Abstract
Type I interferons (IFNs) are innate cytokines with potent antiviral and immunoregulatory activities. It remains unclear how human cytomegalovirus (HCMV) can establish persistence in the face of these strongly antagonistic cytokines. In this study, we confirm that IFN-α efficiently suppresses the penetration of HCMV into susceptible cells, including monocytes, the major cell population in peripheral blood that is highly susceptible to HCMV infection. We further demonstrate that the HCMV-derived interleukin 10 (IL-10) homolog functions similar to cellular IL-10 and broadly inhibits TLR-induced transcriptional activation of IFN-α/β genes in plasmacytoid dendritic cells (PDCs), a major type I IFN-producer in vivo that is highly resistant to HCMV infection in vitro. These results suggest that HCMV subverts innate immunity by suppressing type I IFN production of PDCs during primary viral infection via its IL-10 homolog.
Introduction
Upon primary viral infection, innate immune mechanisms rapidly counteract viral attacks before a prolonged inflammatory response and the onset of severe tissue damage jeopardize the health and/or survival of the host. Type I interferons (IFNs), consisting of a single IFN-β gene and a family of 13 IFN-α genes among others, have long been considered as important effector cytokines that inhibit viral replication by inducing host genes, such as 2’–5’ oligoadenylate synthetase, MxA, and dsRNA-activated protein kinases (Stark et al., 1998; Vilcek and Sen, 1996). Moreover, type I IFNs also play important roles in immunoregulation by activating natural killer (NK) cells (Krug et al., 2004a), modulating the functions of dendritic cells (DCs) (Lapenta et al., 2006; Longman et al., 2007), B and T cells (Aichele et al., 2006; Chang et al., 2007b; Coro, Chang, and Baumgarth, 2006; Le Bon et al., 2006), as well as fine-tuning DC development in the bone marrow (Zuniga et al., 2004).
The plasmacytoid dendritic cells (PDCs) is a unique DC population that regulates both innate and adaptive antiviral immunity (Colonna, Trinchieri, and Liu, 2004). Although most cells can produce type I IFNs, PDCs produce up to 1,000-fold more IFN-α than other cell types in response to viral infection (Cella et al., 1999; Kadowaki et al., 2000; Siegal et al., 1999). PDCs can migrate from the blood to lymphoid tissues, where they secrete high levels of IFN-α to activate other components of the immune system (Siegal et al., 1999). PDCs react to specific viral components via a subset of Toll-like receptors (TLRs) (Gilliet, Cao, and Liu, 2008). They express TLR7 to bind the genomes of RNA viruses, such as influenza and vesicular stomatitis virus (Diebold et al., 2004; Lund et al., 2004), and TLR9 to bind the unmethylated CpG residues of DNA viruses, such as herpesviruses (Krug et al., 2004b; Lund et al., 2003). Both TLR7 and TLR9 signal through the adaptor molecule MyD88 to recruit signaling mediators for the activation of NF-κBF (Takeda, Kaisho, and Akira, 2003). During acute mouse cytomegalovirus (MCMV) infection, IFN-α production by PDCs in blood and spleen depends on the TLR9/ MyD88 signaling pathway (Delale et al., 2005; Krug et al., 2004a).
In mice, a positive feedback loop is initiated by the induction of IFN-β and IFN-α4 through the rapid phosphorylation of interferon regulatory factor 3 (IRF-3), a transcription factor that is constitutively expressed in many cell types. This leads to the accumulation of IRF-7, which subsequently activates the expression of other IFN-α subtypes (Levy, Marie, and Prakash, 2003; Marie, Durbin, and Levy, 1998; Theofilopoulos et al., 2005). Not surprisingly, many viruses have developed mechanisms to disrupt the function of IRF-3, and, thereby, dampen the antiviral effects of type I IFNs (Abate, Watanabe, and Mocarski, 2004; Foy et al., 2003; Lin et al., 2004; Ronco et al., 1998; Talon et al., 2000). The resistance of viruses to type I IFNs is largely attributed to the action of viral components inside the infected cells. Large complex DNA viruses, such as vaccinia virus and other orthopoxviruses, also synthesize soluble IFN-α receptor homologs to counteract type I IFNs (Symons, Alcami, and Smith, 1995).
The induction of IFN-α production occurs much more rapidly in PDCs due to their constitutive expression of IRF-7 (Izaguirre et al., 2003; Prakash et al., 2005). Intact PDC function is essential for the control of infections with many DNA or RNA viruses in vivo, including MCMV, mouse hepatitis virus, and respiratory syncytial virus (Cervantes-Barragan et al., 2007; Dalod et al., 2002; Wang, Peters, and Schwarze, 2006). It has been shown that PDC-derived IFN-α is critical for NK and CD8+ T cell cytotoxicity against MCMV infection (Dalod et al., 2003; Le Bon et al., 2003; Nguyen et al., 2002). It is, therefore, of particular interest to determine whether human cytomegalovirus (HCMV) also suppresses type I IFNs production by PDCs. Our in vitro data demonstrated that, although PDCs were highly resistant to HCMV penetration, their capacity to initiate a robust type I IFN response was markedly reduced by elaboration of the viral IL-10 homolog by HCMV-infected cells. These results establish another aspect of the complex multipotent immunomodulatory function of HCMV IL-10.
