Abstract
We investigated the genomic transcriptional response of female fathead minnows (Pimephales promelas) to an acute (4 day) exposure to 0.1 or 1.0 μg/L of 17β-trenbolone (TB), the active metabolite of an anabolic androgenic steroid used as a growth promoter in cattle and a contaminant of concern in aquatic systems. Our objectives were to investigate the gene expression profile induced by TB, define biomarkers of exposure to TB, and increase our understanding of the mechanisms of adverse effects of TB on fish reproduction. In female gonad tissue, microarray analysis using a 22K oligonucleotide microarray (EcoArray Inc., Gainesville, FL) showed 99 significantly upregulated genes and 741 significantly downregulated genes in response to 1 μg TB/L. In particular, hydroxysteroid (17β) dehydrogenase 12a (hsd17b12a), zona pellucida glycoprotein 2.2 (zp2.2), and protein inhibitor of activated STAT, 2 (pias2) were all downregulated in gonad. Q-PCR measurements in a larger sample set were consistent with the microarray results in the direction and magnitude of these changes in gene expression. However, several novel potential biomarkers were verified by Q-PCR in the same samples, but could not be validated in independent samples. In liver, Q-PCR measurements showed a significant decrease in vitellogenin 1 (vtg1) mRNA expression. In brain, cytochrome P450, family 19, subfamily A, polypeptide 1b (cyp19a1b, previously known as aromatase B) transcript levels were significantly reduced following TB exposure. Our study provides a candidate gene involved in mediating the action of TB, hsd17b12a, and two potential biomarkers sensitive to acute TB exposure, hepatic vtg1 and brain cyp19a1b.
Keywords: 17β-Trenbolone, Fathead minnow, Q-PCR, Microarray
Introduction
Environmental contaminants that interact with the endocrine system have the potential to cause adverse effects on reproduction and development (World Health Organization (WHO), 2002). Among these endocrine-disrupting compounds are those that mimic natural hormones. In particular, environmental contaminants that activate the androgen receptor may be causing adverse effects in wildlife (Gray et al., 2002). For example, investigations of pulp and paper mill effluents from different locations in North America as well as examinations of beef feedlot discharges have associated morphological alterations in fish collected from the field with androgenic activity (Parks et al., 2001; Orlando et al., 2004; Soto et al., 2004; Durhan et al., 2006).
In the United States, approximately 90 % of beef cattle are treated with a growth-promoting mix of estrogens, androgens, and progesterones, including the synthetic androgen trenbolone acetate (17β-hydroxy-estra-4,9,11-trien-3-one-17-acetate) (Balter, 1999). Administration of trenbolone acetate has multiple effects on the endocrine system, including androgenic, progestin, and anti-glucocorticoid activities measured in mammalian systems (Danhaive and Rousseau, 1986; Bauer et al., 2000). The active metabolite of trenbolone acetate is 17β-trenbolone (17β-hydroxy-estra-4,9,11-trien-3-one) (Schiffer et al., 2001), a potent androgen receptor agonist in vertebrates (Wilson et al., 2002; Ankley et al., 2003). 17β-trenbolone is metabolized by oxidation to trendione, followed by reduction to 17α-trenbolone (Schiffer et al., 2001), which has about 20-fold less affinity than 17β-trenbolone for the androgen receptor (Bauer et al., 2000). However, both forms are potent reproductive toxicants in fish, due to reconversion of 17α-trenbolone to 17β-trenbolone by fish following uptake of 17α-trenbolone (Ankley et al., 2003; Jensen et al., 2006).
Runoff from feedlots and fields fertilized with manure has the potential to contaminate aquatic environments with trenbolone. Both the alpha and beta isomers are eliminated from cattle dosed with trenbolone acetate, with the 17α-trenbolone comprising about 95 % of the excreted product (Schiffer et al., 2001). Schiffer et al. (2001) also demonstrated that both metabolites are quite stable in the environment with half-lives of about 270 days in liquid manure. Durhan et al. (2006) documented the occurrence of both 17β-trenbolone and 17α-trenbolone in water with in vitro androgenic activity that had been collected in discharge from a beef feedlot where trenbolone acetate implants had been used. Measured concentrations of 17α- and 17β-trenbolone in samples from the site varied temporally but both were present, at least occasionally, at concentrations greater than 0.01 μg/L (Durhan et al., 2006) concentrations of 17α-trenbolone ranged up to 0.125 μg/L in discharge and 0.05 μg/L in receiving water; while concentrations of 17β-trenbolone ranged up to 0.02 μg/L in discharge and 0.007 μg/L in receiving water (Durhan et al., 2006).
Recent studies have assessed the effects of 17β-trenbolone on fecundity and reproductive endocrinology of various fish species. Peterson et al. (2001) exposed mature Japanese medaka (Oryzias latipes) to 17β-trenbolone for 14 days at nominal water concentrations ranging from 0.002 to 2 μg/L. They found that concentrations ≥ 0.2 μg/L inhibited fecundity of the fish. They also reported an inhibition of vitellogenin production in female medaka exposed to 2 μg/L of 17β-trenbolone. Ankley et al. (2003) conducted a 21-day reproduction test with fathead minnows (Pimephales promelas). They found that a concentration of about 0.027 μg/L of the androgen caused female fathead minnows to develop male secondary sexual characteristics (dorsal nuptial tubercles) and significantly reduced fecundity, plasma vitellogenin, and sex steroid concentrations. An in vitro competitive binding study with fathead minnow androgen receptor (Ar) also demonstrated that 17β-trenbolone had a higher affinity for the receptor than that of the endogenous ligand, testosterone (Ankley et al., 2003). In another study, a 28-day exposure to 17β-trenbolone (1-10 μg/L) induced development of a gonopodium from the female anal fin accompanied by a transient up-regulation of arα and arβ mRNA in adult female mosquitofish (Gambusia affinis affinis) (Sone et al., 2005). Further studies have also demonstrated the potential effects of 17β-trenbolone on various fish species (Ankley et al., 2004; Miller and Ankley, 2004; Holbech et al., 2006; Miracle et al., 2006; Örn et al., 2006; Seki et al., 2006; Hemmer et al., 2008).
Gene expression changes associated with signal pathway activation can provide chemical-specific information on the mechanistic and biological effects of a chemical (Nuwaysir et al., 1999; Bartosiewicz et al., 2001; Hamadeh et al., 2001). We chose to investigate gene expression changes associated with 17β-trenbolone exposure in the fathead minnow (Pimephales promelas), a member of the ecologically important Cyprinidae family, and a standard test species for aquatic toxicology since the 1960s (reviewed by Ankley and Villeneuve, 2006). We chose an acute exposure duration of 96 hours to allow for the assessment of early responses in gene expression following 17β-trenbolone exposure and for consistency with studies of acute effects of other contaminants. Gene expression was analyzed by quantitative PCR (Q-PCR) and by microarray analysis.
The aim of this study was to investigate the gene expression profile induced by 17β-trenbolone and subsequently to understand the molecular mechanism(s) by which 17β-trenbolone causes adverse effects in fish and develop biomarkers of 17β-trenbolone exposure. Since 17β-trenbolone is an androgen and effects on egg production have been previously observed, our study focused on adult female fathead minnows. Exposure levels of 0.1 μg/L and 1 μg/L were chosen based on concentrations of 17β-trenbolone that produced effects in the study by Ankley et al. (2003).
