Abstract
NO production by neuronal nitric oxide synthase (nNOS) requires calmodulin and is enhanced by the chaperone Hsp90, which cycles dynamically with the enzyme. The proteasomal degradation of nNOS is enhanced by suicide inactivation and by treatment with Hsp90 inhibitors, the latter suggesting that dynamic cycling with Hsp90 stabilizes nNOS. Here, we use a purified ubiquitinating system containing CHIP (carboxyl terminus of Hsp70-interacting protein) as the E3 ligase to show that Hsp90 inhibits CHIP-dependent nNOS ubiquitination. Like the established Hsp90 enhancement of NO synthesis, Hsp90 inhibition of nNOS ubiquitination is Ca2+/calmodulin-dependent, suggesting that the same interaction of Hsp90 with the enzyme is responsible for both enhancement of nNOS activity and inhibition of ubiquitination. It is established that CHIP binds to Hsp90 as well as to Hsp70, but we show here the two chaperones have opposing actions on nNOS ubiquitination, with Hsp70 stimulating and Hsp90 inhibiting. We have used two mechanism-based inactivators, guanabenz and NG-amino-L-arginine, to alter the heme/substrate binding cleft and promote nNOS ubiquitination that can be inhibited by Hsp90. We envision that as nNOS undergoes toxic damage, the heme/substrate binding cleft opens exposing hydrophobic residues as the initial step in unfolding. As long as Hsp90 can form even transient complexes with the opening cleft, ubiquitination by Hsp70-dependent ubiquitin E3 ligases, like CHIP, is inhibited. When unfolding of the cleft progresses to a state that cannot cycle with Hsp90, Hsp70-dependent ubiquitination is unopposed. In this way, the Hsp70/Hsp90 machinery makes the quality control decision for stabilization versus degradation of nNOS.
Both the function and turnover of a wide variety of signaling proteins, such as steroid receptors and protein kinases, are regulated by Hsp901 (reviewed in Ref. 1). These Hsp90 ‘client’ proteins are assembled into complexes with the chaperone by a multichaperone machinery in which Hsp90 and Hsp70 function together as essential components (1). Formation of heterocomplexes with Hsp90 stabilizes client proteins, and treatment with an Hsp90 inhibitor such as geldanamycin uniformly triggers their degradation (2). Degradation of the Hsp90-regulated signaling proteins occurs via the ubiquitin-proteasome pathway, which in this case is initiated by Hsp70-dependent E3 ubiquitin ligases, such as CHIP (3) and parkin (4).
The interaction with Hsp90 modulates the ligand binding clefts in client signaling proteins to increase the efficiency of binding of ligands, such as steroids or ATP (reviewed in Refs. 5 and 6), and the proteins constantly undergo cycles of Hsp90 heterocomplex assembly and disassembly in the cytoplasm and nucleoplasm (1). Two types of cycling with Hsp90 have been recognized. The classical client signaling proteins form Hsp90 heterocomplexes that are stable enough to be isolated and analyzed biochemically. For lack of a better term, we call this ‘stable cycling’ with Hsp90, and these proteins are stringently regulated by the chaperone (6). In contrast, other signaling proteins form Hsp90 heterocomplexes that rapidly disassemble such that no (or only trace amounts of) Hsp90 heterocomplexes are recovered from cell lysates. We call this ‘dynamic cycling’, and the activity and turnover of these proteins are not as affected by Hsp90 inhibitors as the classical client proteins (5,6).
The nitric oxide synthases (NOSs), including endothelial NOS (eNOS), neuronal NOS (nNOS) and inducible NOS (iNOS), are signaling proteins whose activity is enhanced by Hsp90, as shown both by studies in intact cells (7–10) and by direct activation assays with purified proteins (7,10–14). NOS activity is Ca2+/calmodulin (CaM)-dependent, and several signaling pathways initiate nNOS and eNOS activity by raising intracellular Ca2+ concentration. Studies with purified proteins show that CaM and Hsp90 increase the binding of each other to both eNOS and nNOS (11,12,14,15). Both direct binding of purified Hsp90 to purified eNOS and nNOS and activation of their activities have been demonstrated in the absence of ATP and Hsp70 (7,14,15). This stands in contrast to the assembly of stable Hsp90 complexes with steroid receptors, for example, where both receptor-bound Hsp70 and Hsp90 must pass through at least one complete ATPase cycle (1). Thus, the observations made with Hsp90 regulation of eNOS and nNOS differ from the classic client signaling proteins, yet, there are similarities between them.
For example, formation of Hsp90 heterocomplexes with the glucocorticoid receptor (GR), the most studied client protein, promotes high affinity ligand binding, thus facilitating response at low concentrations of steroid (1). Similarly, in Sf9 insect cells, which have a low level of endogenous heme, Hsp90 promotes heme binding by apo-nNOS, with concomitant conversion to the enzymatically active holo-nNOS dimer (8,16,17). Also, like the classic client signaling proteins, such as the GR (18), treatment of cells with geldanamycin leads to nNOS degradation via the ubiquitin-proteasome pathway (8,19). In that Hsp90 regulation of client signaling protein function reflects the ability of the chaperone to modulate ligand binding clefts (5,6), it is reasonable to predict that Hsp90 stabilization of client proteins ensues from the same cleft interaction, yet there is no evidence for this.
nNOS is a particularly useful model for exploring a relationship between Hsp90 stabilization of the protein and its interaction with the ligand binding cleft. Certain mechanism-based inactivators, such as NG-amino-L-arginine (NAA) and the antihypertensive drug guanabenz, cause accelerated nNOS degradation (19). Guanabenz is an antihypertensive drug that produces impotence and inhibits nNOS activity, with accompanying loss of immunodetectible enzyme (20). In cultured cells, guanabenz has been shown to enhance the proteasomal degradation of nNOS (19). Guanabenz treatment leads to the oxidation of tetrahydrobiopterin and formation of a pterin-depleted nNOS that is catalytically inactive (21). The loss of tetrahydrobiopterin from its binding site within the heme/substrate binding cleft destabilizes the nNOS dimer and enhances nNOS ubiquitination (22). Many of the other inactivators cross-link heme to the enzyme (23,24), a modification that was shown in a myoglobin model to cause opening of the heme binding cleft (25) to yield a more unfolded state of the protein (26). The reaction of the inactivator in the heme/substrate binding cleft triggers nNOS ubiquitination and proteasomal degradation (19,27). Both CHIP and parkin function as E3 ligases for nNOS ubiquitination (4,28).