Results
Resistance of PDCs to HCMV infection
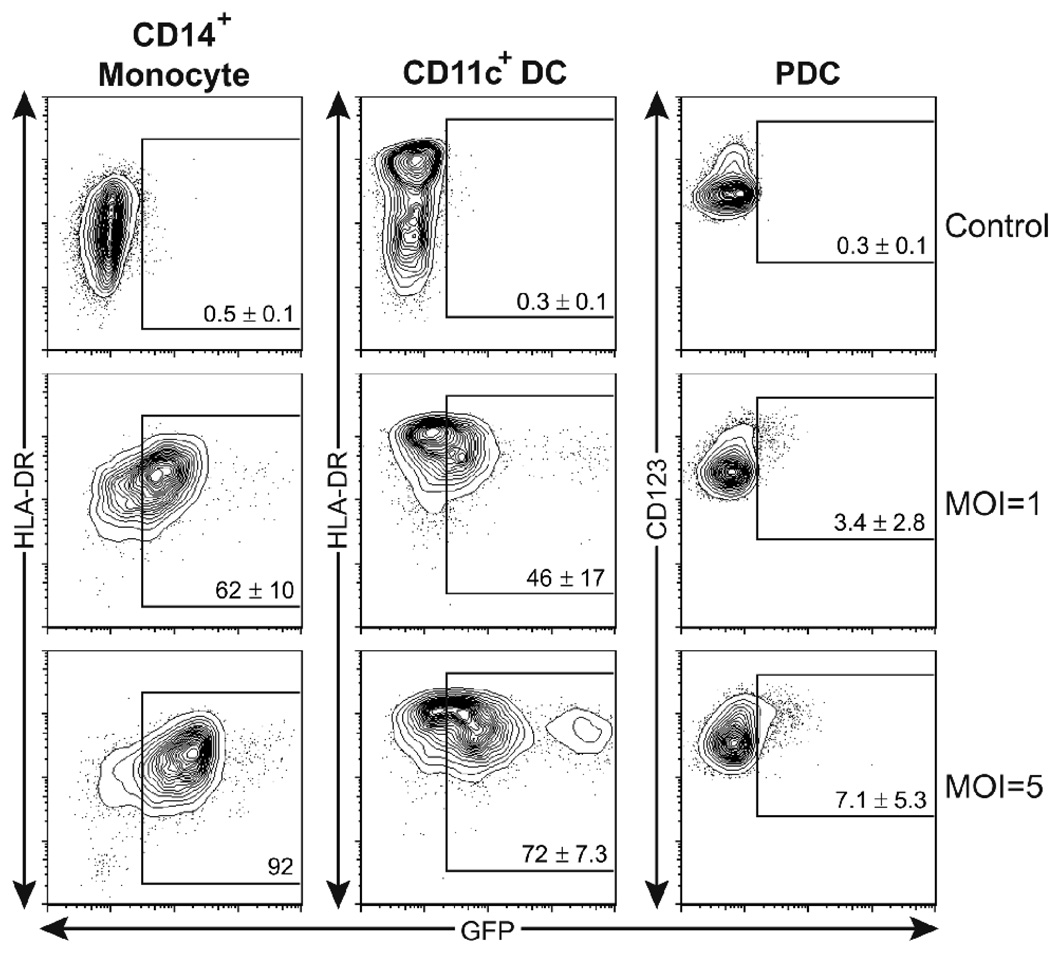
To fully assess the potential target of HCMV infection in the blood, we used an endothelial and epithelial cell tropic HCMV, BADrUL131 (Wang and Shenk, 2005a; Wang and Shenk, 2005b), to infect freshly isolated PBMCs. This HCMV variant contains the intact UL128–UL131 region, which encodes open reading frames crucial for the leukotropism of HCMV (Hahn et al., 2004), and an EGFP expression cassette driven by the constitutive SV40 promoter. At 48 h post infection, FACS analysis was conducted to determine the frequency of the infected cells and their identity. The predominant HCVM-infected cell population was the CD14+ monocyte (data not shown), consistent with a previous report (Soderberg et al., 1993). In addition, a small fraction of non-B non-T cells also became EGFP+, suggesting that these cells either were infected by HCMV or engulfed cell components of other infected cells (data not shown).
Previous reports showed that monocyte-derived DCs (MoDCs) are also susceptible to HCMV infection (Beck et al., 2003; Moutaftsi et al., 2002; Riegler et al., 2000). Therefore, we further focused on HCMV infection in the two major primary DC subsets in human blood: CD11c+ myeloid DCs and CD11c−/CD123+ PDCs (Liu, 2001). CD14+ monocytes, CD11c+ myeloid DCs, and CD123+ PDCs were separately isolated from 3–4 individual donors. 5 × 105 monocytes, 1 × 105 CD11c+ DCs, and 1 × 105 PDCs were inoculated with HCMV at various multiplicities of infection (MOI’s), calculated according to their infectivity titers on MRC-5 fibroblasts. Cells were collected and immunostained with conjugated monoclonal antibodies to CD11c, CD14, CD123, and HLA-DR at 36 h post inoculation, and we identified infected cells by flow cytometry using the EGFP signal from the EGFP-expressing HCMV, driven by a constitutive SV40 promoter. Purified CD14+ monocytes and CD11c+ DCs showed different degrees of susceptibility to HCMV infection, and the surface expression of MHC-II of both cell populations increased after infection (Fig. 1). It was also noted that both cell types were less susceptible to HCMV infection than fibroblasts, as an average of 62% and 46% of CD14+ monocytes and CD11c+ DCs, respectively, became EGFP+ when infected with virus at a dose that caused infection in close to 100% of fibroblasts.
