAG are significantly associated with ARDS development in at-risk males. Our findings should be replicated in other cohorts. Our results also warrant the future basic research to understand the role of EGF, as well as their interactions with sex hormones, in the pathogenesis of ARDS.
Supplementary Material
ACKNOWLEDGEMENTS
Support Statement
This study was supported in part by grants from National Institutes of Health (HL60710, ES00002) and Flight Attendant Medical Research Institute (062459-YCSA).
The authors would like to thank Weiling Zhang, Kelly McCoy, Thomas McCabe, Julia Shin, and Hanae Fujii-Rios for patient recruitment; Andrea Shafer and Starr Sumpter for research support; Maureen Convery for laboratory expertise; Janna Frelich for data management; and the patients and staff of ICUs at Massachusetts General Hospital.
Footnotes
Statement of Interest
None declared.
REFERENCES
- 1.Piantadosi CA, Schwartz DA. The acute respiratory distress syndrome. Ann Intern Med. 2004;141:460–470. doi: 10.7326/0003-4819-141-6-200409210-00012. [DOI] [PubMed] [Google Scholar]
- 2.Ware LB. Pathophysiology of acute lung injury and the acute respiratory distress syndrome. Semin Respir Crit Care Med. 2006;27:337–349. doi: 10.1055/s-2006-948288. [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Wiener-Kronish JP. Intact epithelial barrier function is critical for the resolution of alveolar edema in humans. Am Rev Respir Dis. 1990;142:1250–1257. doi: 10.1164/ajrccm/142.6_Pt_1.1250. [DOI] [PubMed] [Google Scholar]
- 4.Bazley LA, Gullick WJ. The epidermal growth factor receptor family. Endocr Relat Cancer. 2005;12 Suppl 1:S17–S27. doi: 10.1677/erc.1.01032. [DOI] [PubMed] [Google Scholar]
- 5.Desai TJ, Cardoso WV. Growth factors in lung development and disease: friends or foe? Respir Res. 2002;3:2. doi: 10.1186/rr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan RM, Mineo-Kuhn MM, Kramer CM, Finkelstein JN. Growth factors alter neonatal type II alveolar epithelial cell proliferation. Am J Physiol. 1994;266:L17–L22. doi: 10.1152/ajplung.1994.266.1.L17. [DOI] [PubMed] [Google Scholar]
- 7.Van Winkle LS, Isaac JM, Plopper CG. Distribution of epidermal growth factor receptor and ligands during bronchiolar epithelial repair from naphthalene-induced Clara cell injury in the mouse. Am J Pathol. 1997;151:443–459. [PMC free article] [PubMed] [Google Scholar]
- 8.Borok Z, Hami A, Danto SI, Lubman RL, Kim KJ, Crandall ED. Effects of EGF on alveolar epithelial junctional permeability and active sodium transport. Am J Physiol. 1996;270:L559–L565. doi: 10.1152/ajplung.1996.270.4.L559. [DOI] [PubMed] [Google Scholar]
- 9.Danto SI, Borok Z, Zhang XL, Lopez MZ, Patel P, Crandall ED, Lubman RL. Mechanisms of EGF-induced stimulation of sodium reabsorption by alveolar epithelial cells. Am J Physiol. 1998;275:C82–C92. doi: 10.1152/ajpcell.1998.275.1.C82. [DOI] [PubMed] [Google Scholar]
- 10.Sznajder JI, Ridge KM, Yeates DB, Ilekis J, Olivera W. Epidermal growth factor increases lung liquid clearance in rat lungs. J Appl Physiol. 1998;85:1004–1010. doi: 10.1152/jappl.1998.85.3.1004. [DOI] [PubMed] [Google Scholar]
- 11.Puddicombe SM, Polosa R, Richter A, Krishna MT, Howarth PH, Holgate ST, Davies DE. Involvement of the epidermal growth factor receptor in epithelial repair in asthma. FASEB J. 2000;14:1362–1374. doi: 10.1096/fj.14.10.1362. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton LM, Torres-Lozano C, Puddicombe SM, Richter A, Kimber I, Dearman RJ, Vrugt B, Aalbers R, Holgate ST, Djukanovic R, Wilson SJ, Davies DE. The role of the epidermal growth factor receptor in sustaining neutrophil inflammation in severe asthma. Clin Exp Allergy. 2003;33:233–240. doi: 10.1046/j.1365-2222.2003.01593.x. [DOI] [PubMed] [Google Scholar]
- 13.Subauste MC, Proud D. Effects of tumor necrosis factor-alpha, epidermal growth factor and transforming growth factor-alpha on interleukin-8 production by, and human rhinovirus replication in, bronchial epithelial cells. Int Immunopharmacol. 2001;1:1229–1234. doi: 10.1016/s1567-5769(01)00063-7. [DOI] [PubMed] [Google Scholar]
- 14.Lewkowicz P, Tchorzewski H, Dytnerska K, Banasik M, Lewkowicz N. Epidermal growth factor enhances TNF-alpha-induced priming of human neutrophils. Immunol Lett. 2005;96:203–210. doi: 10.1016/j.imlet.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Uddin M, Seumois G, Lau LC, Rytila P, Davies DE, Djukanovic R. Enhancement of neutrophil function by the bronchial epithelium stimulated by EGF. Eur Respir J. 2008;31:714–724. doi: 10.1183/09031936.00144307. [DOI] [PubMed] [Google Scholar]