Materials and methods
Animal culture conditions and exposures
Adult fathead minnows (Pimephales promelas) were cultured at the Columbia Environmental Research Center (CERC), USGS and both sexes were housed separately before the experiment. Fish were held in the test system for one day prior to initiation of the chemical exposure at 20.3 ± 0.5 °C under an 14:10 light:dark photo-period. Fish were exposed to 17β-trenbolone for 4 days using a flow-through exposure system. Nominal water concentrations of 0.1 and 1 μg/L 17β-trenbolone were used. For each exposure, a stock solution was prepared in 100 % ethanol and slowly added to the exposure tanks in a proportional flow-through diluter system (6.5 L tank capacity; filled with 4 L of well water) at a flow rate of 0.5 ml/12.5 min. Control tanks had only ethanol added (solvent control). The final concentration of ethanol was 0.03 %. There were four replicate tanks for each treatment, with six fish per replicate at a ratio of three males to three females, for a total of 12 females per treatment. After 4 days of chemical exposure, fish were removed from the test tanks and anesthetized with MS-222 (tricaine methanesulfonate). The weight of each fish was recorded and liver, brain (without pituitary) and gonad tissues were collected, placed in RNALater (Ambion, Austin, TX), and stored at -20 °C until used for RNA extraction. The female fathead minnows were adults with a mean weight of 1.1 ± 0.25 g. Animals were not fed during exposure. No mortality was observed during the experiment. For Q-PCR analysis, brain RNA was sampled from 12 females per treatment, liver RNA from 6 females per treatment, and gonad RNA from 10 females per treatment. For microarray analysis, gonad RNA was sampled from 4 females each from the control and 1 μg/L treatment groups only.
Chemical analysis
Water concentrations of 17β-trenbolone were determined in samples from each exposure tank at 0-hours and 96-hours using high performance liquid chromatography with photodiode array detection (HPLC-PDA). Water samples were extracted by liquid:liquid partitioning with dichloromethane (DCM). Water concentrations of 17β-trenbolone were stable over the course of the experiment; the concentration means and standard deviations in the 0.1 and in the 1 μg/L treatments were 0.114 ± 0.025 μg/L and 1.47 ± 0.05 μg/L, respectively. The limit of detection was 0.014 μg/L; no analyte was detected in control water.
Microarray analysis of gene expression
Microarray analysis was performed with a commercially available microarray containing 22,000 oligonucleotide probes developed for fathead minnow (EcoArray Inc., Gainesville, FL). Female gonad samples from control and 1 μg/L treatment groups were used for microarray hybridization according to the One-Color Microarray-Based Gene Expression Analysis procedure from Agilent Technologies, Inc. (Santa Clara, CA). Scanned images were processed using Agilent Feature Extraction 9.5.1 software for spot identification and for quantification of the fluorescent signal intensities. The fluorescent signal intensity for each oligo spot was calculated using local background subtraction. Quality control algorithms in the software detected poor quality spots, which were excluded from subsequent analyses. The microarray dataset and MIAME-compliant information have been submitted to the NCBI GEO database, accession number GSE12904.
A statistical analysis was performed in order to identify differentially expressed genes between the control and 1 μg/L treatment groups using SAS 9.1 (SAS Institute, Cary, NC). Data were analyzed using two-stage mixed model ANOVA following the procedure described by Wolfinger et al. (2001). This approach centers around two interconnected ANOVA models, the “normalization” model and the “gene” model. The first stage is to normalize the data by outputting residuals from an ANOVA containing the effects of array and treatment. The treatment effect is fixed and the array effect is random. A second ANOVA uses the residuals from the first ANOVA to test differences among treatments with a separate ANOVA for each gene. The treatment difference for each gene was determined using a one degree of freedom F-test (P < 0.05). Because one aim of this project was to discover novel genes and pathways involved in the response to 17β-trenbolone, we did not use a multiple testing correction. Thus, some of the genes identified as altered are expected to be false positives.
In order to detect changes in groups of genes with related functions, we tested overrepresentation of gene ontology (GO) terms (Ashburner et al., 2000) with the FatiGO (fast assignment and transference of information) web tool (Al-Shahrour et al., 2005; Al-Shahrour et al., 2006). FatiGO uses Fisher’s exact test to compare the GO terms associated with significantly altered probes and the GO terms associated with unaltered probes. It returns a list of GO terms that are significantly over-represented among altered probes, relative to their abundance in the list of unaltered probes. For this analysis, multiple probes assigned to the same gene name are analyzed separately. Since most of the altered genes were downregulated, we tested for significantly enriched GO terms among upregulated and downregulated genes separately. The significance criteria for GO terms were P<0.05 and number of altered genes ≥ 2.
Quantitative PCR (Q-PCR) analysis of gene expression
The expression of selected genes was evaluated by real-time quantitative PCR (Q-PCR), a technique which allows a rapid and reliable quantification of specific mRNAs (Nolan et al., 2006). Total RNA was extracted from female fathead tissues using the AutoGenprep 245, following manufacturer’s instructions (AutoGen, Holliston, MA). Total RNA concentration and A260 nm/A280 nm ratios were measured on a SpectraMax 190 plate-reading spectrophotometer (Molecular Devices, Sunnyvale, CA), and RNA concentrations were adjusted to 0.4 μg/7 μl in order to produce a constant RNA concentration in all reverse transcription reactions. All RNA samples had A260 nm/A280 nm ratios greater than 1.8. Total RNA quality was verified in two randomly selected samples from each tissue by 1 % agarose gel electrophoresis under denaturing conditions with ethidium bromide detection, to confirm the presence of two sharp rRNA bands indicating that the RNA samples were not degraded (Sambrook and Russell, 2001). Procedural blanks run in parallel with tissue samples confirmed that there was no detectable cross-contamination between samples. The quality of mRNA was not independently assessed (Nolan et al., 2006).
Reverse transcription (RT) reactions for Q-PCR assays were carried out in a volume of 20 μl with 0.04 μg of total RNA, primed with random hexamers using the TaqMan® Reverse Transcription Reagents kit following the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA), with the addition of denaturation of the RNA at 75 °C for 10 min prior to the RT reaction. Real-time quantitative PCR was performed with an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using SYBR-Green PCR Master Mix (Applied Biosystems). One-tenth of the cDNA was used for each Q-PCR reaction in a total volume of 25 μl. Duplicates were run for each sample. The expression levels of target genes were normalized using the expression of a housekeeping gene, ribosomal protein L8 (rpl8) (Filby and Tyler, 2005). The mRNA abundance of rpl8 relative to total RNA did not exhibit any significant differences between experimental groups (data not shown). Normalization to rpl8 reduced variation among samples and increased the correlation coefficient for Q-PCR data compared to microarray data in the same samples (data not shown).
Target genes were selected based on array results and by screening the literature. Fathead minnow target gene sequences were found in GenBank (NCBI) by search of annotations or by BLAST (Basic Local Alignment Search Tool) (Altschul et al., 1990) search against an annotated homolog in a related species. Primer sets for Q-PCR were designed using Primer Express 2.0 software according to manufacturer’s guidelines and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) (Table 1). Q-PCR assays were optimized and validated to confirm the specific amplification of the target sequence (Nolan et al., 2006). Primer concentrations were optimized by assaying the same sample with primer concentrations from 100 nM to 800 nM, with four replicates per primer concentration. The optimum primer concentration gives the lowest threshold cycle (CT), lowest variation among replicates, and highest change in fluorescence (ΔRn). No-template controls confirmed the absence of interfering non-specific amplicons such as primer-dimers. Primer sets that resulted in non-specific amplification were redesigned. The absence of non-specific amplification was also confirmed by agarose gel electrophoresis of an amplification from each primer set to confirm the presence of a single amplicon of the expected size, and by examination of dissociation curve profiles for each sample to confirm a single peak at the expected melting temperature.
Table 1.