Fig. 1. HCMV targets monocytes and myeloid DCs in peripheral blood.
Freshly isolated blood CD14+ monocytes (left panels), CD11c+ DCs (middle panels) and CD123+ PDCs (right panels) were infected for 36 h with GFP-recombinant HCMV at MOI’s of 1 and 5. The expression of indicated surface markers and of GFP identifying HCMV-infected cells was assessed by FACS. Shown are 5% contour plots with outliers of cells after gating leukocytes via FSC–SSC and CD14+to identify monocytes, FSC–SSC and CD11c+ to identify CD11c+ DCs, and FSC–SSC and CD11c–/CD123+ to identify PDCs. The percentage of GFP+ cells, among the identified cell populations is shown in each plot, indicated by a box. Numbers are presented as the mean values ± SD of data from 3–4 individual donors.
In contrast, CD11c−/CD123+ PDCs were far more resistant to HCMV infection than monocytes or myeloid DCs. When isolated PDCs were infected with HCMV at a MOI of 5, a titer that infected more than 90% of monocytes, only a small fraction (7.1 ± 5.3%) of PDCs was infected with HCMV (Fig. 1). Although EGFP fluorescence is not a direct indicator of virus replication, our findings of strong PDC resistance to HCMV infection is in accordance with other reports that used different HCMV strains and detection systems (Cederarv, Soderberg-Naucler, and Odeberg, 2009; Kvale et al., 2006; Varani et al., 2007).
Effects of IFN-α on HCMV infection
Since PDCs are strong producers of IFN-α, we next determined whether the presence of type I IFN could directly inhibit the entry of HCMV into susceptible cell types. We pre-treated monolayers of fibroblasts (MRC-5) and epithelial cells (ARPE-19), as well as purified CD14+ monocytes with different concentrations of recombinant IFN-α for 4–6 h before HCMV infection. As these cells exhibited different susceptibility to HCMV attachment/entry and to achieve a similar level of viral infection among different cell types, MRC-5 cells were inoculated at 0.1 MOI and ARPE-19 and CD14+ monocytes were inoculated at 0.2 MOI. The presence of IFN-α significantly reduced the overall frequency of infected cells in a dose-dependent manner (Fig. 2). When a very low dose of IFN-α (10 U/ml) was provided, a more profound reduction of viral entry was observed in epithelial cells (84% reduction) and monocytes (average of 54% reduction in two donors), in comparison to fibroblasts (23% reduction). Together with similar findings in CD11c+ DCs (Kvale et al., 2006), these data suggest that type I IFNR-mediated signals can directly inhibit viral entry not only into adherent cells, but also into the major HCMV targets in the peripheral blood.
Fig. 2. IFN-α reduces the susceptibility of various cell types to HCMV infection.
Monolayer cultures of MRC-5 fibroblasts, ARPE-19 epithelial cells and freshly isolated CD14+ monocytes were pre-treated with indicated doses of IFN-α before HCMV infection. Data represent mean frequencies of infected cells ± SD from triplicate or quadruplicate cultures, as determined by FACS. Statistical comparisons to control samples without IFN-α treatment were performed using one-way ANOVA followed by Dunnett’s post tests (**p < 0.01). The results of monocytes are representative of experiments from two individual donors that gave similar results.
cmvIL-10 released from HCMV-infected cells inhibits IFN-α production
We previously showed that HCMV encodes an IL-10 homolog (cmvIL-10) that inhibits the maturation of MoDCs and alters their cytokine profile (Chang et al., 2004). In this study, we aimed to examine whether cmvIL-10 would also impair IFN-α-production by PDCs. To mimic cmvIL-10 secretion following HCMV infection, PBMCs were treated with conditioned media from fibroblast cultures infected with wild-type HCMV (AD169) or an AD169-derived cmvIL-10 knockout variant (TS359). Addition of AD169- or TS359-conditioned medium alone did not induce a detectable level of IFN-α secretion by PBMCs, despite the presence of viral particles in the conditioned media of both HCMV strains (data not shown). Therefore, we added 5 µg/ml of A-class CpG to induce strong IFN-α production by PBMCs. Supernatants from PBMC cultures were collected after stimulation for 24 h and analyzed by ELISA for IFN-α protein. AD169-conditioned medium significantly suppressed the induction of IFN-α secretion, compared to the control medium (Fig. 3A). In contrast, conditioned medium collected from cmvIL-10 defective mutant HCMV TS359-infected cultures enhanced the CpG-induced IFN-α production to a level significantly higher than the control group. When 5 ng/ml of recombinant cmvIL-10 was introduced to the culture, a dose equivalent to the level of HCMV-encoded cmvIL-10 in the conditioned medium (Chang et al., 2004), the wild-type phenotype of TS359-conditioned medium was restored (Fig. 3A). Thus, supernatants from virus expressing cmvIL-10 but not those lacking it could suppress IFN-α production.