To date, it has not been established whether Hsp90 stabilizes proteins by inhibiting their ubiquitination or by inhibiting proteasomal degradation of the ubiquitinated protein. In this paper we use a purified ubiquitinating system with CHIP as the E3 ligase to show that Hsp90 inhibits nNOS ubiquitination. Like the established Hsp90 enhancement of NO synthesis from nNOS, Hsp90 inhibition of nNOS ubiquitination is promoted by Ca2+/calmodulin, consistent with the two Hsp90 effects ensuing from the same interaction with the enzyme. The proposal that the Hsp90 interaction is with the heme/substrate binding cleft is supported by the observation that nNOS ubiquitination triggered by the mechanism-based inactivators NAA and guanabenz is prevented by Hsp90. We propose that, although nNOS·Hsp90 heterocomplexes are not assembled by the multichaperone machinery that forms heterocomplexes with steroid receptors and a variety of other client signaling proteins (17), the interaction of Hsp90 with nNOS and its effects on the enzyme are similar to its interaction with and its effects on the classic client proteins.
EXPERIMENTAL PROCEDURES
Materials
Untreated rabbit reticulocyte lysate was from Green Hectares (Oregon, WI). (6R)-5,6,7,8-Tetrahydro-L-biopterin was purchased from Dr. Schirck’s Laboratory (Jona, Switzerland). Protein A-Sepharose, ubiquitin, ATP, creatine phosphokinase, rabbit polyclonal anti-nNOS, purified bovine calmodulin, and L- and D- isomers of NG-nitro-arginine (L-NNA, D-NNA) were purchased from Sigma. GST-tagged ubiquitin, ubiquitin aldehyde, and ubiquitin activating enzyme (E1) were from Boston Biochem (Cambridge, MA). Geldanamycin and NG-amino-L-arginine were from Alexis Biochemicals (San Diego, CA). Guanabenz was purchased from Research Biochemicals International (Natick, MA). Nickel-nitrilotriacetic acid (Ni-NTA)-agarose was from Qiagen Inc (Valencia, CA).
Expression and Purification of nNOS, Hsp90, Hsp70, Hsp40, CHIP, and GST-tagged UbcH5a
Rat nNOS was expressed in Sf9 insect cells using a recombinant baculovirus and purified by 2′,5′-ADP Sepharose and gel-filtration chromatography as described previously (8). Heme was added as an albumin conjugate during the expression to convert all of the nNOS to the holo-nNOS dimer (8). His-CHIP was bacterially expressed and purified by Ni-NTA affinity chromatography as previously described (28). GST-tagged UbcH5a (E2, ubiquitin carrier protein) was bacterially expressed and purified by GSH-Sepharose affinity chromatography as described (29). Hsp90 and Hsp70 were purified from rabbit reticulocyte lysate by sequential chromatography on DE52, hydroxylapatite, and ATP-agarose as described previously (30). YDJ-1, the yeast ortholog of Hsp40, was expressed in bacteria and purified by sequential chromatography on DE52 and hydroxylapatite as described previously (31).
In Vitro Ubiquitination of nNOS by Purified Ubiquitination System
Prior to ubiquitination, 2 μM purified nNOS was preincubated for 15 min at 30 °C with 4.4 μM Hsp90, 4.2 μM Hsp70, 0.1 μM Hsp40/YDJ-1, the indicated concentration of calmodulin with 0.5 mM CaCl2, and 2 μl of an ATP-generating system (50 mM ATP, 250 mM creatine phosphate, 20 mM magnesium acetate, and 100 units/ml creatine phosphokinase) in a total volume of 20 μl of HKD buffer (10 mM Hepes, pH 7.4, 100 mM KCl, and 5 mM DTT). The reaction mixture was placed on ice and diluted 2-fold with HKD buffer. An aliquot (5 μl) of this reaction mixture was added to an ubiquitination reaction mixture containing ubiquitin activating enzyme (0.1 μM), GST-tagged UbcH5a (0.6 μM), His-tagged CHIP (4.0 μM), GST-tagged ubiquitin (8.3 μM), 1 mM DTT, and 10 mM ATP. For the experiment shown in Figure 4, an aliquot (5 μl) of the first reaction mixture was added to reticulocyte lysate DE52 Fraction A (30) at a final concentration of 5.5 mg/ml, 0.3 mg/ml BSA, 0.8 μM ubiquitin-aldehyde, 0.66 mM N-Acetyl-Leu-Leu-Nle-CHO, 8.3 μM GST-tagged ubiquitin, 1 mM DTT, and 10 mM ATP. The mixtures were incubated for 1 h at 30 °C in a total volume of 20 μl of 50 mM Tris-HCl, pH 7.5. After incubation, 20 μl of sample buffer was added and an aliquot (20 μl) was loaded for Western blotting.
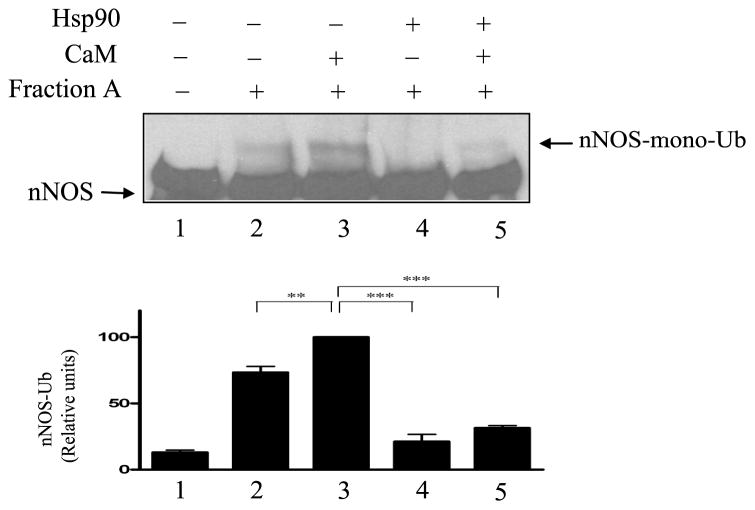
FIGURE 4.
Calmodulin does not inhibit and Hsp90 does inhibit nNOS ubiquitination by Fraction A of reticulocyte lysate. nNOS was preincubated with 30 μM CaM and/or Hsp90 as indicated and then incubated with Hsp90-free Fraction A of reticulocyte lysate. The samples were Western blotted by probing with anti-nNOS.
In experiments examining the effect of guanabenz, a mechanism-based inactivator, on nNOS ubiquitination, the enzyme was preincubated with the drug under conditions required for enzyme activity, but with low biopterin, necessitating the following incubation protocol. Purified nNOS (0.75 μM) was preincubated at 30 °C for 20 min with 1.7 μM Hsp70, 0.04 μM Hsp40, 0.1 μM tetrahydrobiopterin, 0.2 mM CaCl2, 8 μM calmodulin, an NADPH-generation system (consisting of 1 mM NADP+, 25 mM glucose-6-phosphate, and 2.5 unit/ml of glucose-6-phosphate dehydrogenase, expressed as final concentrations), and the ATP-generation system in the presence or absence of 2.6 μM Hsp90, 100 μM guanabenz, 100 μM D-NNA, or 100 μM L-NNA adjusted to a total volume of 50 μl with HKD buffer. Guanabenz inactivates nNOS by oxidation of enzyme-bound tetrahydrobiopterin. Thus, we used low concentrations of the biopterin as excess tetrahydrobiopterin, which can replace the oxidized pterin, greatly slows the inactivation process (21). In experiments examining the mechanism-based inactivator NAA, the enzyme was preincubated under the same conditions except that biopterin was present at 10 μM. Ten microliter of the preincubation was added to 20 μl of the in vitro ubiquitination mixture for a further incubation of 1 hour at 30 °C as described above, and the ubiquitin conjugates of nNOS were detected by Western blotting.
