Abstract
The cellular uptake of a series of dipyridophenazine (dppz) complexes of Ru(II) was examined by flow cytometry. The complexes, owing to their facile synthesis, stability, and luminescence, provide a route to compare and contrast systematically factors governing cellular entry. Substituting the ancillary ligands in the dppz complexes of Ru(II) permits variation in the overall complex charge, size, and hydrophobicity. In HeLa cells, cellular uptake appears to be facilitated by the lipophilic 4,7-diphenyl-1,10-phenanthroline (DIP) ligand. Despite the large size of Ru(DIP)2dppz2+ (20 Å diameter), this complex is readily transported inside the cell compared to smaller and more hydrophilic complexes such as Ru(bpy)2dppz2+. Accumulation in the cellular interior is confirmed by confocal microscopy.
The cellular uptake characteristics of a small molecule are critical to its application as a therapeutic or diagnostic agent. However, our understanding of the chemical rules governing uptake is rudimentary.1,2 Although transition metal complexes have increasingly been applied for biological applications,3–5 their uptake properties are even less well developed. Here, we exploit flow cytometry to provide statistics on the uptake of ruthenium complexes into HeLa cells. These ruthenium complexes, owing to their facile synthesis, stability, and luminescence, provide a route to compare and contrast factors governing cellular uptake.
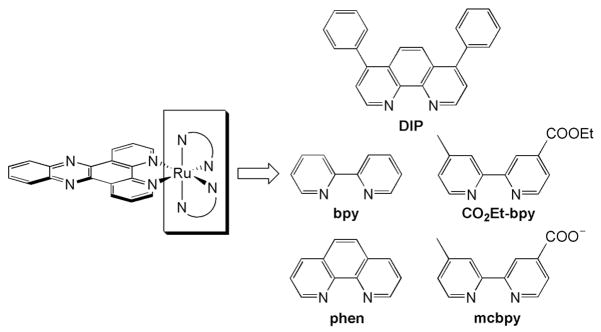
A series of dipyridophenazine (dppz) complexes of Ru(II) was synthesized for systematic comparison.6–8 Substituting the ancillary ligands on the dppz complex permits variation in the overall complex charge, size, and hydrophobicity (Figure 1). Furthermore since these dppz complexes all act as molecular light switches, showing minimal luminescence in aqueous solution and intense luminescence when bound to DNA or otherwise protected from water, they provide a sensitive cellular probe (Table 1).9–11
Figure 1.
Dipyridophenazine complexes of Ru(II).
Table 1.
Characteristics of Ru complexes
| Ancillary Ligand of RuL2dppz | Relative emission intensity in CH3CNa | Relative emission intensity w/DNAa,b | Octanol/H2O partition coefficient (log P)c | Diameter (Å)d |
|---|---|---|---|---|
| bpy | 1.0 | 1.0 | −2.50 | 16.2 |
| CO2Et-bpy | 1.8 | 1.1 | −0.76 | 20.4 |
| mcbpy | 1.2 | 0.6 | −0.43 | 18.2 |
| phen | 1.2 | 2.3 | −1.48 | 16.2 |
| DIP | 2.7 | 2.7 | 1.30 | 20.4 |
Excited at 488 nm; integrated emission at 600–620 nm. 10 μM Ru was used, except for Ru(DIP)2dppz2+ in Tris buffer, where a lower concentration was used due to poor solubility; emission values were scaled accordingly.