Fig. 3. cmvIL-10 inhibits IFN-α production by PBMCs.
Freshly isolated PBMCs were treated with CpG and conditioned medium from HCMV-infected cultures (A), CpG or heat-inactivated influenza virus in the presence or absence of indicated concentrations hIL-10 or cmvIL-10 (ng/ml) (B). The supernatants were collected after stimulation for 24 h and IFN-α concentrations were determined by ELISA. Data represent mean values ± SD. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post tests (***p < 0.001) (A) or Dunnett’s post tests in comparison with the control samples without IL-10 treatment (**p < 0.01) (B). Shown are representative of data from one of two individual donors.
To directly test the effects of cmvIL-10 on IFN-α production, PBMCs were treated with purified recombinant protein. Cells were stimulated through TLR9 with 50 µg/ml of A-Class CpG, or TLR7/8 with heat-inactivated influenza virus A/Mem/71 (Doucett et al., 2005). Parallel cultures were established with human IL-10 (hIL-10) to compare the potency of cmvIL-10 and hIL-10. The results showed that cmvIL-10 significantly suppressed PDC-derived IFN-α production, in a dose-dependent fashion (Fig. 3B). As little as 0.5 ng/ml of cmvIL-10 was sufficient to reduce IFN-α production by ∼50%. cmvIL-10 and hIL-10, used at approximately equimolar concentrations, showed that they were similarly effective in suppressing type I IFN secretion. These data established that infection-induced cmvIL-10 could inhibit IFN-α production by PBMCs.
Direct impacts of cmvIL-10 on PDC functionality
We next investigated whether cmvIL-10 reduces IFN-α secretion by acting directly on PDCs. Because HCMV activates PDCs via TLR9 (Varani et al., 2007), we used the TLR9 agonist A-class CpG as a stimulus and compared the effects of cmvIL-10 and hIL-10 (5 ng/ml) on IFN-α production by freshly isolated primary PDCs. A high dose of CpG (50 µg/ml) was added to induce an average of 57,145 ± 23,980 pg/ml of IFN-α and 5,288 ± 1,644 pg/ml of IFN-β secretion by 5 × 104 PDCs isolated from 3–5 individual donors. Neither cmvIL-10 nor hIL-10 inhibited the CpG-induced maturation of PDCs, as determined by increased expression of CD83, CD86, and MHC-II (Fig. 4A). Statistical analyses demonstrated that cmvIL-10 or hIL-10 did not significantly alter the expression levels of these surface molecules (Table 1). However, both IL-10 markedly suppressed production of IFN-α and IFN-β by an average of 75% (Fig. 4B). Similar trends were observed following stimulation of PDCs with the TLR7/8 ligands, heat-inactivated influenza virus or imiquimod-R837 (Invivogen) (data not shown).
Fig. 4. cmvIL-10 inhibits IFN-expression but not maturation of PDCs.
Freshly isolated PDCs were left untreated or stimulated with CpG in the presence or absence of 5 ng/ml hIL-10 or cmvIL-10 for 18 h. The surface expression of indicated molecules on PDCs was assessed by FACS and is presented with overlaid histograms. Shown are representative data from one of five individual donors that gave similar results (A). IL-10 significantly reduces type I IFN production by CpG-activated PDCs. IFN-α (upper panel, n = 5) or IFN-β (lower panel, n = 3) protein concentrations in supernatants were measured by ELISA. Each dot represents the relative concentrations (%) in the cultures compared to CpG-only stimulation from individual donors. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post tests (***p < 0.001) (B).
Table 1.
Effects of IL-10 treatments on the surface marker expression of PDCs.a
| Treatment |
Untreated |
A-class CpG (50 µg/ml) |
R837 (5 µg/ml) |
||||
|---|---|---|---|---|---|---|---|
| IL-10b | – | – | hIL-10 | cmvIL-10 | – | hIL-10 | cmvIL-10 |
| Marker | |||||||
| CD83 | 37±6.8 | 350±4.7 | 339±11 | 352±14 | 250±6.7 | 281±16 | 287±18 |
| CD86 | 201±7.6 | 547±9.7 | 525±1.5 | 527±14 | 536±25 | 529±24 | 536±22 |
| MHC-II | 493±34 | 697±27 | 734±33 | 735±28 | 705±50 | 719±37 | 715±37 |
| CD123 | 646±12 | 608±12 | 658±18 | 655±14 | 629±37 | 641±31 | 636±37 |
Data presented here are the mean fluorescent intensity (MFI) of indicated surface markers on gated CD123+ PDCs. Shown are the mean value ± SD of data from 3 individual donors assessed by FACS at the same time with the identical equipment setting.
