Abstract
One challenge in aging research concerns identifying physiological parameters or biomarkers that can reflect the physical health of an animal and predict its lifespan. In C. elegans, a model organism widely used in aging research, motor deficits develop in old worms. Here we employed machine vision to quantify worm locomotion behavior throughout lifespan. We confirm that aging worms undergo a progressive decline in motor activity, beginning in early life. Importantly, the rate of motor activity decline rather than the absolute motor activity in the early-to-mid life of individual worms in an isogenic population inversely correlates with their lifespan, and thus may serve as a lifespan predictor. Long-lived mutant strains with deficits in insulin/IGF-1 signaling or food intake display a reduction in the rate of motor activity decline, suggesting that this parameter might also be used for across-strain comparison of healthspan. Our work identifies an endogenous physiological parameter for lifespan prediction and healthspan comparison.
Introduction
Aging is characterized by progressive declines in multiple physiological functions leading to death. Studies in the past decade using model organisms (e.g. worms, flies and mice) have demonstrated that aging can be modulated by both genetic and environmental factors [1,7,10,19]. It also becomes clear that stochastic factors affect longevity, as animals bearing the same genetic background and reared under the same environment show variation in lifespan and rate of tissue aging [11,12]. This makes the chronological age an unreliable parameter to reflect the physical health or the biological age of an animal. Thus, to better understand how genes and environmental cues modulate the rate of aging, it would be important to search for physiological parameters or biomarkers that can be correlated with the biological age of an individual [17]. In particular, it would be highly valuable if such parameters can also be used to predict lifespan [17]. Clearly, it is more desirable to make lifespan predictions at the early rather than the late stage of aging. However, no reliable lifespan predictor has been identified in mammals [17].
C. elegans has emerged as a powerful model organism for the study of aging. A recent study has reported an ectopically-expressed marker that can predict worm lifespan at the early stage of aging [29]. Nevertheless, no similar endogenous lifespan predictors have been identified in worms. Locomotion behavior represents one of the most prominent behaviors in C. elegans [4]. It is known that old worms develop motor deficits and are less active than young worms [5,9,13,14,16,18]. Indeed, the expression of motor activity has been used as a common standard in lifespan analysis to score whether a worm is alive [20]. Previous studies have attempted to correlate the behavioral states of worm locomotion with the biological age of the animal, and some qualitative or semi-quantitative parameters have been proposed to predict lifespan at the late or mid-to-late rather than early stage of aging [13,14,18]. For example, there appears to be a correlation between the lifespan of a worm and the number of days during which it can maintain active movement [14]. However, because these methods all rely on human observation/description rather than machine vision, it is difficult to extract reliable behavioral parameters in a quantitative, objective manner. In addition, motor activity displays great variation between different strains and some strains are less active than others, thus making it difficult to conduct across-strain comparison.
Here we employed machine vision to quantify locomotion behavior of aging worms throughout lifespan. We identify the rate of motor activity decay as an endogenous physiological parameter that can predict lifespan at the early-to-mid stage of aging. We also present evidence that this parameter may potentially be used for across-strain comparison of healthspan.
Methods
Worms were analyzed for spontaneous locomotion behavior on NGM plates every other day throughout lifespan using an automated worm tracking system as previously described [6,24]. In brief, NGM plates were spread with a thin layer of freshly-grown bacteria (OP50) five minutes prior to tracking. Tracking was performed at 20°C and at a relative humidity of ∼40% with the lid off. The tracking system consists of a stereomicroscope (Zeiss Stemi 2000C) mounted with a digital camera (Cohu 7800) and a digital motion system (Parker Automation) that follows worm movement. The system is controlled by laboratory-developed software. To record locomotion, worm images were captured at 2 Hz for 5 mins and compressed into AVI files, and the mean centroid velocity of each worm was quantified using laboratory-developed software.