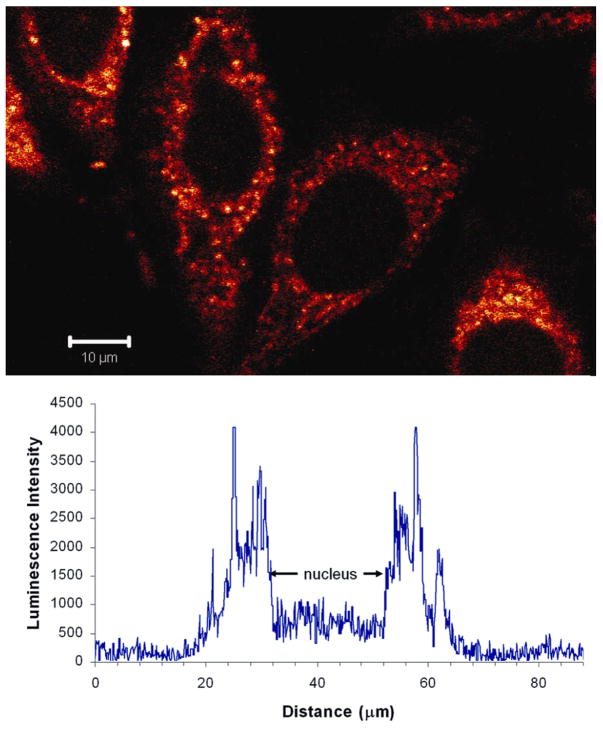
Luminescence values with DNA were obtained at saturation.
Cl− salt.
Diameters were estimated using Titan.
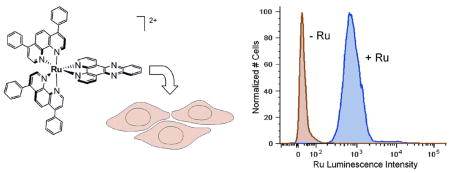
HeLa cells were prepared for flow cytometry analysis after incubation with the Ru complexes at various concentrations and times.12 Flow cytometry was performed on a BD FACS Aria using ~20,000 cells per sample. The ruthenium complexes were excited at 488 nm, with the emission observed at 600–620 nm. Live cells were distinguished by their low To-Pro-3 emission.
Figure 2 illustrates results of the flow cytometry. Cells not treated with complex exhibit some background luminescence. Incubation with 10 μM Ru(bpy)2dppz2+ or Ru(phen)2dppz2+ for 2 h causes only a small change in the luminescence profile. When cells are incubated with 10 μM Ru(DIP)2dppz2+, however, the luminescence intensity of the cell population increases dramatically.
Figure 2.
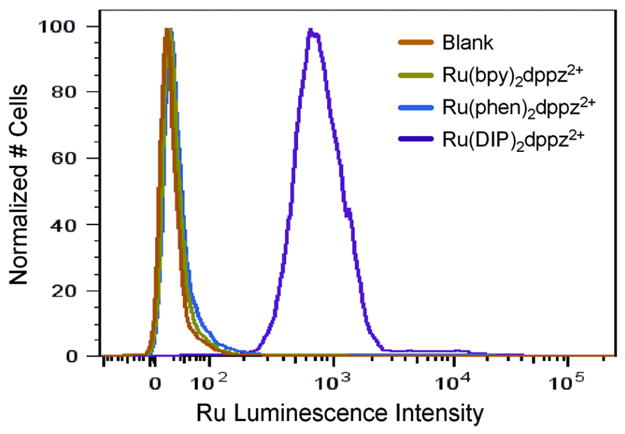
Flow cytometry analysis of HeLa cells incubated with 10 μM ruthenium complex for 2 h. Luminescence data were obtained by excitation at 488 nm with emission at 600–620 nm using a light scatter gate to exclude debris and To-Pro-3 (exciting at 633 nm and observing at 650–670 nm) to exclude dead cells.
Uptake for the different Ru complexes may be compared based upon the mean luminescence intensity of the cell population (Table 2). Below 1 μM, Ru(DIP)2dppz2+ is taken up appreciably above background. At higher concentrations, Ru(bpy)2dppz2+, Ru(CO2Et-bpy)2dppz2+, and Ru(phen)2dppz2+ are taken up to some extent, but even at 20 μM Ru, little luminescence is evident for Ru(mcbpy)2dppz.13 Washing with buffer reduces luminescence by 20–50%, suggesting that, while some Ru is non-specifically adhered to the surface or rapidly exported, the bulk of Ru remains.
Table 2.