Both hIL-10 and cmvIL-10 were given at the concentration of 5 ng/ml upon PDC activation.
cmvIL-10 suppresses expression of type I IFN genes
To determine whether the reduction of type I IFNs was caused by suppression of expression of all or only a subset of the type I IFN subtype genes, a comprehensive analysis on the transcriptional profiles of type I IFN genes in PDCs was conducted using a nested, multiplex, real-time PCR assay (Szubin et al., 2008). hIL-10 and cmvIL-10 effectively suppressed IFN-β gene transcription in activated PDCs (Fig. 5A). Moreover, both hIL-10 and cmvIL-10 broadly reduced the steady-state mRNA levels of all 13 IFN-α subtypes by 87–96% (Fig. 5B). The impact of hIL-10 and cmvIL-10 on these genes was similar, reducing expression levels to, or in some instances below, the threshold of detection. Although hIL-10 appeared to have slightly stronger suppressive effect on gene expression of some IFN-α subtypes (for example subtypes 1/13, 4 and 8), the levels of transcription were so low that this unlikely constitutes a biologically relevant difference, particularly as these small expression differences did not translate into a distinguishable difference at the overall IFN-α protein level (Fig. 4B, upper panel). In summary, these data confirm and expand results from our earlier study (Chang et al., 2004) and demonstrate the high degree of functional overlap between viral and human IL-10 and their impacts on type I IFN gene regulation.
Fig. 5. Effects of cmvIL-10 on the transcription of type I IFN genes in activated PDCs.
IL-10 suppresses IFN-β gene expression induced through TLR signals. Shown are fold-changes in gene expression compared to the untreated cells after normalization (A). Impacts of hIL-10 and cmvIL-10 on the expression of all IFN-α subtypes in CpG-activated PDCs. Shown are the relative mRNA copy numbers after normalization to the expression of an internal control gene. Data presented here are the mean values of the results from 2 individual donors (B). These data are part of a large analysis on IFN-α subset responses of PDCs that we have published previously (Szubin et al.), in which we did not distinguish between the effects of hIL-10 and cmvIL-10.
Discussion
PDCs are a critical IFN-α-producing cell type that protects cells from viral infection, inhibits viral replication in infected cells, and optimizes the innate and adaptive anti-viral immune responses. The broad cell tropism enables HCMV to infect various innate immune components and endothelial cells, the cell type that is closely associated with the cells of both innate and adaptive immune system. Unlike monocytes and myeloid DCs, the susceptibility of PDCs to HCMV infection is very low. Our results suggest that HCMV can efficiently quell robust type I IFN antiviral responses via cmvIL-10 secretion, by targeting PDCs to influence their functions in blood vessels or lymph nodes.
In this study, we demonstrate that IFN-α boosts the resistance of leukocytes, particularly monocytes, against HCMV infection, implying the importance of systemic production of type I IFNs in containing viral propagation and dissemination at the early stage of infection. While it is currently unclear why PDCs are refractory to HCMV infection, the blockade of viral entry does not seem to be due to the autocrine effects of type I IFNs, as PDCs remained resistant to HCMV when IFN-α activity is inhibited (Kvale et al., 2006). Recent evidence suggests that monocytes/macrophages, myeloid DCs, and PDCs are derived from macrophage and DC progenitors (MDPs) (Liu et al., 2007; Waskow et al., 2008). Since HCMV establishes latency within the common myeloid progenitor CD34+ cells (Hahn, Jores, and Mocarski, 1998; Zhuravskaya et al., 1997), i.e. the precursors of MDPs, PDCs seem to lose susceptibility to HCMV infection after their maturation.
Together with other reports (Cederarv, Soderberg-Naucler, and Odeberg, 2009; Varani et al., 2007), our findings confirm that HCMV infects a small fraction of PDCs in vitro. This infection is abortive, as late viral proteins are not detected in infected cells (Varani et al., 2007). Although pp65, the major CMV structural protein, can prevent the activation of IRF-3 (Abate, Watanabe, and Mocarski, 2004), this mechanism is unlikely to affect type I IFN production by PDCs because (i) pp65 is a late protein and (ii) the rapid and strong induction of IFN-α by PDCs relies on their high constitutive expression of IRF-7, an unique characteristic of PDCs (Izaguirre et al., 2003; Prakash et al., 2005). Whether HCMV encodes genes that target IRF-7, like Kaposi’s sarcoma-associated herpesvirus (Zhu et al., 2002), remains to be further studied.
The viral IL-10 homolog released from HCMV-infected cells potently inhibited IFN-α secretion by TLR9-activated PDCs while not impairing their phenotypic maturation (Fig. 4A, Table 1). Thus, IL-10-exposed PDCs should likely remain functional antigen-presenting cells. However, the outcomes of their interaction with other leukocytes might be modulated, as they produce a substantially lower level of type I IFNs than the normal activated PDCs.
It is broadly assumed that HCMV enters PDCs and elicits IFN-α production. We did not assess whether cmvIL-10 suppresses IFN-α expression in HCMV-infected PDCs because primary isolated PDCs are highly resistant to HCMV infection in vitro. Nevertheless, utilizing a synthetic TLR9 ligand for the stimulation of PDCs should be considered biological relevant as the induction of IFN-α production during CMV infection has been shown to be dependent on a pathway mediated through TLR9 in vivo (Delale et al., 2005; Krug et al., 2004a).