Primer sequences for quantitative PCR. When possible, primer sets were designed over exon boundaries to prevent amplification of genomic DNA. Under Primer sequences, the primers used for quantitation are labeled F=forward and R=reverse. Position refers to numbering within coding sequence (CDS) or expressed sequence tag (EST). Exon boundaries are predicted based on homology with zebrafish (Danio rerio).
| Gene name (Symbol, accession number) |
Primer sequences | Position | Exon boundary |
|---|---|---|---|
|
vitellogenin 1 (vtg1, AF130354) |
F 5′- GCTGCAGAGGCCATTTCTAAGA -3′ R 5′- AGCATTGCCCAGAACTTTCAG -3′ |
1393- 1461 |
none |
|
cytochrome P450, family 19, subfamily A, polypeptide 1b (cyp19a1b, AJ277866) |
F 5′- GGACGTTTCCAATAGACTCTTCCTAA -3′ R 5′- ATAGCGATGGATCTTTATCAGCAA -3′ |
585-657 | 625 |
|
cytochrome P450, family 19, subfamily A, polypeptide 1a (cyp19a1a, AF288755) |
F 5′- GGAGAGCTGAGCGCTGAGA -3′ R 5′- GGAGCCGCGATCAACATCT -3′ |
181-239 | none |
|
estrogen receptor 1 (esr1, AY727528) |
F 5′- AACTCATCTTTGCTCAGGATCTCA -3′ R 5′- AGCCATCCCCTCGACACAT -3′ |
1109- 1173 |
1142 |
|
androgen receptor (ar, AY727529) |
F 5′- GTGCCATGCGCTTCCAA -3′ R 5′- CTGACCTTTGTGGGCAAGGA -3′ |
2568- 2718 |
2647 |
|
protein inhibitor of activated STAT, 2 (pias2, DT238876.1) |
F 5′- CAGACCCTGACAGTGAAATTGC -3′ R 5′- GCTCGACAGGGTACCGTAAGG -3′ |
94-184 | 149 |
|
hydroxysteroid (17β)dehydrogenase 12a (hsd17b12a, DT161033.1) |
F 5′- AATTCTTCCTCCACATCCCTGAT dehydrogenase -3′ R 5′- TCATTTGGCACACTGAGGTGAT -3′ |
260-340 | 291 |
|
zona pellucida glycoprotein 2.2 (zp2.2, DT088130.1) |
F 5′- ACGAGGAGGTCATTCTGTTTTCA -3′ R 5′- TGGGATCCACACCTTTCTTTG -3′ |
636-746 | 682 |
|
hypothetical protein LOC565514 (loc565514, DT346207 (5′ EST)) |
F 5′- GCTGTGTTTGTGGAGCCGTTA -3′ R 5′- GTCTGGGACTCTTGGTCATCATC -3′ |
207-287 | none |
|
hypothetical protein LOC565514 (loc565514, DT306961 (3′ EST)) |
F 5′- ACACAATCTCATTCAGTGAAATCCTT -3′ R 5′- TGAGTGCTTGGGTATGAGAAAACT -3′ |
328-408 | none |
|
hypothetical protein LOC565514 (loc565514, DT306961 (3′ EST)) |
F 5′- GCAGAGCATCTGCATACATCTTAATT -3′ R 5′- TGTGAGCCCCTTGAGAGAGAGTA -3′ |
34-156 | none |
|
clusterin (clu, UniGene Ppr.6588) |
F 5′- ACGAGGATGTTGTTATATCGAAACC -3′ R 5′- AATTGCGGCGGATTTCC -3′ |
957- 1025 |
990 |
|
prostaglandin D2 synthase (ptgds, UniGene Ppr.12268) |
F 5′- TTGGACACCGGCATCCTT -3′ R 5′- TTTAAGAGACCCTCAGGCATCTG -3′ |
540-620 | 589 |
|
zgc:55572 (zgc:55572, UniGene Ppr.6860) |
F 5′- TGCGCAAGGCTGATATGCT -3′ R 5′- GACTGTCTGCCTCCACAAGGA -3′ |
385-465 | 417 |
|
ribosomal protein L8 (rpl8, AY919670) |
F 5′- CATACCACAAGTACAAGGCCAAGA -3′ R 5′- ACCGAAGGGATGCTCAACAG -3′ |
516-598 | 577 |
The linearity of the Q-PCR assays and the absence of inhibitors in the RNA samples were confirmed using dilution series. Four two-fold dilutions of a randomly selected total RNA sample were prepared using bacterial total RNA to maintain a constant RNA concentration and assayed in parallel with the rest of the RNA samples and controls. This procedure reveals any deviation from linearity in either the reverse transcription or the real-time PCR reactions. The dilution series also reveals the presence of inhibitors in the RNA sample, because inhibitory components will result in anomalously low CT values and increasing PCR efficiency as the dilution increases (Nolan et al., 2006). Because the endpoints of interest are changes in expression relative to the control group, standard curves with artificial templates to define the copy number of expressed genes were not performed.
Statistical differences in relative mRNA expression between experimental groups were assessed by one-way analysis of variance (ANOVA) followed by Fisher’s test. Some data were log-transformed in order to satisfy the assumptions of normality and homogeneity. All statistical analyses were performed using SAS 9.1 (SAS Institute, Cary, NC). All data are shown as the mean ± SEM. Differences were considered statistically significant at P < 0.05.
Results
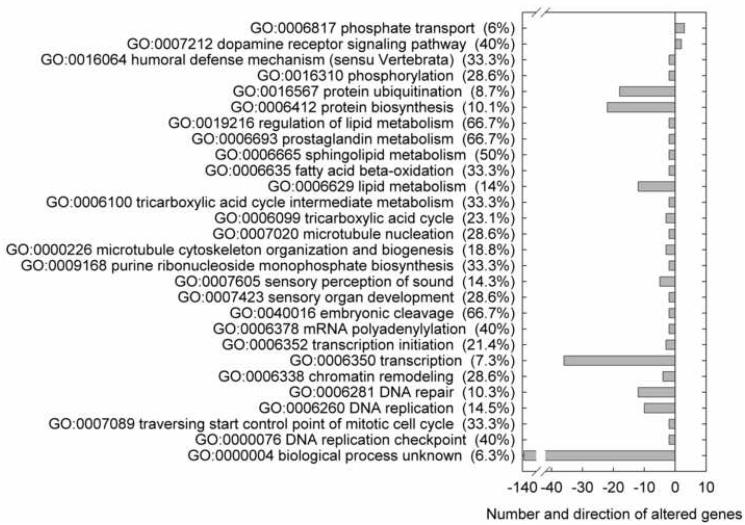
Results obtained from the microarray analysis reflect transcriptional changes in gonad tissues of female fathead minnows exposed for 4 days to 1 μg/L of 17β-trenbolone. There were 14,714 genes out of 21,495 probes on the microarray which were expressed in gonad tissue at levels greater than the limit of detection of the microarray. 17β-trenbolone exposure caused 840 genes in female gonad tissue to be significantly altered (P < 0.05) in expression relative to control fish. Among those, 99 genes (0.67 % of expressed genes) were up-regulated following 17β-trenbolone exposure while 741 (5.04 % of expressed genes) exhibited a down-regulation relative to control fish. Gene ontology (GO) terms for biological process were examined to determine the function of genes with altered patterns of expression (Fig. 1). GO information for fathead minnow was limited, in that among altered genes, 33.1% had no biological process GO annotation and 16.2% were annotated as biological process unknown (GO:0000004). This is in part a consequence of the fathead minnow genome being less characterized in comparison to other fishes such as the zebrafish (Danio rerio) and Japanese medaka (Oryzias latipes), although a fully sequenced genome is not necessarily a prerequisite for understanding biological function. Genes annotated as biological process unknown (GO:0000004) were significantly over-represented among down-regulated genes; 6.3 % of the expressed genes annotated as biological process unknown were downregulated (Fig. 1). Other GO terms significantly enriched among down-regulated genes included terms associated with protein synthesis, lipid metabolism, and synthesis of DNA and RNA (Fig. 1).