Lifespan analysis was conducted at 20°C as described previously [20]. In all experiments, the first day of adulthood was scored as day 1. When analyzing N2 worms individually, worms were grown separately on NGM plates (one per plate). Each worm was scored for its viability every day, and the same worm was assayed for behavioral analysis every other day until death. In experiments involving multiple strains, 360 worms of each strain were grown on NGM plates (12 per plate), and one animal from each plate (total of 30 plates) was randomly picked for behavioral analysis. Worms that crawled off the plate, exploded or bagged were censored at the time of the event. All statistical analyses were done using the Statistica (StatSoft, Inc.) and Statview 5.01 (SAS) software.
Results and discussion
Worms exhibit a progressive decline in motor activity, beginning in early life
The observation that old worms are less active than young worms suggests a correlation between locomotion behavior and aging [5,9,13,14,16,18]. In addition, the stochastic nature of age-associated motor activity decline makes it possible to predict lifespan by defining physiological parameters in locomotion behavior [12]. To explore this possibility, we quantified spontaneous locomotion behavior of 169 wild-type worms individually every other day throughout their lifespan. To do so, we utilized an automated worm tracking system that records worm movement and provides quantitative measurement of its activity such as locomotion speed in real time [6,24]. We found that the locomotion speed of these worms exhibited an exponential decay beginning as early as day 5 of adulthood (Figure 1A). These results indicate that aging worms undergo a progressive decline in motor activity, beginning in early life.
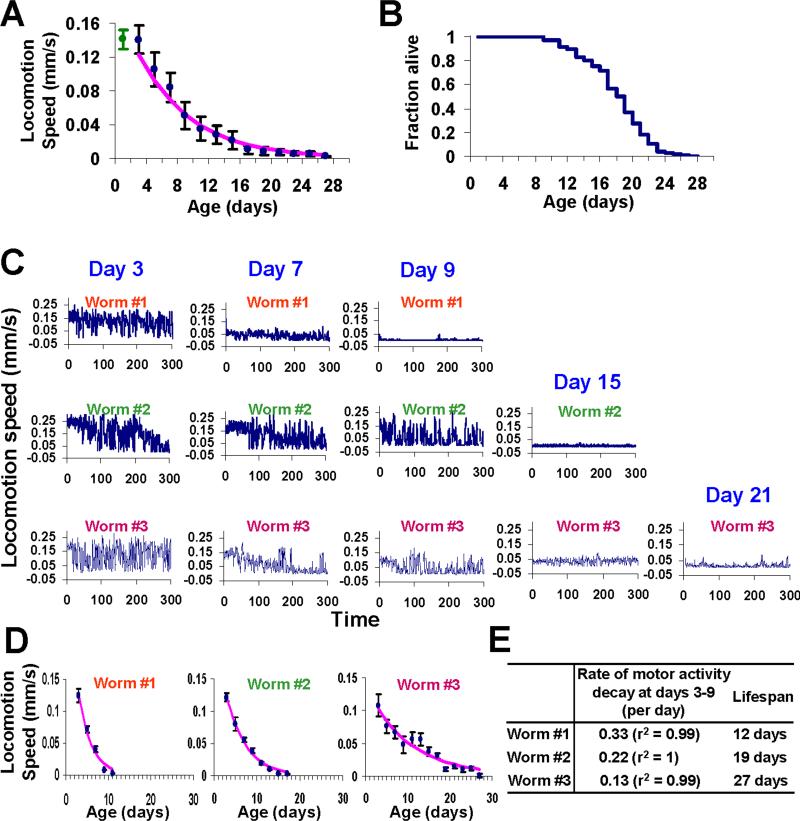
Figure 1. Worms undergo progressive decay in motor activity throughout lifespan.
(A) Progressive decay in the speed of spontaneous locomotion in aging worms. Locomotion behavior of 169 wild-type worms (N2) was quantified every other day until death, and the mean locomotion speed of these worms was plotted as a function of age. Beginning at day 3, worms display a progressive decline in locomotion speed, which can be best fit by first-order exponential decay. Censored worms were not included for analysis. Error bars represent SD.
(B) Lifespan curve (Kaplan–Meier test) of the same N2 worms assayed in (A). Mean lifespan: 19.5 ± 0.3 (SEM) days; 75% lifespan: 22 days.