We demonstrate that cmvIL-10 and cellular IL-10 are equally potent suppressers of IFN-β and the entire spectrum of IFN-α gene expression by PDCs. While IL-10 is known to induce cell death of CD40L- or CpG-activated PDCs (Duramad et al., 2003; Rissoan et al., 1999), our data indicated that the reduction of type I IFNs secretion may be largely attributed to IL-10-mediated suppression of type I IFN gene transcription. Impaired IFN-α production, along with a moderate reduction in Bcl-2 expression (Chang et al., 2007a), may synergistically contribute to the IL-10-accelerated cell death, because IFN-α is a known autocrine survival factor for activated PDCs (Kadowaki et al., 2000).
In summary, this study shows that cmvIL-10 secreted by HCMV-infected cells can directly and broadly affect type I IFN production by PDCs, providing another example of how HCMV establishes chronic infection despite the presence of fully functional immune surveillance by the host. Compromised systemic/local production of pleiotropic IFN-α/β may facilitate the dissemination of HCMV during primary infection and, suppress effective antiviral immunity. Once the well-orchestrated type I IFN network is disrupted, it may be too late to boost the resistance of target cells to HCMV infection and to promote the activity of NK cells DCs, B cells, and CD8+ T cells, allowing HCMV to establish persistence.
Materials and methods
Cell culture and virus
MRC-5 (human embryonic lung fibroblasts) were purchased from the American Type Culture Collection (ATCC) and cultured in complete DMEM (Invitrogen) (Chang et al., 2004). ARPE-19 (human retinal pigmented epithelial cells) were purchased from ATCC and cultured in complete RPE medium (DMEM/F12 supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 U penicillin/ml, 100 µg streptomycin/ml, 1 mM sodium pyruvate, 0.1 mM MEM non-essential amino acids and 1 M HEPES, all from Invitrogen). HCMV strains AD169 and TS359 (an AD169-derived cmvIL-10 knockout variant) (Yu, Silva, and Shenk, 2003) were used in this study to generate HCMV-infected conditioned medium. The conditioned medium was prepared as described previously (Chang et al., 2004). In brief, MRC-5 cultures were infected with AD169 or TS359 at a MOI of 0.02, and the supernatants were collected seven days later, filtered through a 0.45 µm filter, and stored at −80oC until use. BADrUL131, an endothelial and epithelial cell tropic AD169 variant of HCMV (Wang and Shenk, 2005a; Wang and Shenk, 2005b), was used for monitoring HCMV infection. The BADrUL131 stock used for experiments was propagated in ARPE-19, purified by high-speed centrifugation (Chang et al., 2002), and titrated in MRC-5.
Primary cell isolation
Leukocyte-enriched buffy coats from healthy individuals without donor identifiers were obtained from the Stanford Blood Center. PBMCs were isolated by Ficoll-Paque (GE Healthcare) gradient centrifugation. CD14+ monocytes, CD11c+ DCs, and PDCs were positively selected using the CD14 microbeads, BDCA-1 (CD1c), and BDCA-4 (CD304) Cell Isolation kits, respectively (Miltenyi Biotec) with an autoMACS separator with programs recommended by the manufacturer (Chang et al., 2007a). Reanalysis of monocytes (CD14+), CD11c+ DCs (CD1c+/CD11c+), and PDCs (CD11c−/CD123+/CD304+) showed purities of 97% (±0.044), 94% (±0.042), and 90% (±0.013%), respectively.
Flow cytometry
Human PBMCs and isolated leukocytes were stained with directly conjugated mAbs against CD3, CD4, CD8, CD11c, CD14, CD20, CD83, CD86, CD123, and HLR-DR (all purchased from BD Biosciences) for phenotypic analysis. Data acquisition was done using a FACSCalibur or FACSAria (BD Biosciences). Data analysis and illustration were done by using FlowJo software (Tree Star).
Cell treatment and activation
For HCMV infectivity tests, cells were pre-treated with recombinant IFN-α (PBL InterferonSource) for 4–6 h before infection. For induction of type I IFN expression, PBMCs (plated at a concentration of 1 × 107 cells/ml) or isolated PDCs (plated at a concentration of 5 × 105 cells/ml) were activated with 5 or 50 µg/ml A-class CpG oligodeoxynucleotides (ODN 2336, Coley Pharmaceutical Group), 5 µg/ml imiquimod (R837, InvivoGen), or heat-inactivated influenza virus A/Mem/71 equivalent to a MOI of 0.01. For IL-10 treatment, recombinant hIL-10 or cmvIL-10 (both from R&D Systems) was added to cultures concomitantly with the appropriate stimuli.