Fig. 1.
Significantly over-represented gene ontology (GO) biological process categories for the genes upregulated (positive numbers) or downregulated (negative numbers) in female fathead minnow gonads following 4 days exposure to 1 μg/L of 17β-trenbolone. The y-axis labels include the GO term, description, and percent of genes associated with the term that were up- or down-regulated. Columns denote the number of up- or down-regulated genes associated with each GO term.
Each gene may be associated with many biological process GO terms. Several of the GO terms significantly over-represented among down-regulated genes are associated with overlapping sets of genes (Fig. 1). For example, lipid metabolism (GO:0006629) overlaps with sphingolipid metabolism (GO:0006665), fatty acid beta-oxidation (GO:0006635) and prostaglandin metabolism (GO:0006693). DNA replication (GO:0006260) has genes in common with DNA repair (GO:0006281), chromatin remodeling (GO:0006338), DNA replication checkpoint (GO:0000076), traversing start control point of mitotic cell cycle (GO:0007089) and transcription (GO:0006350). Sensory perception of sound (GO:0007605) overlaps with transcription (GO:0006350) and DNA repair (GO:0006281).
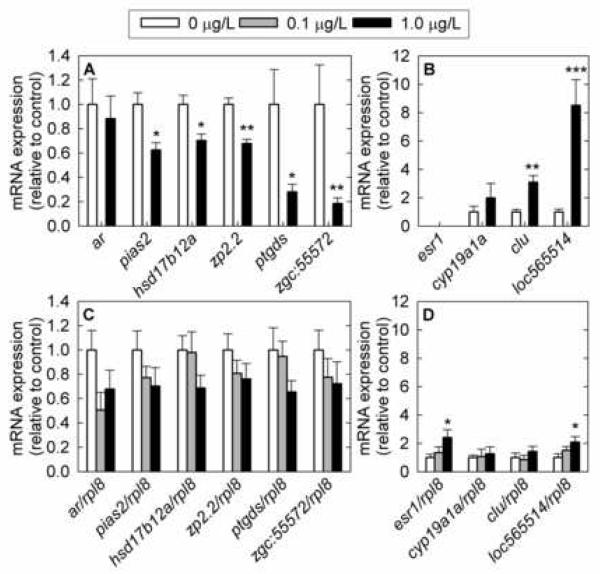
A subset of the differentially expressed genes identified by microarray analysis was selected for validation by Q-PCR: hydroxysteroid (17β) dehydrogenase 12a (hsd17b12a), zona pellucida glycoprotein 2.2 (zp2.2), and protein inhibitor of activated STAT, 2 (pias2) (Fig. 2). The fold change, the variation within each treatment, the level of statistical significance, the overall expression level, and the gene’s function in reproduction were all factors considered in selection of genes for Q-PCR. Three additional candidate genes which were not significantly altered based on microarray analysis, cytochrome P450, family 19, subfamily A, polypeptide 1a (cyp19a1a, previously known as aromatase A); androgen receptor (ar); and estrogen receptor 1 (esr1, previously known as estrogen receptor alpha) were selected by screening the literature and their expression levels were also evaluated by Q-PCR in female gonad tissue. Six of the selected genes fall within three groups of interest: receptors and transcription factors (ar, esr1, and pias2); steroid metabolism (cyp19a1a and hsd17b12a); and egg maturation (zp2.2). Four additional genes were also selected for further analysis based on large fold changes and highly significant p-values in the microarray data, without regard to gene annotation: similar to zebrafish hypothetical protein LOC565514 (loc565514), clusterin (clu), prostaglandin D2 synthase (ptgds), and similar to zebrafish zgc:55572, which has homology to human serine/threonine kinase 38 (Fig. 2).
Fig. 2.
Cross-validation of gene expression changes in selected genes measured by microarray (A, B) and validated in a larger set of samples by Q-PCR (C, D) in female fathead minnow gonads following exposure to 17β-trenbolone for 4 days. A) Microarray data for downregulated genes. B) Microarray data for upregulated genes. C) Q-PCR data for downregulated genes. D) Q-PCR data for upregulated genes. ar: androgen receptor; pias2: protein inhibitor of activated STAT, 2; hsd17b12a: hydroxysteroid (17β) dehydrogenase 12a; zp2.2: zona pellucida glycoprotein 2.2; ptgds: prostaglandin D2 synthase; zgc:55572: similar to zebrafish zgc:55572 (homologous to human serine/threonine kinase 38); esr1: estrogen receptor 1; cyp19a1a: cytochrome P450, family 19, subfamily A, polypeptide 1a; clu: clusterin; loc565514: similar to zebrafish hypothetical protein LOC565514; rpl8: ribosomal protein L8. The Q-PCR data are normalized to rpl8 expression and the array data by residuals. For microarray analysis, n=4 fish; for Q-PCR analysis, n=10 fish (4 fish measured by microarray and 6 additional fish exposed in the same experiment) at doses 0 μg/L and 1 μg/L; n=6 fish at dose 0.1 μg/L. Error bars denote the standard error of the mean. Statistically significant differences in the expression levels of fathead minnow genes between control and treated fish are denoted by stars (* P < 0.05; ** P < 0.01; *** P < 0.001).
For genes involved in reproduction, agreement between Q-PCR data and microarray data was good as to the direction of gene change and the magnitude of difference between control and exposed groups (Table 2, Fig. 2). The expression of pias2, hsd17b12a, and zp2.2 genes determined by Q-PCR was decreased following 17β-trenbolone exposure, in accordance with microarray data. However, in the case of significant expression changes based on microarray data, only the hsd17b12a gene was close to significantly altered (Table 2). Significant changes in gene expression between treatment and control groups may not have been observed due to the small number of replicates, a small magnitude in the changes of expression, and to a large degree of variation among individual fish. Additionally, variation is not unexpected in female gonad tissue, due to the heterogeneous nature of ovarian tissues and differential reproductive staging that may have occurred among females. The difference in significance by these two techniques may also reflect the greater power of normalization to residuals for array data. Lastly, in gonads, ar gene expression was reduced in the treated groups compared to controls, while esr1 and cyp19a1a transcript levels tended to increase and also showed increased variation in treated groups (Fig. 2). However, the changes in ar and cyp19a1a were not statistically significant (P > 0.05).
Table 2.
Comparison between array data and Q-PCR data.
| Gene | Array (n=4) | Q-PCR (n=10) | ||
|---|---|---|---|---|
| Fold change |
p-Value | Fold change |
p-Value | |
| Genes selected based on function in reproduction | ||||
| androgen receptor (ar) | -1.1 | 0.6599 | -1.5 | 0.2240 |
|
protein inhibitor of activated STAT, 2 (pias2) |
-1.6 | 0.0107 | -1.4 | 0.2071 |
| estrogen receptor 1 (esr1) | ND | -- | 2.4 | 0.0157 |
|
hydroxysteroid (17β) dehydrogenase 12a (hsd17b12a) |
-1.4 | 0.0137 | -1.5 | 0.0510 |
|
cytochrome P450, family 19, subfamily A, polypeptide 1a (cyp19a1a) |
2.0 | 0.2848 | 1.3 | 0.6896 |
|
zona pellucida glycoprotein 2.2 (zp2.2) |
-1.5 | 0.0017 | -1.3 | 0.2230 |
| Genes selected based on large fold changes in microarray data | ||||
|
similar to zebrafish zgc:55572 (zgc:55572) |
-5.4 | 0.0054 | -1.4 | 0.1887 |
| prostaglandin D2 synthase (ptgds) | -3.6 | 0.0119 | -1.5 | 0.2561 |
| clusterin (clu) | 3.1 | 0.0028 | 1.4 | 0.2859 |
|
similar to zebrafish hypothetical protein LOC565514 (loc565514) |
8.5 | 0.0002 | 2.1 | 0.0134 |
ND = not detected.