In Vitro Ubiquitination of nNOS by DE52-retained Fraction of Reticulocyte Lysate
The DE52-retained fraction of rabbit reticulocyte lysate was prepared as described previously (32). Purified nNOS (2 μM) was prebound with calmodulin at the indicated concentrations in the presence of 200μM CaCl2 in a total volume of 100 μl of HKD buffer. An aliquot (5 μl) of this mixture was incubated for 1 h at 37 °C with 4.5 μl of DE52-retained fraction (final concentration 7 mg protein/ml), 0.3 mg/ml bovine serum albumin, 8.3 μM GST-tagged ubiquitin, 1 mM dithiothreitol, 10 mM ATP/Mg2+, 1 μl of Complete Mini protease inhibitor cocktail, 0.6 mM N-Acetyl-Leu-Leu-Nle-CHO, and 0.8 μM ubiquitin aldehyde (deubiquitination inhibitor), adjusted to a final volume of 20 μl with 1 mM Tris, pH 7.5. Incubations were terminated by boiling with an equal volume of SDS-sample buffer containing 8 M urea and 2 M thiourea.
Assay of nNOS Activity
NO formation was assayed by the NO-mediated conversion of oxyhemoglobin to methemoglobin. Conversion of oxyhemoglobin was assayed by adding 8 μl of nNOS preincubation mixture to an assay solution containing 200 μM CaCl2, 250 μM L-arginine, 100 μM tetrahydrobiopterin, 100 units/ml catalase, 5 μg/ml calmodulin, 25 μM oxyhemoglobin, and an NADPH-generating system composed of 0.4 mM NADP+, 10 mM glucose 6-phosphate, and 1 unit of glucose 6-phosphate dehydrogenase/ml (expressed as final concentrations) in a total volume of 180 μl of 50 mM potassium phosphate, pH 7.4. The mixture was incubated 4 min at 37 °C and the rate of oxidation of oxyhemoglobin was monitored by measuring the absorbance at λ401–411 nm with a microtiter plate reader.
Gel Electrophoresis and Western Blotting
Aliquots (10 μl) from the ubiquitination reactions were boiled in SDS sample buffer (3.75% SDS, 15% glycerol, 6 mg/ml DTT, and 0.02% bromophenol blue in 125 mM Tris-HCl, pH 6.8), resolved on 5% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with anti-nNOS (1:8000). Immunoreactive bands were visualized with the use of enhanced chemiluminescence reagent (Super Signal, Pierce) and X-Omat film (Eastman Kodak Co.). Either the monoubiquitinated nNOS bands (experiments with reticulocyte lysate) or the polyubiquitinated nNOS bands (experiments with purified ubiquitinating system) were scanned and the relative densities were determined with ImageJ software (http://rsb.info.nih.gov/ij/). Relative densities for 3 experiments are presented in bar graphs as percent of control or percent of the condition with the greatest ubiquitination ± S.E. Significance of difference was determined by one-way ANOVA (Tukey’s Multiple Comparison Test). Statistical probability is expressed as *p<0.05, **p<0.01, ***p<0.001.
RESULTS
Calmodulin Inhibits nNOS Ubiquitination by the DE52-retained Fraction of Reticulocyte Lysate
We have previously reported that nNOS is ubiquitinated in vivo and that the predominant nNOS-ubiquitin conjugate detected in human embryonic kidney cells and in rat brain cytosol is the monoubiquitinated form (27). This ubiquitination was mimicked in vitro by incubating purified nNOS with an extract of rabbit reticulocyte lysate, ubiquitin and ATP (27). Studies with 125I-ubiquitin showed that the mono-ubiquitinated nNOS is the major product in these in vitro experiments (27). The extract of reticulocyte lysate consists of all of the lysate material that is retained by a DE52 column, and the DE52-retained fraction is the same as the lysate ‘fraction II’ that has been extensively used to study protein ubiquitination (32). The DE52-retained fraction contains Hsp90, Hsp70 and Hsp40, as well as the ubiquitinating enzymes, with all the components being present in the same ratios as exist in reticulocyte lysate (8,27).
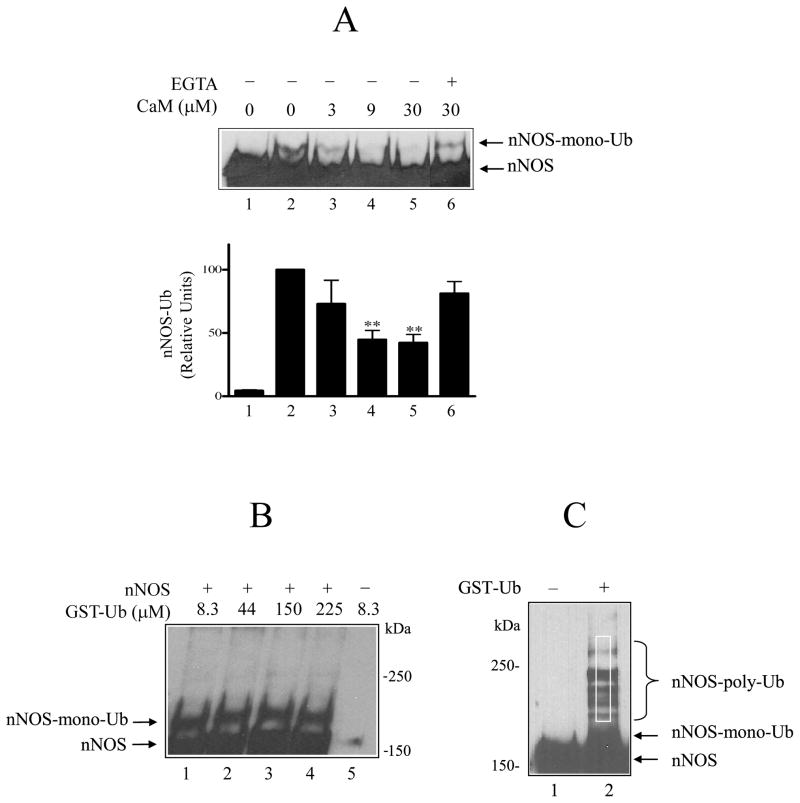
Because of the mutual interaction between calmodulin and Hsp90 reported for the NOS enzymes (11,12,14,15), we asked whether CaM would affect nNOS ubiquitination by the DE52-retained fraction of reticulocyte lysate. As shown in Fig. 1A, incubation of purified nNOS with the DE52-retained fraction and GST-ubiquitin yields a slower migrating band (lane 2) that has been previously demonstrated to be the major monoubiquitinated nNOS product (27). Addition of calmodulin inhibits nNOS ubiquitination (lanes 3, 4, and 5) in a calcium-dependent manner, as indicated by the increased ubiquitination seen in the presence of EGTA (lane 6). The amount of ubiquitinated product relative to the control without added calmodulin is presented for several experiments in the bar graph of Fig. 1A. It should be noted that because only a small fraction of nNOS is ubiquitinated, the assay contains a lot of the purified enzyme to serve as substrate, as is indicated by the broad bands of unmodified nNOS in Fig. 1A. Given the large amount of nNOS, we have tested the effect of CaM over a range of concentrations that reflect a reasonable stoichiometry with the enzyme. To give a point of reference, the ratio of CaM to nNOS in our enzyme activity assay is 11:1 and the highest concentration of CaM (30 μM) in the ubiquitination mixture yields a ratio of 15:1. As shown in Fig. 1B, we have verified that the mono-ubiquitinated nNOS is the predominant form even when the ubiquitin concentration is increased (lanes 1–4) and that the bands are specific for nNOS (lane 5).