Quantitative real-time PCR analysis
Total RNA from PDCs was isolated using the RNeasy Mini kit (Qiagen) after treatment for 18 h. RNA processing and cDNA synthesis were performed as described previously (Chang et al., 2004). Steady state mRNA levels of IFN-α subtypes, IFN-β, and the internal control gene were measured by quantitative real-time PCR as described previously (Szubin et al., 2008). Primer and probe sequences to distinguish 12 of the 13 IFN-α subtypes (IFN-α1 and 13 differ by only one nucleotide and are amplified together) have been published previously (Szubin et al., 2008). For detection of IFN-β mRNA, the outer primers were : 5’-ACTTACAGGTTACCTCCGAAACTGA-3’, 5’-TAGCCATCAGTCACTTAAACAGCAT-3’, the inner primers were: 5’-ACTTACAGGTTACCTCCGAAACTGAA-3’, 5’-GGTTGAAGAATGCTTGAAGCAA-3’, and the probe was: 5’-TGTCCAGTCCCAGAGGCACAGGCT-3’. Raw data were normalized to an internal control gene, ubiquitin conjugating enzyme, and the relative copy number of IFN-α mRNA was calculated according to the standard curve generated from serial dilutions of plasmid DNA containing the amplicon of each tested IFN-α subtype.
ELISA
The quantities of total IFN-α or IFN-β in supernatants collected from PBMC or PDC cultures were measured by commercial ELISA kits (PBL InterferonSource) following the manufacturer’s instructions. The VeriKine Human IFN-α Multi-Subtype ELISA Kit is specified to measure the following IFN subtypes: 1, 2, 4, 5, 6, 7, 8, 10, 14, 16, and 17.
Statistical analysis
Statistics were done by one-way ANOVA followed by Tukey’s multiple comparison tests or by Dunnett’s multiple comparison tests using Prism software (GraphPad).
Acknowledgments
We are grateful to T. Shenk at Princeton University for kindly providing the HCMV variants. We also thank A. Spinner and K.-W. Yang for technical assistance. This study is supported in part by grants from the National Institutes of Health PO1DE07946 and U01AI057264 to N.B. and AI49342 and RR000169 to P.A.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 2004;78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichele P, Unsoeld H, Koschella M, Schweier O, Kalinke U, Vucikuja S. CD8 T cells specific for lymphocytic choriomeningitis virus require type I IFN receptor for clonal expansion. J. Immunol. 2006;176:4525–4529. doi: 10.4049/jimmunol.176.8.4525. [DOI] [PubMed] [Google Scholar]
- Beck K, Meyer-Konig U, Weidmann M, Nern C, Hufert FT. Human cytomegalovirus impairs dendritic cell function: a novel mechanism of human cytomegalovirus immune escape. Eur. J. Immunol. 2003;33:1528–1538. doi: 10.1002/eji.200323612. [DOI] [PubMed] [Google Scholar]
- Cederarv M, Soderberg-Naucler C, Odeberg J. HCMV infection of PDCs deviates the NK cell response into cytokine-producing cells unable to perform cytotoxicity. Immunobiology. 2009 doi: 10.1016/j.imbio.2008.10.009. In press. [DOI] [PubMed] [Google Scholar]
- Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Baumgarth N, Eberhardt MK, Lee CY, Baron CA, Gregg JP, Barry PA. Exposure of myeloid dendritic cells to exogenous or endogenous IL-10 during maturation determines their longevity. J. Immunol. 2007a;178:7794–7804. doi: 10.4049/jimmunol.178.12.7794. [DOI] [PubMed] [Google Scholar]
- Chang WL, Baumgarth N, Yu D, Barry PA. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 2004;78:8720–8731. doi: 10.1128/JVI.78.16.8720-8731.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Coro ES, Rau FC, Xiao Y, Erle DJ, Baumgarth N. Influenza virus infection causes global respiratory tract B cell response modulation via innate immune signals. J. Immunol. 2007b;178:1457–1467. doi: 10.4049/jimmunol.178.3.1457. [DOI] [PubMed] [Google Scholar]
- Chang WL, Tarantal AF, Zhou SS, Borowsky AD, Barry PA. A recombinant rhesus cytomegalovirus expressing enhanced green fluorescent protein retains the wild-type phenotype and pathogenicity in fetal macaques. J. Virol. 2002;76:9493–9504. doi: 10.1128/JVI.76.18.9493-9504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- Coro ES, Chang WL, Baumgarth N. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J. Immunol. 2006;176:4343–4351. doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- Dalod M, Hamilton T, Salomon R, Salazar-Mather TP, Henry SC, Hamilton JD, Biron CA. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 2003;197:885–898. doi: 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalod M, Salazar-Mather TP, Malmgaard L, Lewis C, Asselin-Paturel C, Briere F, Trinchieri G, Biron CA. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 2002;195:517–528. doi: 10.1084/jem.20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delale T, Paquin A, Asselin-Paturel C, Dalod M, Brizard G, Bates EE, Kastner P, Chan S, Akira S, Vicari A, Biron CA, Trinchieri G, Briere F. MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J. Immunol. 2005;175:6723–6732. doi: 10.4049/jimmunol.175.10.6723. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis eSousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Doucett VP, Gerhard W, Owler K, Curry D, Brown L, Baumgarth N. Enumeration and characterization of virus-specific B cells by multicolor flow cytometry. J. Immunol. Methods. 2005;303:40–52. doi: 10.1016/j.jim.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Duramad O, Fearon KL, Chan JH, Kanzler H, Marshall JD, Coffman RL, Barrat FJ. IL-10 regulates plasmacytoid dendritic cell response to CpG-containing immunostimulatory sequences. Blood. 2003;102:4487–4492. doi: 10.1182/blood-2003-07-2465. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Hahn G, Jores R, Mocarski ES. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 2004;78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre A, Barnes BJ, Amrute S, Yeow WS, Megjugorac N, Dai J, Feng D, Chung E, Pitha PM, Fitzgerald-Bocarsly P. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 2003;74:1125–1138. doi: 10.1189/jlb.0603255. [DOI] [PubMed] [Google Scholar]