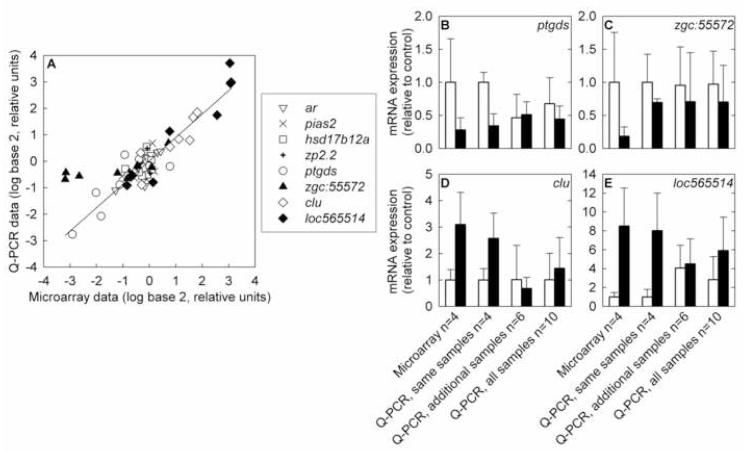
The genes selected based on large fold changes in the microarray data were not closely matched by Q-PCR data (Table 2, Fig. 2). In particular, the magnitudes of the changes in gene expression were much less in the Q-PCR data set than in the microarray data set (Table 2, Fig. 2). This unexpected result was not caused by technical differences in Q-PCR and microarray measurements; for individual samples, the Q-PCR and microarray measurements were in close agreement, with the exception of zgc:55572 (Fig. 3). The differences between the results based on Q-PCR and microarray are likely to be the result of sample size. The microarray analysis used samples from 4 individual fish per treatment, while the Q-PCR analysis used the same samples run on microarray and 6 additional fish exposed in the same experiment, for a total of 10 samples per treatment (Fig. 3). The deviation between Q-PCR and microarray results for zgc:55572 could be due to positional effects, because the microarray probe and Q-PCR primer set targeted different regions of the mRNA; or could be due to differential reverse transcription efficiency, because the microarray experiment used oligo-dT primers and the Q-PCR experiment used random hexamers.
Fig. 3.
Comparison of gene expression measurements by microarray and Q-PCR. A) Correlation of microarray results with Q-PCR results from the same RNA samples. Data for each gene is expressed relative to the average of the untreated samples for that gene. ar: androgen receptor; pias2: protein inhibitor of activated STAT, 2; hsd17b12a: hydroxysteroid (17β) dehydrogenase 12a ; zp2.2: zona pellucida glycoprotein 2.2; ptgds: prostaglandin D2 synthase; zgc:55572: similar to zebrafish zgc:55572; clu: clusterin; loc565514: similar to zebrafish hypothetical protein LOC565514. The regression line shown is for the entire data set, excluding zgc:55572 (R2 = 0.84, slope = 0.88, intercept = 0.016). If zgc:55572 is included, the regression parameters are R2 = 0.71, slope = 0.70, intercept = 0.094. B-E) Comparisons of microarray results with Q-PCR results from the same samples, samples of additional fish from the same exposure tanks, and all samples. White bars represent the control group and black bars represent the 1 μg/L 17β-trenbolone treatment group. Error bars denote one standard deviation. B) prostaglandin D2 synthase, C) similar to zebrafish zgc:55572, D) clusterin, E) similar to zebrafish hypothetical protein LOC565514.
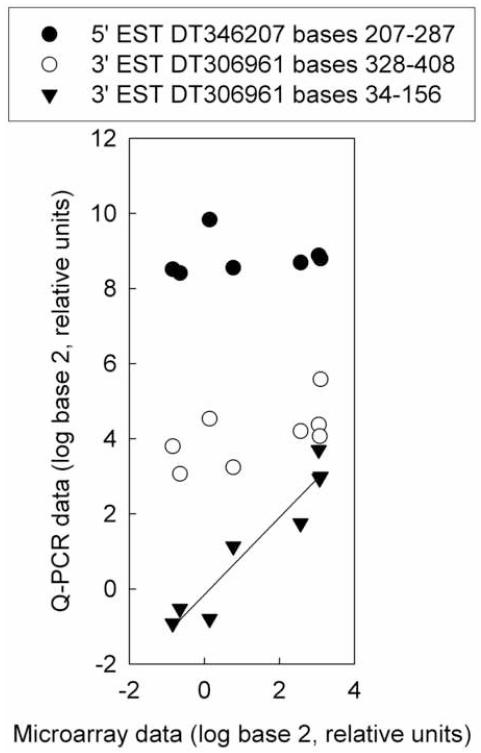
One gene (loc565514) measured by both Q-PCR and microarray exhibited positional effects on its measured expression levels (Fig. 4). A Q-PCR primer set that overlapped the position of the microarray probe in the mRNA sequence closely matched the microarray data, but two primer sets targeted to other locations in the mRNA sequence deviated from the expression pattern shown by microarray (Fig. 4). Positional effects have previously been observed for microarray probes of the fathead minnow vtg1 gene (Larkin et al., 2007).
Fig. 4.
Position effects shown by three Q-PCR primer sets for hypothetical protein LOC565514 (loc565514). The microarray probe targeted bases 101-160 of 3′ EST DT306961. Q-PCR data was normalized to the abundance of ribosomal protein L8 (rpl8). Microarray data are expressed relative to the average of the untreated samples. Q-PCR data for all three primer sets are expressed relative to the average of the 3′ EST DT306961 bases 34-156 primer set for the untreated samples. Regression analysis did not show significant relationships for primer sets 5′ EST DT346207 bases 207-287 or 3′ EST DT306961 bases 328-408. The regression parameters for primer set 3’ EST DT306961 bases 34-156 are R2 = 0.96, slope = 1.0, intercept = -0.15.
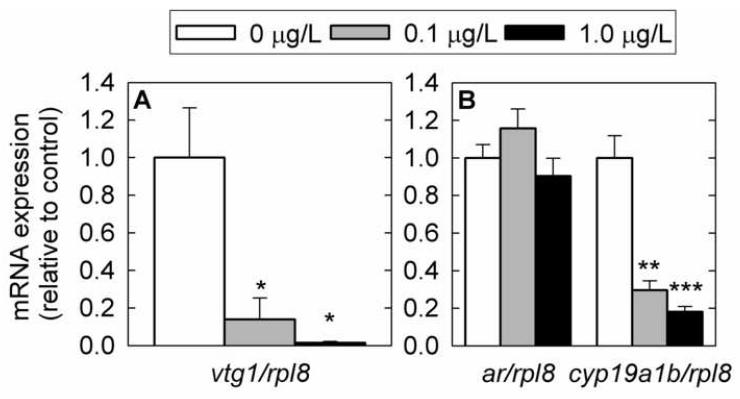
We investigated gene expression changes in brain and liver by Q-PCR. In liver, vtg1 mRNA was strongly down-regulated in a dose-dependent manner (Fig. 5A). In brain, cyp19a1b mRNA was also down-regulated in a dose-dependent manner, but ar mRNA was unchanged with treatment (P > 0.05) (Fig. 5B). Exposure to 1 μg 17β-trenbolone/L caused a 6-fold decrease (P < 0.001) in mean cyp19a1b mRNA expression.