FIGURE 1.
Calmodulin inhibition of nNOS ubiquitination by the DE52-retained fraction of rabbit reticulocyte lysate. A, nNOS monoubiquitin conjugates (nNOS-mono-Ub) were detected by Western blot. nNOS was incubated for 1 h at 37 °C with a DE52-retained fraction of rabbit reticulocyte lysate, ATP, GST-ubiquitin and the indicated concentrations of calmodulin as described under Methods. Samples were Western blotted by probing with anti-nNOS. Lane 1, incubation time 0; lanes 2–6, incubation time 1 h. For bar graph, the relative amount of nNOS-Ub in replicate experiments as in A was determined by scanning and expressed as % of the 1 h control without calmodulin. The values are the mean ± S.E. (n=3). ** denotes significantly (p<0.01) lower nNOS-Ub conjugates relative to the 1 h control without CaM. B, the effect of increasing the ubiquitin concentration on the ubiquitination of nNOS. The studies were as in A, except that the indicated concentrations of GST-ubiquitin were used (lanes 1–4). Lane 5, nNOS was omitted. The data are representative of three identical experiments. C, nNOS polyubiquitin conjugates (nNOS-poly-Ub) produced by the purified ubiquitinating enzyme mixture without (lane 1) or with (lane 2) GST-ubiquitin. The white rectangle above the monoubiquitinated nNOS represents the region scanned to determine relative densities of polyubiquitination in experiments with the purified ubiquitination system.
Hsp90 Inhibits nNOS Ubiquitination by the Purified Ubiquitination System
We have previously shown that the Hsp70-dependent E3 ubiquitin ligase CHIP promotes nNOS ubiquitination both in human embryonic kidney cells and in a purified ubiquitination system (28). In contrast to reticulocyte lysate, where we see predominantly monoubiquitinated nNOS, the purified ubiquitination system is more active and produces easily detected, higher mass ubiquitinated nNOS products in addition to the monoubiquitinated form (28). As illustrated in Fig. 1C, the relative amounts of these polyubiquitinated products can be determined by comparing densities of scans of the area demarcated by white rectangle in lane 2. The monoubiquitinated nNOS lies below the scanned region, and it is fused with the unmodified nNOS at exposure times used to detect polyubiquitinated nNOS. Thus, conditions yielding high level of polyubiquitination will appear to have thicker unubiquitinated/monoubiquitinated nNOS bands than conditions where there is less polyubiquitination, even though the amount of nNOS loaded on each lane is the same.
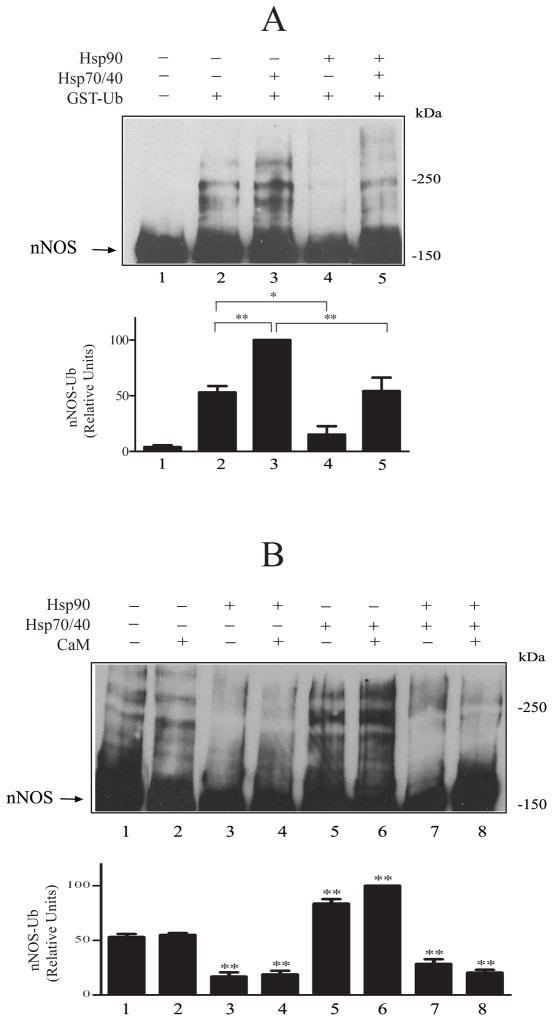
In Fig. 2, we use the purified system to assess the effects of both the protein chaperones and Ca2+/calmodulin on CHIP-dependent polyubiquitination. A small amount of insect Hsp70 copurifies with nNOS (8), and this may account for the basal level of ubiquitination that occurs in the absence of added Hsp70 (e.g., Fig. 2A, lane 2). Nevertheless, addition of Hsp70 increases nNOS ubiquitination (Fig. 2A, lane 3), and the presence of Hsp90 in the preincubation mix inhibits both basal (lane 4) and Hsp70-stimulated (lane 5) ubiquitination. It is important to note that the concentrations of purified chaperone proteins used here are the same as those used in our five-chaperone system for steroid receptor activation, and these are the concentrations of Hsp90 and Hsp70 that are present in reticulocyte lysate (33).
FIGURE 2.