- Kadowaki N, Antonenko S, Lau JY, Liu YJ. Natural interferon alpha/beta-producing cells link innate and adaptive immunity. J. Exp. Med. 2000;192:219–226. doi: 10.1084/jem.192.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004a;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004b;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- Kvale EO, Dalgaard J, Lund-Johansen F, Rollag H, Farkas L, Midtvedt K, Jahnsen FL, Brinchmann JE, Olweus J. CD11c+ dendritic cells and plasmacytoid DCs are activated by human cytomegalovirus and retain efficient T cell-stimulatory capability upon infection. Blood. 2006;107:2022–2029. doi: 10.1182/blood-2005-05-2016. [DOI] [PubMed] [Google Scholar]
- Lapenta C, Santini SM, Spada M, Donati S, Urbani F, Accapezzato D, Franceschini D, Andreotti M, Barnaba V, Belardelli F. IFN-alpha-conditioned dendritic cells are highly efficient in inducing cross-priming CD8(+) T cells against exogenous viral antigens. Eur. J. Immunol. 2006;36:2046–2060. doi: 10.1002/eji.200535579. [DOI] [PubMed] [Google Scholar]
- Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, Tough DF. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 2003;4:1009–1015. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- Le Bon A, Thompson C, Kamphuis E, Durand V, Rossmann C, Kalinke U, Tough DF. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol. 2006;176:2074–2078. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- Levy DE, Marie I, Prakash A. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 2003;15:52–58. doi: 10.1016/s0952-7915(02)00011-0. [DOI] [PubMed] [Google Scholar]
- Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- Liu YJ. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell. 2001;106:259–262. doi: 10.1016/s0092-8674(01)00456-1. [DOI] [PubMed] [Google Scholar]
- Longman RS, Braun D, Pellegrini S, Rice CM, Darnell RB, Albert ML. Dendritic-cell maturation alters intracellular signaling networks, enabling differential effects of IFN-alpha/beta on antigen cross-presentation. Blood. 2007;109:1113–1122. doi: 10.1182/blood-2006-05-023465. [DOI] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. Embo J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaftsi M, Mehl AM, Borysiewicz LK, Tabi Z. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood. 2002;99:2913–2921. doi: 10.1182/blood.v99.8.2913. [DOI] [PubMed] [Google Scholar]
- Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, Biron CA. Coordinated and distinct roles for IFN-alpha beta IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- Prakash A, Smith E, Lee CK, Levy DE. Tissue-specific positive feedback requirements for production of type I interferon following virus infection. J. Biol Chem. 2005;280:18651–18657. doi: 10.1074/jbc.M501289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler S, Hebart H, Einsele H, Brossart P, Jahn G, Sinzger C. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 2000;81:393–399. doi: 10.1099/0022-1317-81-2-393. [DOI] [PubMed] [Google Scholar]
- Rissoan MC, Soumelis V, Kadowaki N, Grouard G, Briere F, de Waal Malefyt R, Liu YJ. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Soderberg C, Larsson S, Bergstedt-Lindqvist S, Moller E. Definition of a subset of human peripheral blood mononuclear cells that are permissive to human cytomegalovirus infection. J. Virol. 1993;67:3166–3175. doi: 10.1128/jvi.67.6.3166-3175.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Szubin R, Chang WL, Greasby T, Beckett L, Baumgarth N. Rigid interferon-alpha subtype responses of human plasmacytoid dendritic cells. J. Interferon Cytokine Res. 2008;28:745–759. doi: 10.1089/jir.2008.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Varani S, Cederarv M, Feld S, Tammik C, Frascaroli G, Landini MP, Soderberg-Naucler C. Human cytomegalovirus differentially controls B cell and T cell responses through effects on plasmacytoid dendritic cells. J. Immunol. 2007;179:7767–7776. doi: 10.4049/jimmunol.179.11.7767. [DOI] [PubMed] [Google Scholar]
- Vilcek J, Sen GC. Interferons and other cytokines. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 3rd ed. Philadelphia: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 2005a;79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 2005b;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peters N, Schwarze J. Plasmacytoid dendritic cells limit viral replication pulmonary inflammation, and airway hyperresponsiveness in respiratory syncytial virus infection. J. Immunol. 2006;177:6263–6270. doi: 10.4049/jimmunol.177.9.6263. [DOI] [PubMed] [Google Scholar]
- Waskow C, Liu K, Darrasse-Jeze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Silva MC, Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravskaya T, Maciejewski JP, Netski DM, Bruening E, Mackintosh FR, St Jeor S. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood. 1997;90:2482–2491. [PubMed] [Google Scholar]
- Zuniga EI, McGavern DB, Pruneda-Paz JL, Teng C, Oldstone MB. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat. Immunol. 2004;5:1227–1234. doi: 10.1038/ni1136. [DOI] [PMC free article] [PubMed] [Google Scholar]