Fig. 5.
Gene expression changes in female fathead minnow liver (A) and brain (B) measured by Q-PCR following exposure to 17β-trenbolone for 4 days. A) vitellogenin 1 (vtg1) in liver tissue. B) cytochrome P450, family 19, subfamily A, polypeptide 1b (cyp19a1b) and androgen receptor (ar) in brain tissue. mRNA expression was normalized to the abundance of ribosomal protein L8 (rpl8). Experimental groups consisted of six fish (liver) or twelve fish (brain) and each fish was analyzed in duplicate. Error bars denote the standard error of the mean. Statistically significant differences in expression levels between control and treated fish are denoted by stars (* P < 0.05; ** P < 0.01; *** P < 0.001).
Discussion
The results of the present study are consistent with the previously observed declines in both estradiol and testosterone levels in female fish in response to 17β-trenbolone exposure (Ankley et al., 2003; Ankley et al., 2004). In the microarray analysis of gonadal gene expression, the preponderance of down-regulated genes relative to up-regulated genes suggests that the many estrogen-dependent functions of ovarian tissue are declining in response to the drop in endogenous estradiol levels caused by exposure to 17β-trenbolone. The GO terms associated with the greatest numbers of down-regulated genes, protein biosynthesis and ubiquitination, lipid metabolism, transcription, and DNA repair and replication, are all basic functions required for cell growth and maintenance. Thus, the effects of 17β-trenbolone exposure on reproduction may be due to an overall loss of ovarian function. The results for several specific down-regulated genes are consistent with this hypothesis.
Protein inhibitor of activated STAT, 2 (pias2) encodes a transcriptional co-regulatory protein that recognizes the zinc finger region (ZFR) of Ar and modulates Ar-dependent transcription (Moilanen et al., 1999). While the expression of androgen receptor in brain was unaffected by the 17β-trenbolone treatment in the present study, it tended to decrease in female gonads, although the difference was not statistically significant. A similar decrease was observed for gonadal pias2 expression. In contrast, although gonadal esr1 expression was not detected in either control or treated samples on the microarray, Q-PCR analysis was able to detect significantly increased gonadal esr1 expression in response to 17β-trenbolone exposure. The observations of decreased ar and pias2 and increased esr1 are suggestive of negative feedback controls on expression of these transcription factors (Shupnik et al., 1989; Nastiuk and Clayton, 1994). Results of the GO term analysis of microarray results also showed down-regulation of several categories associated with transcription.
Cyp19a1a and cyp19a1b encode members of the cytochrome P450 superfamily that catalyze the critical reaction converting C19 androgens to C18 estrogens in gonad and brain tissues, respectively (Callard et al., 1978; Callard et al., 2001; Kishida and Callard, 2001). In brain tissue, exposure to 17β-trenbolone resulted in a strong and significant decrease in cyp19a1b expression. The results for gonad indicated a small increase in average cyp19a1a transcript level and a large increase in variation among individual fish. Thus, loss of aromatase activity is unlikely to be the cause of the previously observed drop in plasma estrogen levels in response to 17β-trenbolone exposure (Ankley et al., 2003). However, loss of cyp19a1b expression in the brain may have behavioral consequences (Forlano et al., 2006).
The contrast between the brain and ovary results is consistent with reports in the literature indicating that aromatase isoforms are regulated differently (Kazeto et al., 2001; Kishida and Callard, 2001; Villeneuve et al., 2006). The promoter region of cyp19a1a contains a SF-1 (steroidogenic factor-1) regulatory element but no estrogen response elements (EREs), while the cyp19a1b promoter contains EREs but no SF-1 regulatory element (Callard et al., 2001; Kazeto et al., 2001). Based on the difference in promoters, one could hypothesize that expression of cyp19a1a transcript levels would be primarily influenced by gonadotropin-mediated regulation of steroidogenesis facilitated through SF-1, while cyp19a1b expression would be more closely tied to endogenous 17β-estradiol concentrations. Thus, the strong down-regulation of cyp19a1b observed in female brain tissue may be reflective of the decrease in plasma concentrations of 17β-estradiol reported by Ankley et al. (2003) in fathead minnows exposed to 17β-trenbolone.
Activation of transcription of vitellogenin isoforms in the liver is also controlled by EREs (Brock and Shapiro, 1983). The results for liver indicated a strong and significant decrease in vitellogenin 1 (vtg1 mRNA expression, with exposure to 1 μg 17β-trenbolone/L causing a 64-fold decrease (P < 0.01) in expression relative to controls. This observation is in total agreement with existing data on the down-regulation of hepatic vitellogenin gene expression and plasma vitellogenin levels in female fish exposed to steroidal androgens (Ankley et al., 2004; Miracle et al., 2006). Similar to vitellogenins, egg envelope precursor proteins, also known as zona pellucida or choriogenin proteins, are produced in response to circulating estrogen. In teleosts, depending on the species zona pellucida proteins are expressed in either liver (e.g., winter flounder, seabream, and cod) or gonad (e.g., zebrafish, goldfish, and carp), or in both (medaka) (Conner and Hughes, 2003; Litscher and Wassarman, 2007). In fathead minnow, zona pellucida proteins appear to be mostly expressed in gonad tissues (NCBI Unigene Expression Profile, Ppr.12560). In the present study, down-regulation of zp2.2 in gonads of female fathead minnow after exposure to 17β-trenbolone was observed, as measured by microarray. The decreases in vtg1 and zp2.2 observed in liver and gonad, respectively, are likely to reflect decreased concentrations of plasma 17β-estradiol and testosterone, as previously observed (Ankley et al., 2003; Miracle et al., 2006). Decreases in vtg1 expression have direct implications for population-level effects on exposed fish, because fish require vitellogenin production in order to reproduce (Miller et al., 2007).
The mechanism leading to decreased concentrations of the steroid hormones 17β-estradiol and testosterone in response to 17β-trenbolone exposure is apparently not androgen-receptor dependent, because co-treatment with 17β-trenbolone and androgen receptor antagonists flutamide or vinclozolin does not restore expression of vitellogenin 1 mRNA . (Ankley et al., 2004; Martinović et al., 2008). In Atlantic croaker ovary tissue culture, non-aromatizable androgens had direct, non-genomic, antiandrogen-insensitive inhibitory effects on estrogen production (Braun and Thomas, 2003). Thus, the effects of 17β-trenbolone exposure on plasma levels of the steroid hormones 17β-estradiol and testosterone are likely mediated by mechanisms other than the classical genomic androgen receptor (Heinlein and Chang, 2002).
One potential candidate gene involved in the decrease in sex steroid concentrations of of fish exposed to the strong exogenous androgen 17-trenbolone is hydroxysteroid (17β) dehydrogenase 12a (hsd17b12a). In our study, hsd17b12a was significantly down-regulated in gonads as measured by microarray. Q-PCR measurement validated the direction and magnitude of the change in expression shown by microarray, and was very close to statistical significance. Hydroxysteroid (17β) dehydrogenase 12a catalyzes the conversion of androstenedione to testosterone which, in turn, is converted to 17β-estradiol by the aromatase enzymes Cyp19a1a and Cyp19a1b. Thus, down-regulation of hsd17b12a is expected to lead to declines in both testosterone and estradiol, as observed by Ankley et al. (2003). In summary, it is plausible that down-regulation of hsd17b12a caused decreases in testosterone, leading to depressed estrogen levels which, in turn, decreased vtg1 expression through decreased activation of the estrogen receptor, and caused reproductive impairment.