Mean Luminescence Intensity of HeLa Cells Incubated with Ruthenium Complex by Flow Cytometrya
| Ancillary Ligands of RuL2dppz |
|||||
|---|---|---|---|---|---|
| Conc. (μM) | bpy | CO2Et-bpy | mcbpy | phen | DIP |
| 0.5 | n.d. | n.d. | n.d. | n.d. | 60 |
| 1 | n.d. | n.d. | n.d. | n.d. | 99 |
| 5 | 38 | 45 | 20 | 52 | 597 |
| 10b | 38 | 45 | 21 | 58 | 974 (571) |
| 20b | 48 (27) | 51 (29) | 26 (19) | 111 (50) | n.d. |
Cells were incubated with ruthenium complex for 2 h at ambient temperature. Ruthenium complexes were excited at 488 nm, with emission observed at 600–620 nm. The mean luminescence intensity of cells not treated with complex is 23. Data are an average of two independent experiments, and data not determined are indicated by n.d.
Samples washed after incubation are shown in parenthesis.
That the complexes are actually transported into the cellular interior rather than associating solely at the membrane surface is evident by confocal microscopy (Figure 3).14 For Ru(DIP)2dppz2+, intense luminescence in the interior is apparent within 2 h. For Ru(phen)2dppz2+ and Ru(bpy)2dppz2+, microscopy experiments also show uptake into the cellular interior but, consistent with the flow cytometry data, on a slower time scale (≥ 4 h for phen). For all complexes, greatest luminescence is evident in the cytoplasm, likely associated with the mitochondria and endoplasmic reticulum based upon costaining experiments with organelle-specific dyes;15 without protection from water through macromolecular binding, Ru quenching in the cytosol is expected.
Figure 3.
Confocal microscopy of HeLa cells with Ru(DIP)2dppz2+. (top) Microscopy after incubation with 5 μM Ru for 2 h at 37 °C. Excitation wavelength = 488 nm. (bottom) Intensity profile of ruthenium luminescence across a HeLa cell after incubation for 12 h with 10 μM Ru.
Interestingly, significantly less luminescence is apparent in the nucleus. Quantitation by line plots (Figure 3) does show nuclear uptake but of diminished intensity in the nucleus compared to other regions.16 Flow cytometry experiments on nuclear preparations isolated after incubation of cells with Ru(DIP)2dppz2+ provide consistent evidence of Ru uptake (Supporting Information).
These data establish that the ruthenium complexes are indeed taken up inside HeLa cells. If the complexes are entering the cell by passive diffusion, one would predict that neutral charge, smaller size, and greater hydrophobicity should aid uptake. Here, however, cellular uptake appears to be facilitated by the lipophilic DIP ligand, despite the larger size of the complex. It is surprising that the large expanse of the DIP complex does not limit its uptake. Also of interest, changing the overall charge from +2 to neutral does not improve uptake, based upon the low luminescence results for Ru(mcbpy)2dppz.17
There have been few systematic studies on the cellular uptake of transition metal complexes reported.18–20 Our results are in agreement with studies on cisplatin analogues, where the complexes with the greatest lipophilicity exhibit the highest uptake; note that for the Pt complexes, all were hydrophilic, with octanol/water partition coefficients of <1.20 Importantly, these data also establish that the Ru complexes are stable to the intracellular environment; no degradation in luminescence is evident, as would be expected based upon changes in complex coordination.
Flow cytometry, traditionally used to examine organic fluorophores, thus provides an opportunity to examine the cellular uptake of transition metal complexes. Ruthenium analogues in particular can be readily tested without special instrumentation or complicated synthesis. Statistics on thousands of cells of varied cell type and using a range of metal complexes can be generated to provide a powerful complement in the design of metal complexes for biological application.
Supplementary Material
Acknowledgments
We are grateful to the NIH (GM33309) for their financial support. We also thank the Caltech Flow Cytometry Facility and the Caltech Biological Imaging Center.