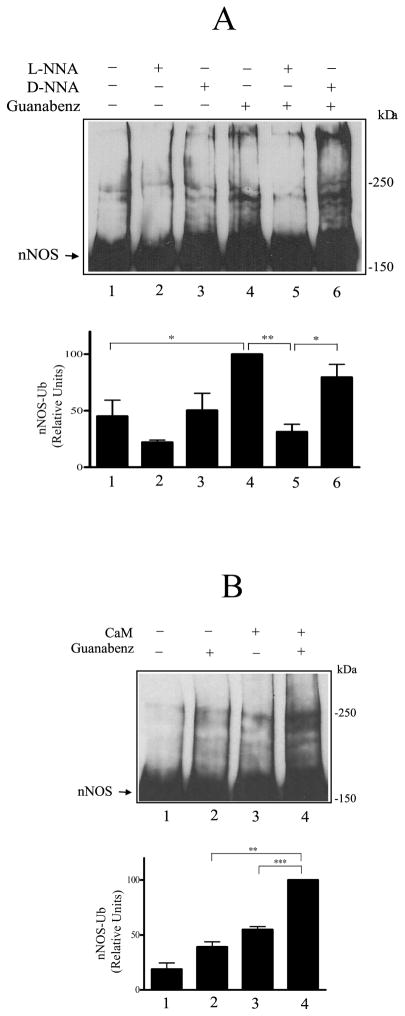
Hsp90 inhibits basal and Hsp70-stimulated nNOS ubiquitination. A, Hsp90 inhibition of ubiquitination. nNOS was preincubated with Hsp70, Hsp40 and Hsp90 as indicated and then incubated with the ubiquitinating enzyme mixture. The samples were Western blotted by probing with anti-nNOS. Arrow points to the unubiquitinated/monoubiquitinated nNOS band. The relative densities in the bar graph are presented as a % of the Hsp70-stimulated value in lane 3. B, Hsp90 inhibits ubiquitination in the absence and presence of calmodulin. nNOS was preincubated with Hsp70/Hsp40, Hsp90 and 30 μM calmodulin as indicated and then incubated with the ubiquitination mixture, followed by immunoblotting as above. The relative densities in the bar graph are presented as a % of the highest condition in lane 6.
In Fig. 2B, inhibition of ubiquitination by Hsp90 is again examined on basal and Hsp70-stimulated nNOS ubiquitination, but in the presence and absence of 30 μM CaM. Although CaM alone does not inhibit nNOS ubiquitination (lanes 2 and 6), Hsp90 alone causes extensive inhibition (lanes 3 and 7). Because Hsp90 inhibition of ubiquitination in this case is essentially complete in the absence of CaM (lanes 3 and 7), no additional effect of CaM on Hsp90-dependent inhibition can be seen. As shown in Fig. 3, however, when the inhibition by Hsp90 is somewhat less (cf. lanes 1 and 2), increasing concentrations of CaM yield increased inhibition (lanes 3–5) that is calcium-dependent, as indicated by the increased ubiquitination seen in the presence of EGTA (lane 6). The extent of inhibition by Hsp90 is consistent as long as the same preparation of Hsp70 and Hsp90 are used, however, the extent of inhibition can vary between 35–65% depending on the preparation of purified protein used.
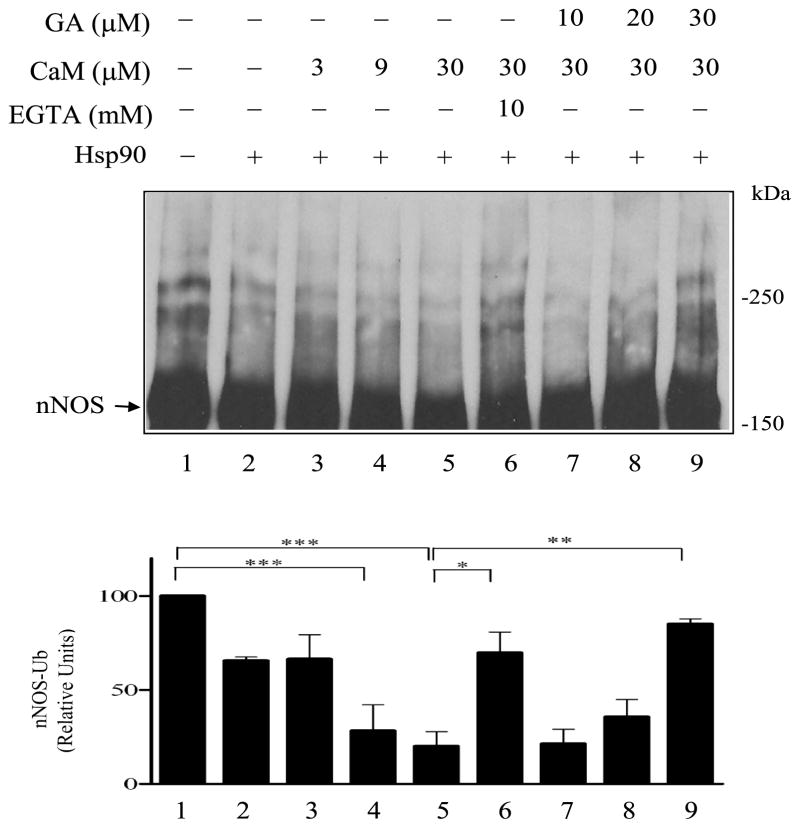
FIGURE 3.
Calmodulin enhances Hsp90 inhibition of nNOS ubiquitination. nNOS was preincubated with Hsp70/Hsp40 in the presence or absence of Hsp90, CaM, EGTA, or geldanamycin (GA) as noted at the top of the figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted by probing with anti-nNOS.
It has been shown that the enhancement of NO synthesis by purified nNOS caused by purified Hsp90 is inhibited by geldanamycin (15). As shown in Fig. 3 (lanes 7–9), the Hsp90 inhibitor geldanamycin also reduces Hsp90 inhibition of nNOS ubiquitination. Although previous studies of binding of purified Hsp90 to eNOS and nNOS and the activation of eNOS and nNOS activity by purified Hsp90 were carried out in the absence of ATP (7,14,15), both Hsp70 and the ubiquitination system require ATP; thus, our experiments are performed in the presence of an ATP generating system.
The results with the purified ubiquitination system in Figs. 2 and 3 suggest that the inhibition of nNOS ubiquitination observed when CaM is added to the DE52 fraction of reticulocyte lysate in Fig. 1 is caused by Hsp90. To demonstrate this, we prepared an Hsp90-free pool of reticulocyte lysate fractions eluted from DE52 with a gradient of KCl, as we described in Dittmar et al. (30). The pooled fractions that elute before Hsp90 contain Hsp70, Hsp40 and nNOS ubiquitinating activity (Fig. 4). We have called this Hsp90-free pool Fraction A (30). As shown in Fig. 4, addition of CaM to Fraction A increases nNOS ubiquitination (cf. lanes 2 and 3), whereas addition of Hsp90 inhibits ubiquitination (lanes 4 and 5).