The Q-PCR data and array data were in close agreement for individual samples, and were in general agreement for genes involved in reproduction, as to the direction of gene change and the magnitude of difference between control and exposed groups. The link between genes with altered expression and mode of toxic action, and the consistency between array and Q-PCR results supports the use of microarrays for investigations of biomarkers and mode of toxic action of environmental contaminants. However, the differences between the subsample selected for microarray analysis and the larger sample set measured by Q-PCR for genes selected based on large fold changes on the microarray emphasize the need for cross-validation of microarray results in independent samples, particularly when working with small sample sizes.
Conclusions
The gene expression profile for 17β-trenbolone defined here suggests a general down-regulation of the expression of estrogen-responsive genes (hepatic vtg1, brain cyp19a1b, and gonad zp2.2 as suggested by the interference of 17β-trenbolone with the biosynthesis of endogenous estrogens in fish. Our study also provides a potential candidate gene mediating the action of 17β-trenbolone, the hydroxysteroid (17β) dehydrogenase 12a, involved in the production of testosterone, the precursor of 17β-estradiol. Finally, the expression of hepatic vtg1 and brain cyp19a1b appear to be highly responsive to 4-days exposure to 17β-trenbolone, and are potential biomarkers of exposure to 17β-trenbolone or other contaminants that act to lower endogenous estrogen levels.
Acknowledgements
This work was supported by grant number 2R44ES011882-03A1-02 from the National Institute of Environmental Health Sciences (NIEHS), NIH. Its contents are solely the responsibility of the authors and do not necessarily reflect the official views of the NIEHS, NIH. Jennifer Dorts is a PhD student at the F.R.S.-FNRS, Fonds National de la Recherche Scientifique (Belgium). Diane Nicks, Mandy Annis, Marie Pope, Mike Tanner, and James Candrl provided technical assistance. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Defoe DL, Kahl MD, Jensen KM, Makynen EA, Miracle A, Hartig P, Gray LE, Cardon M, Wilson V. Evaluation of the model anti-androgen flutamide for assessing the mechanistic basis of responses to an androgen in the fathead minnow (Pimephales promelas) Environ. Sci. Technol. 2004;38:6322–6327. doi: 10.1021/es040022b. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Jensen KM, Makynen EA, Kahl MD, Korte JJ, Hornung MW, Henry TR, Denny JS, Leino RL, Wilson VS, Cardon MC, Hartig PC, Gray LE. Effects of the androgenic growth promoter 17-β-trenbolone on fecundity and reproductive endocrinology of the fathead minnow. Environ. Toxicol. Chem. 2003;22:1350–1360. [PubMed] [Google Scholar]
- Ankley GT, Villeneuve DL. The fathead minnow in aquatic toxicology: Past, present and future. Aquat. Toxicol. 2006;78:91–102. doi: 10.1016/j.aquatox.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balter M. Scientific cross-claims fly in continuing beef war. Science. 1999;284:1453–1455. doi: 10.1126/science.284.5419.1453. [DOI] [PubMed] [Google Scholar]
- Bartosiewicz M, Penn S, Buckpitt A. Applications of gene arrays in environmental toxicology: Fingerprints of gene regulation associated with cadmium chloride, benzo(a)pyrene, and trichloroethylene. Environ. Health Perspect. 2001;109:71–74. doi: 10.1289/ehp.0110971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer ERS, Daxenberger A, Petri T, Sauerwein H, Meyer HHD. Characterisation of the affinity of different anabolics and synthetic hormones to the human androgen receptor, human sex hormone binding globulin and to the bovine progestin receptor. APMIS. 2000;108:838–846. doi: 10.1111/j.1600-0463.2000.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Braun AM, Thomas P. Androgens inhibit estradiol-17β synthesis in Atlantic croaker (Micropogonias undulatus) ovaries by a nongenomic mechanism initiated at the cell surface. Biol. Reprod. 2003;69:1642–1650. doi: 10.1095/biolreprod.103.015479. [DOI] [PubMed] [Google Scholar]
- Brock ML, Shapiro DJ. Estrogen regulates the absolute rate of transcription of the Xenopus laevis vitellogenin genes. J. Biol. Chem. 1983;258:5449–5455. [PubMed] [Google Scholar]
- Callard GV, Petro Z, Ryan KJ. Phylogenetic distribution of aromatase and other androgen-converting enzymes in the central nervous system. Endocrinology. 1978;103:2283–2290. doi: 10.1210/endo-103-6-2283. [DOI] [PubMed] [Google Scholar]
- Callard GV, Tchoudakova AV, Kishida M, Wood E. Differential tissue distribution, developmental programming, estrogen regulation and promoter characteristics of cyp19 genes in teleost fish. J. Steroid Biochem. Mol. Biol. 2001;79:305–314. doi: 10.1016/s0960-0760(01)00147-9. [DOI] [PubMed] [Google Scholar]
- Conner SJ, Hughes DC. Analysis of fish ZP1/ZPB homologous genes--evidence for both genome duplication and species-specific amplification models of evolution. Reproduction. 2003;126:347–352. doi: 10.1530/rep.0.1260347. [DOI] [PubMed] [Google Scholar]
- Danhaive PA, Rousseau GG. Binding of glucocorticoid antagonists to androgen and glucocorticoid hormone receptors in rat skeletal muscle. J. Steroid Biochem. 1986;24:481–487. doi: 10.1016/0022-4731(86)90109-3. [DOI] [PubMed] [Google Scholar]
- Durhan EJ, Lambright CS, Makynen EA, Lazorchak J, Hartig PC, Wilson VS, Gray LE, Ankley GT. Identification of metabolites of trenbolone acetate in androgenic runoff from a beef feedlot. Environ. Health Perspect. 2006;114S:65–68. doi: 10.1289/ehp.8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filby AL, Tyler CR. Molecular characterization of estrogen receptors 1, 2a, and 2b and their tissue and ontogenic expression profiles in fathead minnow (Pimephales promelas) Biol. Reprod. 2005;73:648–662. doi: 10.1095/biolreprod.105.039701. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Schlinger BA, Bass AH. Brain aromatase: New lessons from non-mammalian model systems. Front. Neuroendocrinol. 2006;27:247–274. doi: 10.1016/j.yfrne.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Gray LE, Jr., Ostby J, Wilson V, Lambright C, Bobseine K, Hartig P, Hotchkiss A, Wolf C, Furr J, Price M, Parks L, Cooper RL, Stoker TE, Laws SC, Degitz SJ, Jensen KM, Kahl MD, Korte JJ, Makynen EA, Tietge JE, Ankley GT. Xenoendocrine disrupters-tiered screening and testing: filling key data gaps. Toxicology. 2002;181-182:371–382. doi: 10.1016/s0300-483x(02)00469-9. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel P, Paules R, Afshari CA. Discovery in toxicology: mediation by gene expression array technology. J. Biochem. Mol. Toxicol. 2001;15:231–242. doi: 10.1002/jbt.10006. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. The roles of androgen receptors and androgen-binding proteins in nongenomic androgen actions. Mol. Endocrinol. 2002;16:2181–2187. doi: 10.1210/me.2002-0070. [DOI] [PubMed] [Google Scholar]