- 16.Araujo A, Ribeiro R, Azevedo I, Coelho A, Soares M, Sousa B, Pinto D, Lopes C, Medeiros R, Scagliotti GV. Genetic polymorphisms of the epidermal growth factor and related receptor in non-small cell lung cancer--a review of the literature. Oncologist. 2007;12:201–210. doi: 10.1634/theoncologist.12-2-201. [DOI] [PubMed] [Google Scholar]
- 17.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, Hutchinson PE, Osborne JE, Lear JT, Smith AG, Hutchinson IV. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 18.Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 19.Klein JM, Nielsen HC. Androgen regulation of epidermal growth factor receptor binding activity during fetal rabbit lung development. J Clin Invest. 1993;91:425–431. doi: 10.1172/JCI116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruo T, Ohara N, Wang J, Matsuo H. Sex steroidal regulation of uterine leiomyoma growth and apoptosis. Hum Reprod Update. 2004;10:207–220. doi: 10.1093/humupd/dmh019. [DOI] [PubMed] [Google Scholar]
- 21.Rosenblum DA, Volpe MV, Dammann CE, Lo YS, Thompson JF, Nielsen HC. Expression and activity of epidermal growth factor receptor in late fetal rat lung is cell- and sex-specific. Exp Cell Res. 1998;239:69–81. doi: 10.1006/excr.1997.3888. [DOI] [PubMed] [Google Scholar]
- 22.Guh JY, Chen HC, Chuang LY, Yang CY, Tsai JH, Lai YH. Significance of salivary epidermal growth factor in peptic ulcer disease in hemodialysis patients. Nephron. 2001;87:134–138. doi: 10.1159/000045901. [DOI] [PubMed] [Google Scholar]
- 23.Nava A, Barton K, Monroy DC, Pflugfelder SC. The effects of age, gender, and fluid dynamics on the concentration of tear film epidermal growth factor. Cornea. 1997;16:430–438. [PubMed] [Google Scholar]
- 24.Wingren U, Brown TH, Watkins BM, Larson GM. Delayed gastric ulcer healing after extirpation of submandibular glands is sex-dependent. Scand J Gastroenterol. 1989;24:1102–1106. doi: 10.3109/00365528909089262. [DOI] [PubMed] [Google Scholar]
- 25.Anttila S, Illi A, Kampman O, Mattila KM, Lehtimaki T, Leinonen E. Association of EGF polymorphism with schizophrenia in Finnish men. Neuroreport. 2004;15:1215–1218. doi: 10.1097/00001756-200405190-00027. [DOI] [PubMed] [Google Scholar]
- 26.Hanninen K, Katila H, Anttila S, Rontu R, Maaskola J, Hurme M, Lehtimaki T. Epidermal growth factor a61g polymorphism is associated with the age of onset of schizophrenia in male patients. J Psychiatr Res. 2007;41:8–14. doi: 10.1016/j.jpsychires.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Gong MN, Thompson BT, Williams P, Pothier L, Boyce PD, Christiani DC. Clinical predictors of and mortality in acute respiratory distress syndrome: potential role of red cell transfusion. Crit Care Med. 2005;33:1191–1198. doi: 10.1097/01.ccm.0000165566.82925.14. [DOI] [PubMed] [Google Scholar]
- 28.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol. 2005;28:261–272. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;51:289–300. [Google Scholar]
- 30.Burgess AW. Epidermal growth factor and transforming growth factor alpha. Br Med Bull. 1989;45:401–424. doi: 10.1093/oxfordjournals.bmb.a072331. [DOI] [PubMed] [Google Scholar]
- 31.Parries G, Chen K, Misono KS, Cohen S. The human urinary epidermal growth factor (EGF) precursor. Isolation of a biologically active 160-kilodalton heparin-binding pro-EGF with a truncated carboxyl terminus. J Biol Chem. 1995;270:27954–27960. doi: 10.1074/jbc.270.46.27954. [DOI] [PubMed] [Google Scholar]
- 32.Aybay C, Karakus R, Yucel A. Characterization of human epidermal growth factor in human serum and urine under native conditions. Cytokine. 2006;35:36–43. doi: 10.1016/j.cyto.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Tanabe KK, Lemoine A, Finkelstein DM, Kawasaki H, Fujii T, Chung RT, Lauwers GY, Kulu Y, Muzikansky A, Kuruppu D, Lanuti M, Goodwin JM, Azoulay D, Fuchs BC. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299:53–60. doi: 10.1001/jama.2007.65. [DOI] [PubMed] [Google Scholar]
- 34.Carey MA, Card JW, Voltz JW, Germolec DR, Korach KS, Zeldin DC. The impact of sex and sex hormones on lung physiology and disease: lessons from animal studies. Am J Physiol Lung Cell Mol Physiol. 2007;293:L272–L278. doi: 10.1152/ajplung.00174.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.