Footnotes
Supporting Information Available. Flow cytometry for cell nuclei. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Footnotes
- 1.DeVito SC. In: Handbook of Property Estimation Methods for Chemicals: Environmental and Health Sciences. Boethling RS, Mackay D, editors. CRC Press; Boca Raton, FL: 2000. pp. 261–278. [Google Scholar]
- 2.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CX, Lippard SJ. Curr Opin Chem Biol. 2003;7:481–489. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 4.Boerner LJK, Zaleski JM. Curr Opin Chem Biol. 2005;9:135. doi: 10.1016/j.cbpa.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 5.(a) Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]; (b) Hart JR, Glebov O, Ernst RJ, Kirsch IL, Barton JK. Proc Nat Acad Sci. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickeson JE, Summers LA. Aust J Chem. 1970;23:1023–1027. [Google Scholar]
- 7.Ru(phen)2Cl2, and Ru(DIP)2Cl2 were synthesized in analogous fashion to Ru(bpy)2Cl2 described in Sullivan BP, Salmon DJ, Meyer TJ. Inorg Chem. 1978;17:3334–3341.. The dppz ligand was then added by refluxing in ethanol-water for > 3 h.
- 8.Ru(CO2Et-bpy)2Cl2 was synthesized using a modification of Leasure RM, Ou W, Moss JA, Linton RW, Meyer TJ. Chem Mater. 1996;8:264–273.. 2:1 DME-ethanol was used as the reaction solvent. The dppz ligand was added by refluxing in EtOH for 24 h. Ru(mcbpy)2dppz was formed by hydrolysis of the ester.
- 9.(a) Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960. [Google Scholar]; (b) Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochem. 1992;31:10809–16. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]
- 10.Olofsson J, Onfelt B, Lincoln P. J Phys Chem A. 2004;108:4391. [Google Scholar]
- 11.Ardhammer M, Lincoln P, Norden B. J Phys Chem B. 2001;105:11363–11368. [Google Scholar]
- 12.HeLa cells were detached from monolayer culture with EDTA, resuspended in Hank’s Balanced Salt Solution supplemented with bovine serum albumin fraction 5, and diluted to 1×106 cells/mL. The ruthenium complexes were added to the cell suspensions at concentrations of 0.5–10 μM and incubated for 2 h at ambient temperature. Dead cells were stained with 1 μM To-Pro-3 (Molecular Probes), which enters cells having compromised membranes.
- 13.The low luminescence from Ru(mcbpy)2dppz may be in part due to poor nucleic acid binding by the neutral complex.
- 14.Confocal microscopy of cells incubated with a Ru dimer has been reported. See Önfelt B, Gostring L, Lincoln P, Nordén B, Önfelt A. Mutagenesis. 2002;17:317–320. doi: 10.1093/mutage/17.4.317.
- 15.Haugland RP. In: Handbook of Fluorescent Probes and Research Products. 9. Gregory J, Spence M, editors. Molecular Probes; Eugene, OR: 2002. pp. 473–488.pp. 496–502. [Google Scholar]
- 16.After incubation of HeLa cells for 2 h with 5 μM Ru(DIP)2dppz2+, line plot quantitation shows an average of 11% luminescence in the nucleus compared to cytoplasm; for 10 μM Ru after 12 h, as in Figure 3b, the ratio is ~ 30%. Some contribution to fluorescence in the nucleus could arise from cellular autofluorescence.
- 17.The lower luminescence of Ru(mcbpy)2dppz with nucleic acids contributes to its relatively poor luminescence in cells but cannot fully account for it.
- 18.Jonas SK, Riley PA. Cell Biochem Function. 1991;9:245–253. doi: 10.1002/cbf.290090406. [DOI] [PubMed] [Google Scholar]
- 19.Kalayda GV, Fakih S, Bertram H, Ludwig T, Oberleithner H, Krebs B, Reedijk J. J Inorg Biochem. 2006;100:1332–1338. doi: 10.1016/j.jinorgbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Ghezzi A, Aceto M, Cassino C, Gabano E, Osella D. J Inorg Biochem. 2004;98:73–78. doi: 10.1016/j.jinorgbio.2003.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.