- Hemmer MJ, Cripe GM, Hemmer BL, Goodman LR, Salinas KA, Fournie JW, Walker CC. Comparison of estrogen-responsive plasma protein biomarkers and reproductive endpoints in sheepshead minnows exposed to 17β-trenbolone. Aquat. Toxicol. 2008;88:128–136. doi: 10.1016/j.aquatox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Holbech H, Kinnberg K, Petersen GI, Jackson P, Hylland K, Norrgren L, Bjerregaard P. Detection of endocrine disrupters: evaluation of a Fish Sexual Development Test (FSDT) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006;144:57–66. doi: 10.1016/j.cbpc.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Jensen KM, Makynen EA, Kahl MD, Ankley GT. Effects of the feedlot contaminant 17α-trenbolone on reproductive endocrinology of the fathead minnow. Environ. Sci. Technol. 2006;40:3112–3117. doi: 10.1021/es052174s. [DOI] [PubMed] [Google Scholar]
- Kazeto YK, Ijiri S, Place AR, Zohar Y, Trant JM. The 5 ′-flanking regions of CYP19A1 and CYP19A2 in zebrafish. Biochem. Biophys. Res. Commun. 2001;288:503–508. doi: 10.1006/bbrc.2001.5796. [DOI] [PubMed] [Google Scholar]
- Kishida M, Callard GV. Distinct cytochrome P450 aromatase isoforms in zebrafish (Danio rerio) brain and ovary are differentially programmed and estrogen regulated during early development. Endocrinology. 2001;142:740–750. doi: 10.1210/endo.142.2.7928. [DOI] [PubMed] [Google Scholar]
- Larkin P, Villeneuve DL, Knoebl I, Miracle AL, Carter BJ, Liu L, Denslow ND, Ankley GT. Development and validation of a 2,000-gene microarray for the fathead minnow (Pimephales promelas) Environ. Toxicol. Chem. 2007;26:1497–1506. doi: 10.1897/06-501r.1. [DOI] [PubMed] [Google Scholar]
- Litscher ES, Wassarman PM. Egg extracellular coat proteins: from fish to mammals. Histol. Histopathol. 2007;22:337–347. doi: 10.14670/HH-22.337. [DOI] [PubMed] [Google Scholar]
- Martinović D, Blake LS, Durhan EJ, Greene KJ, Kahl MD, Jensen KM, Makynen EA, Villeneuve DL, Ankley GT. Reproductive toxicity of vinclozolin in the fathead minnow: confirming an anti-androgenic mode of action. Environ. Toxicol. Chem. 2008;27:478–488. doi: 10.1897/07-206R.1. [DOI] [PubMed] [Google Scholar]
- Miller DH, Ankley GT. Modeling impacts on populations: fathead minnow (Pimephales promelas) exposure to the endocrine disruptor 17β-trenbolone as a case study. Ecotoxicol. Environ. Saf. 2004;59:1–9. doi: 10.1016/j.ecoenv.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Miller DH, Jensen KM, Villeneuve DL, Kahl MD, Makynen EA, Durhan EJ, Ankley GT. Linkage of biochemical responses to population-level effects: a case study with vitellogenin in the fathead minnow (Pimephales promelas) Environ. Toxicol. Chem. 2007;26:521–527. doi: 10.1897/06-318r.1. [DOI] [PubMed] [Google Scholar]
- Miracle A, Ankley G, Lattier D. Expression of two vitellogenin genes (vg1 and vg3) in fathead minnow (Pimephales promelas) liver in response to exposure to steroidal estrogens and androgens. Ecotoxicol. Environ. Saf. 2006;63:337–342. doi: 10.1016/j.ecoenv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Moilanen AM, Karvonen U, Poukka H, Yan W, Toppari J, Janne OA, Palvimo JJ. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J. Biol. Chem. 1999;274:3700–3704. doi: 10.1074/jbc.274.6.3700. [DOI] [PubMed] [Google Scholar]
- Nastiuk KL, Clayton DF. Seasonal and tissue-specific regulation of canary androgen receptor messenger ribonucleic acid. Endocrinology. 1994;134:640–649. doi: 10.1210/endo.134.2.8299561. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protocols. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol. Carcinog. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Orlando EF, Kolok AS, Binzcik GA, Gates JL, Horton MK, Lambright CS, Gray LE, Jr., Soto AM, Guillette LJ., Jr. Endocrine-disrupting effects of cattle feedlot effluent on an aquatic sentinel species, the fathead minnow. Environ. Health Perspect. 2004;112:353–358. doi: 10.1289/ehp.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örn S, Yamani S, Norrgren L. Comparison of vitellogenin induction, sex ratio, and gonad morphology between zebrafish and Japanese medaka after exposure to 17α-ethinylestradiol and 17β-trenbolone. Arch. Environ. Contam. Toxicol. 2006;51:237–243. doi: 10.1007/s00244-005-0103-y. [DOI] [PubMed] [Google Scholar]
- Parks LG, Lambright CS, Orlando EF, Guillette LJ, Jr., Ankley GT, Gray LE., Jr. Masculinization of female mosquitofish in kraft mill effluent-contaminated Fenholloway River water is associated with androgen receptor agonist activity. Toxicol. Sci. 2001;62:257–267. doi: 10.1093/toxsci/62.2.257. [DOI] [PubMed] [Google Scholar]
- Peterson BN, Foran CM, Benson WH. Reproductive and physiological effects of aquatic exposure to trenbolone, an environmental androgen. 22nd Annual Meeting SETAC.2001. p. 333. [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Schiffer B, Daxenberger A, Meyer K, Meyer HHD. The fate of trenbolone acetate and melengestrol acetate after application as growth promoters in cattle: environmental studies. Environ. Health Perspect. 2001;109:1145–1151. doi: 10.1289/ehp.011091145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Fujishima S, Nozaka T, Maeda M, Kobayashi K. Comparison of response to 17β-estradiol and 17β-trenbolone among three small fish species. Environ. Toxicol. Chem. 2006;25:2742–2752. doi: 10.1897/05-647r.1. [DOI] [PubMed] [Google Scholar]
- Shupnik MA, Gordon MS, Chin WW. Tissue-specific regulation of rat estrogen receptor mRNAs. Mol. Endocrinol. 1989;3:660–665. doi: 10.1210/mend-3-4-660. [DOI] [PubMed] [Google Scholar]
- Sone K, Hinago M, Itamoto M, Katsu Y, Watanabe H, Urushitani H, Tooi O, Guillette LJ, Jr., Iguchi T. Effects of an androgenic growth promoter 17β-trenbolone on masculinization of Mosquitofish (Gambusia affinis affinis) Gen. Comp. Endocrinol. 2005;143:151–160. doi: 10.1016/j.ygcen.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Soto AM, Calabro JM, Prechtl NV, Yau AY, Orlando EF, Daxenberger A, Kolok AS, Guillette LJ, Jr., le Bizec B, Lange IG, Sonnenschein C. Androgenic and estrogenic activity in water bodies receiving cattle feedlot effluent in Eastern Nebraska, USA. Environ. Health Perspect. 2004;112:346–352. doi: 10.1289/ehp.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Knoebl I, Kahl MD, Jensen KM, Hammermeister DE, Greene KJ, Blake LS, Ankley GT. Relationship between brain and ovary aromatase activity and isoform-specific aromatase mRNA expression in the fathead minnow (Pimephales promelas) Aquat. Toxicol. 2006;76:353–368. doi: 10.1016/j.aquatox.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Lambright C, Ostby J, Gray LE., Jr. In vitro and in vivo effects of 17β-trenbolone: A feedlot effluent contaminant. Toxicol. Sci. 2002;70:202–211. doi: 10.1093/toxsci/70.2.202. [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) In: Global Assessment of the State-of-the-Science of Endocrine Disruptors. Damstra T, Barlow S, Bergman A, Kavlock R, van der Kraak G, editors. International Programme on Chemical Safety; Geneva: 2002. [Google Scholar]