Hsp90 Inhibits Guanabenz-induced nNOS Ubiquitination
To determine if Hsp90 inhibits ubiquitination triggered by specific attack within the heme/substrate binding cleft, we looked at guanabenz-induced ubiquitination. In Fig. 5A, purified nNOS was preincubated with Hsp70/40 and guanabenz under conditions required for enzyme activity. The slowly reversible nNOS inhibitor L-NNA or its inactive isomer D-NNA were present in samples to demonstrate stereospecific competition for guanabenz stimulation of ubiquitination. As shown in Fig. 5A, guanabenz increased nNOS ubiquitination (cf. lanes 1 and 4), and L-NNA prevented the guanabenz effect (lane 5) whereas D-NNA did not (lane 6). This shows that guanabenz must enter the heme/substrate binding cleft to cause an increase in ubiquitination.
FIGURE 5.
Guanabenz causes a mechanism-based ubiquitination of nNOS. A, guanabenz acts in the heme/substrate binding cleft. nNOS was preincubated with Hsp70/40 under conditions required for enzyme activity (see Methods) and in the presence of 100 μM guanabenz, 100 μM D-NNA, or 100 μM L-NNA as noted at the top of the figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted with anti-nNOS. B, guanabenz-dependent ubiquitination requires nNOS enzymatic activity. nNOS was preincubated with Hsp70/40 under conditions for full enzyme activity or minus calmodulin (no enzyme activity) in the presence or absence of guanabenz as noted at the top of the figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted.
CaM is required for nNOS enzymatic activity, and in Fig. 5B, nNOS was preincubated as in Fig. 5A with or without CaM to yield catalytically active and inactive nNOS, respectively. Guanabenz does not cause an increase in nNOS ubiquitination when the enzyme is catalytically inactive (Fig. 5B, lane 2) but it increases ubiquitination when it is catalytically active (lane 4). These observations show that guanabenz must enter the heme/substrate binding cleft where it causes a mechanism-based change that leads to nNOS ubiquitination.
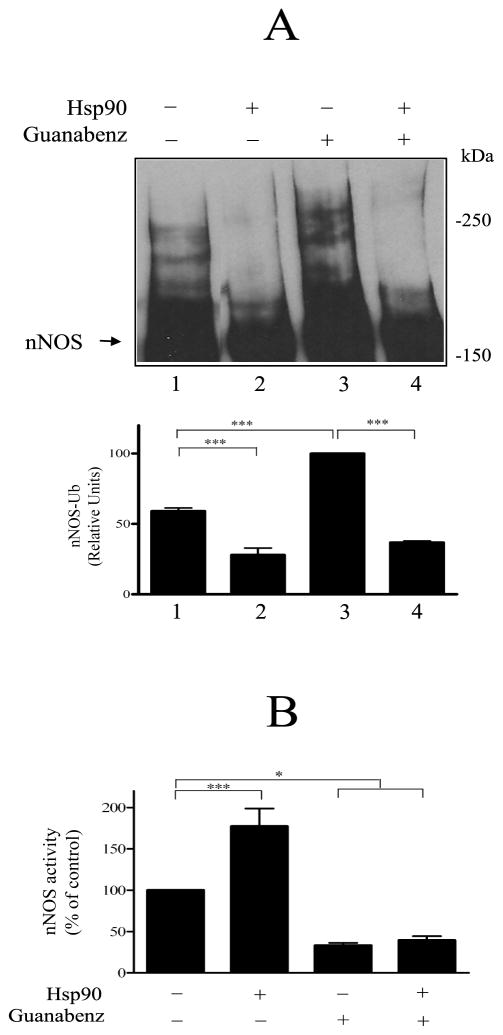
As shown in Fig. 6A, both basal ubiquitination (lane 1) and the increased ubiquitination that ensues from guanabenz-mediated alteration within the heme/substrate binding cleft (lane 3) are inhibited by Hsp90 (lanes 2 and 4). In contrast, Hsp90 does not affect guanabenz-mediated inhibition of nNOS enzymatic activity. As shown in Fig. 6B, in the absence of guanabenz, Hsp90 causes the previously demonstrated (14,15) enhancement of nNOS activity (lanes 1 and 2). However, preincubation with guanabenz inhibits nNOS activity whether or not Hsp90 is present (Fig. 6B, lanes 3 and 4).
FIGURE 6.
Hsp90 inhibits the guanabenz-triggered ubiquitination of nNOS. A, inhibition of ubiquitination. nNOS was preincubated with Hsp70/40 under conditions required for enzyme activity and in the presence of guanabenz and/or Hsp90 as noted at the top of figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted for nNOS. B, guanabenz inhibition of nNOS activity. nNOS was preincubated as in A and nNOS activity was assayed by measuring the NO-mediated conversion of oxyhemoglobin to methemoglobin as described under Methods. Bars represent mean ± S.E. for three experiments.
Hsp90 Inhibits NAA-induced nNOS Ubiquitination
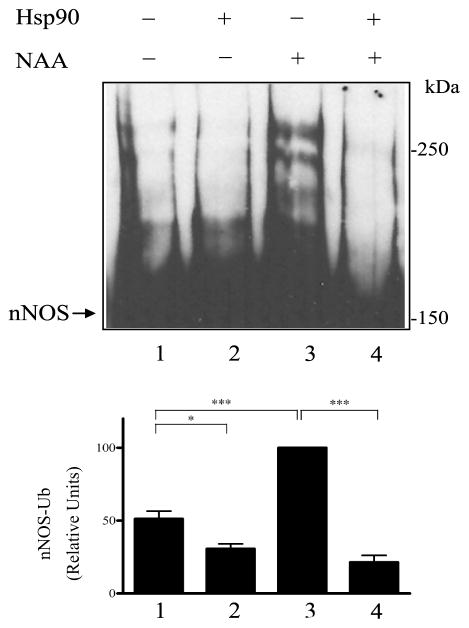
NAA (NG-amino-L-arginine) is a metabolism-based inactivator of nNOS that targets the heme moiety (34). Covalent alteration of the heme by NAA leads both to heme adducts that are dissociable and to heme adducts that are irreversibly bound to a residue in the nNOS oxygenase domain (23). To determine if Hsp90 inhibits nNOS ubiquitination triggered in this manner, nNOS was preincubated under catalytic conditions with NAA in the presence or absence of Hsp90. As shown in Fig. 7, nNOS incubated with NAA (lane 3) was ubiquitinated much more than the untreated enzyme (lane 1) and NAA-induced ubiquitination was prevented by Hsp90 (lane 4). Taken together, the data of Figs. 6 and 7 show that alteration of the heme/substrate binding cleft by either loss of the tetrahydrobiopterin cofactor or heme adduct formation leads to nNOS ubiquitination that is inhibited by Hsp90.
FIGURE 7.
Hsp90 inhibits NAA-triggered ubiquitination of nNOS. nNOS was preincubated with Hsp70/40 under conditions required for enzyme activity (see Methods) in the presence of 100 μM NAA and/or Hsp90 as noted at the top of the figure. The samples were then incubated with the ubiquitinating enzyme mixture and Western blotted for nNOS.
DISCUSSION
It is clear from Fig. 1A that nNOS ubiquitination by the reticulocyte lysate system is inhibited by Ca2+/calmodulin. However, addition of CaM alone to the purified CHIP-dependent ubiquitination system does not yield inhibition of ubiquitination (Fig. 2B). Hsp90 inhibits ubiquitination by the purified ubiquitinating system when added alone (Fig. 2), and the simultaneous presence of CaM increases this inhibition (Fig. 3). This suggests that the inhibition of nNOS ubiquitination observed in the reticulocyte lysate system when CaM is added is due to Hsp90. When nNOS is ubiquitinated by an Hsp90-free fraction of reticulocyte lysate, addition of CaM does not inhibit nNOS ubiquitination but addition of purified Hsp90 does inhibit (Fig. 4). The fact that CaM interaction with nNOS enhances both Hsp90 stimulation of nNOS activity (14,15) and Hsp90 inhibition of nNOS ubiquitination is consistent with a model in which the two effects of Hsp90 are caused by the same interaction of the chaperone with the enzyme.
It is established that Hsp90 binds to the oxygenase domain of eNOS (35). This domain contains the heme/substrate binding cleft, and it is the likely site of Hsp90 interaction with nNOS as well. Inasmuch as CaM enhances electron flux from flavin bound to the reductase domain to heme bound within the cleft (36), CaM binding is likely to affect the state of the cleft. Although it is unclear exactly how CaM affects cleft structure/mobility, it is clear that CaM and Hsp90 affect both eNOS and nNOS in such a manner as to increase the binding of the other (11,12,14,15).
Here, we have used two mechanism-based inactivators, guanabenz and NAA, to alter the heme/substrate binding cleft and promote nNOS ubiquitination (Figs. 5–7). Both basal and inactivator-stimulated ubiquitination are inhibited by Hsp90 (Figs 6 and 7). It is this ability to inhibit ubiquitination that likely accounts for Hsp90 stabilization of a wide variety of proteins to proteasomal degradation. We have previously proposed that Hsp90 acts to stabilize an open state of the ligand binding cleft, an effect that both facilitates substrate access to increase enzyme activity and prevents further cleft unfolding that triggers Hsp70-dependent ubiquitination (5,6).
U-box E3 ligases, such as CHIP, are thought to act as bridging proteins to bring the ubiquitin-charged E2 enzyme into the vicinity of the substrate. It is not known if CHIP itself contacts the substrate and it is thought that CHIP is targeted to the unfolding protein by the chaperone (37). CHIP binds via an amino-terminal TPR domain to both Hsc/Hsp70 and Hsp90 (38,39), and it has been suggested that both chaperones may have the ability to target substrates for degradation (3,39). Several reports have concluded that an Hsp90-CHIP complex selectively degrades various Hsp90 client proteins. It is quite clear, however, that Hsp70 promotes CHIP-dependent ubiquitination of nNOS (28, Fig. 2) and that Hsp90 inhibits ubiquitination (Figs. 2,3,6,7). This Hsp90 inhibition is consistent with hundreds of reports that treatment of cells with Hsp90 inhibitors, such as geldanamycin, promotes the proteasomal degradation of various Hsp90-regulated proteins. Thus, inhibition of Hsp90 cycling with a protein permits unopposed activity of chaperone-dependent E3 ligases that bind to substrate-bound Hsp70 (5.6).
It has not been known how proteins that have undergone oxidative or toxic damage are recognized and shunted to the ubiquitin-proteasome pathway of degradation. The effects of guanabenz and NAA serve as examples of such toxic damage that is targeted to the ligand binding cleft and triggers ubiquitination of nNOS. Other examples of mechanism-based protein damage triggering ubiquitination and proteasomal degradation have been discussed in a review (5). It is reasonable to propose that, as proteins undergo such toxic damage, ligand binding clefts open, exposing hydrophobic residues as the initial step in unfolding. As long as Hsp90 can form even transient complexes with the opening cleft, ubiquitination by Hsp70-dependent ubiquitin E3 ligases, like CHIP, is inhibited. But a point is reached where unfolding of the cleft progresses to a state that cannot cycle with Hsp90, and ubiquitination directed by Hsp70-dependent E3 ligases is unopposed. In this way, the Hsp70/Hsp90 chaperone machinery may be the major mechanism by which the quality control decision is made for degradation of damaged proteins via the ubiquitin-proteasome pathway.
Footnotes
Abbreviations: Hsp, heat shock protein; NOS, nitric oxide synthase; nNOS, neuronal NOS; eNOS, endothelial NOS; iNOS, inducible NOS; CHIP, carboxyl terminus of Hsp70-interacting protein; E1, ubiquitin activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; CaM, Ca2+/calmodulin; NNA, NG-nitro-L-arginine; NAA, NG-amino-L-arginine; Ni-NTA, nickel-nitrilotriacetic acid; GST, glutathione-S-transferase; GA, geldanamycin.
This work was supported by National Institutes of Health Grants GM077430 and DA022354 (to Y.O.)
References
- 1.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;222:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 3.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 4.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pratt WB, Morishima Y, Osawa Y. The Hsp90 Chaperone Machinery Acts at Protein Folding Clefts to Regulate Both Signaling Protein Function and Protein Quality Control. In: Calderwood SK, Sherman MY, Ciocca DR, editors. Heat Shock Proteins in Cancer. Springer-Verlag; Dordrecht, The Netherlands: 2007. pp. 1–30. [Google Scholar]
- 6.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 8.Bender AT, Silverstein AM, Demady DR, Kanelakis KC, Noguchi S, Pratt WB, Osawa Y. Neuronal nitric oxide synthase is regulated by the hsp90-based chaperone system in vivo. J Biol Chem. 1999;274:1472–1478. doi: 10.1074/jbc.274.3.1472. [DOI] [PubMed] [Google Scholar]
- 9.Brouet A, Sonveaux P, Dessy C, Ballingand JL, Feron O. Hsp90 ensures the transition from the early Ca2+-dependent to the late phosphorylation-dependent activation of the endothelial nitric-oxide synthase in vascular endothelial growth factor-exposed endothelial cells. J Biol Chem. 2001;276:32663–32669. doi: 10.1074/jbc.M101371200. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida M, Xia Y. Heat shock protein 90 as an endogenous protein enhancer of inducible nitric oxide synthase. J Biol Chem. 2003;278:36953–36958. doi: 10.1074/jbc.M305214200. [DOI] [PubMed] [Google Scholar]
- 11.Gratton JP, Fontana J, O’Connor DS, Garcia-Cardena G, McCabe TJ, Sessa WC. Reconstitution of an endothelial nitric-oxide synthase (eNOS), hsp90, and caveolin-1 complex in vitro, evidence that hsp90 facilitates calmodulin stimulated displacement of eNOS from caveolin-1. J Biol Chem. 2000;275:22268–22272. doi: 10.1074/jbc.M001644200. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S, Mendelsohn ME. Calmodulin-dependent and -independent activation of endothelial nitric-oxide synthase by heat shock protein 90. J Biol Chem. 2003;278:9339–9344. doi: 10.1074/jbc.M212651200. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi S, Mendelsohn ME. Synergistic activation of endothelial nitric-oxide synthase (eNOS) by HSP90 and Akt. J Biol Chem. 2003;278:30821–30827. doi: 10.1074/jbc.M304471200. [DOI] [PubMed] [Google Scholar]
- 14.Song Y, Zweier JL, Xia Y. Heat shock protein 90 augments neuronal nitric oxide synthase by enhancing Ca2+/calmodulin binding. Biochem J. 2001;355:357–360. doi: 10.1042/0264-6021:3550357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Zweier JL, Xia Y. Determination of the enhancing action of HSP90 on neuronal nitric oxide synthase by EPR spectroscopy. Am J Physiol. 2001;281:C1819–C1824. doi: 10.1152/ajpcell.2001.281.6.C1819. [DOI] [PubMed] [Google Scholar]
- 16.Billecke SS, Bender AT, Kanelakis KC, Murphy PJM, Lowe ER, Kamada Y, Pratt WB, Osawa Y. Hsp90 is required for heme binding and activation of apo-neuronal nitric-oxide synthase, geldanamycin-mediated oxidant generation is unrelated to any action of hsp90. J Biol Chem. 2002;277:20504–20509. doi: 10.1074/jbc.M201940200. [DOI] [PubMed] [Google Scholar]
- 17.Billecke SS, Dragonov DI, Morishima Y, Murphy PJM, Dunbar AY, Pratt WB, Osawa Y. The role of hsp90 in the heme-dependent activation of apo-neuronal nitric-oxide synthase. J Biol Chem. 2004;279:30252–30258. doi: 10.1074/jbc.M403864200. [DOI] [PubMed] [Google Scholar]
- 18.Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 19.Noguchi S, Jianmongkol S, Bender AT, Kamada Y, Demady DR, Osawa Y. Guanabenz-mediated inactivation and enhanced proteolytic degradation of neuronal nitric oxide synthase. J Biol Chem. 2000;275:2376–2380. doi: 10.1074/jbc.275.4.2376. [DOI] [PubMed] [Google Scholar]
- 20.Nakatsuka M, Nakatsuka K, Osawa Y. Metabolism based inactivation of penile nitric oxide synthase activity by guanabenz. Drug Metab Disp. 1998;26:497–501. [PubMed] [Google Scholar]
- 21.Dunbar AY, Jenkins GJ, Jianmongkol S, Nakatsuka M, Lowe ER, Lau M, Osawa Y. Tetrahydrobiopterin protects against guanabenz-mediated inhibition of neuronal NO synthase in vitro and in vivo. Drug Metab Disp. 2006;34:1448–1456. doi: 10.1124/dmd.106.009951. [DOI] [PubMed] [Google Scholar]
- 22.Kamada Y, Jenkins GJ, Lau M, Dunbar AY, Lowe ER, Osawa Y. Tetrahydrobiopterin depletion and ubiquitylation of neuronal NO-synthase. Mol Brain Res. 2005;142:19–27. doi: 10.1016/j.molbrainres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Vuletich JL, Lowe ER, Jianmongkol S, Kamada Y, Kent UM, Bender AT, Demady DR, Hollenberg PF, Osawa Y. Alteration of the heme prosthetic group of neuronal nitric-oxide synthase during inactivation by NG-amino-L-arginine in vitro and in vivo. Mol Pharmacol. 2002;62:110–118. doi: 10.1124/mol.62.1.110. [DOI] [PubMed] [Google Scholar]
- 24.Osawa Y, Lowe ER, Everett AC, Dunbar AY, Billecke SS. Proteolytic degradation of nitric oxide synthase: effect of inhibitors and role of hsp90-based chaperones. J Pharmacol Exptl Ther. 2003;304:493–497. doi: 10.1124/jpet.102.035055. [DOI] [PubMed] [Google Scholar]
- 25.Osawa Y, Darbyshire JF, Steinbach PJ, Brooks BR. Metabolism-based transformation of myoglobin to an oxidase by BrCCl3 and molecular modeling of the oxidase form. J Biol Chem. 1993;268:2953–2959. [PubMed] [Google Scholar]
- 26.Osawa Y, Pohl LR. Covalent bonding of the prosthetic heme to protein: a potential mechanism for the suicide inactivation or activation of hemoproteins. Chem Res Toxicol. 1989;2:131–141. doi: 10.1021/tx00009a001. [DOI] [PubMed] [Google Scholar]
- 27.Bender AT, Demady DR, Osawa Y. Ubiquitination of neuronal nitric oxide synthase in vitro and in vivo. J Biol Chem. 2000;275:17407–17411. doi: 10.1074/jbc.M000155200. [DOI] [PubMed] [Google Scholar]
- 28.Peng HM, Morishima Y, Jenkins GJ, Dunbar AY, Lau M, Patterson C, Pratt WB, Osawa Y. Ubiquitylation of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J Biol Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Pickart CM. A HECT domain E3 enzyme assembles novel polyubiquitin chains. J Biol Chem. 2001;276:19871–19878. doi: 10.1074/jbc.M100034200. [DOI] [PubMed] [Google Scholar]
- 30.Dittmar KD, Hutchison KA, Owens-Grillo JK, Pratt WB. Reconstitution of the steroid receptor·hsp90 heterocomplex assembly system of reticulocyte lysate. J Biol Chem. 1996;271:12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- 31.Dittmar KD, Banach M, Galigniana MD, Pratt WB. The role of DnaJ-like proteins in glucocorticoid receptor·hsp90 heterocomplex assembly by the reconstituted hsp90·p60·hsp70 foldosome complex. J Biol Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 32.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system: resolution, affinity purification and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 33.Morishima Y, Kanelakis KC, Murphy PJM, Lowe ER, Jenkins GJ, Osawa Y, Sunahara RK, Pratt WB. The hsp90 cochaperone p23 is the limiting component of the multiprotein hsp90/hsp70-based chaperone system in vivo where it acts to stabilize the client protein·hsp90 complex. J Biol Chem. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- 34.Bryk R, Lubeskie A, Wolff DJ. Studies of neuronal nitric oxide synthase inactivation by diverse suicide inhibitors. Arch Biochem Biophys. 1999;369:243–251. doi: 10.1006/abbi.1999.1340. [DOI] [PubMed] [Google Scholar]
- 35.Fontana J, Fulton D, Chen Y, Fairchild TA, McCabe TJ, Fujita N, Tsuruo T, Sessa WC. Molecular mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ Res. 2002;90:866–873. doi: 10.1161/01.res.0000016837.26733.be. [DOI] [PubMed] [Google Scholar]
- 36.Abu-Soud HM, Stuehr DJ. Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc Natl Acad Sci USA. 1993;90:10769–10772. doi: 10.1073/pnas.90.22.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hohfeld J, Cyr DM, Patterson C. From cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]