(C-E) Individual worms with different lifespan exhibit variation in the rate of locomotion activity decay.
(C) Sample traces from three representative worms with different lifespan. Each sample trace shows the locomotion speed of a worm in real time recorded at the indicated age by an automated worm tracking system.
(D) The speed of spontaneous locomotion of each representative worm plotted as a function of age beginning day 3. Locomotion speed decayed throughout lifespan and can be best fitted by first-order exponential decay, and the rate of the decay can be described by the equation: ΔV/Δt = -k*V, where V denotes the speed of locomotion of the animal at a given age, t denotes age and k is a constant. The kinetic constant k is thus used to quantify the rate of motor activity decay.
(E) A table showing the rate of locomotion activity decay and lifespan of the three representative worms.
The rate of motor activity decay in the early-to-mid life of aging worms inversely correlates with lifespan
In analyzing data from individual worms, we made a preliminary observation that short-lived worms seemed to exhibit a faster rate of decay in the speed of locomotion throughout lifespan than long-lived worms (Figure 1C-E). This implies a possible correlation between the rate of motor activity decay and lifespan.
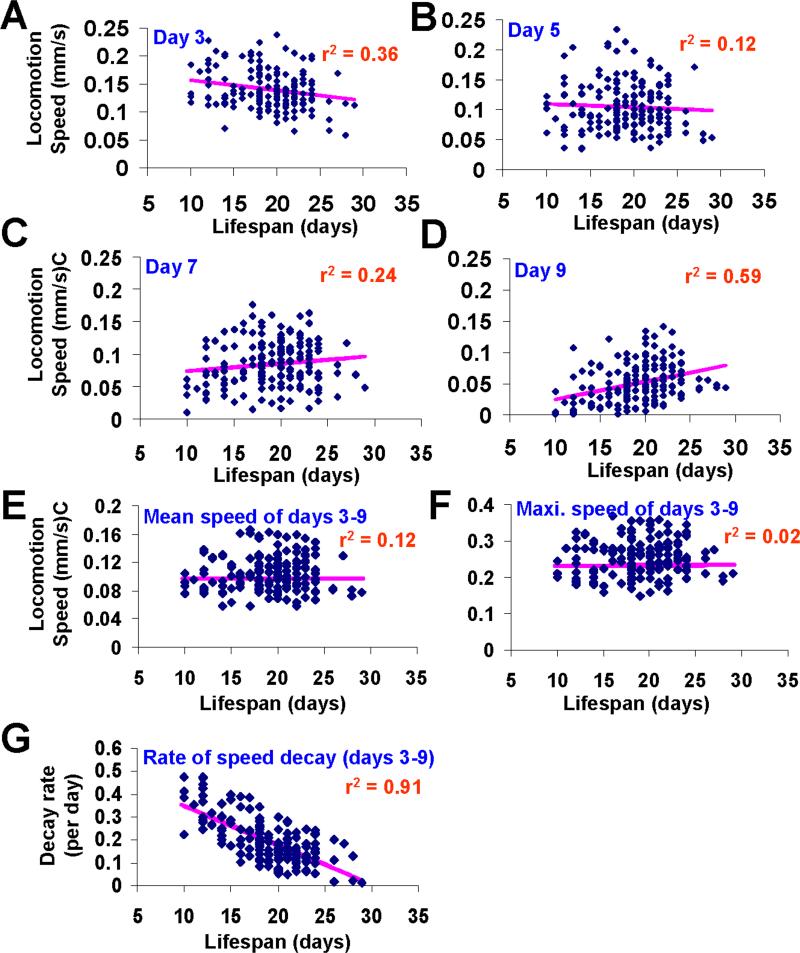
We then asked whether a lifespan prediction can be made at the early-to-mid stage of aging. As the mean lifespan of these worms was 19.5 days, we decided to focus our attention on events occurring between day 3 and day 9, a period that spans the early-mid life of these worms. By plotting out the lifespan value of individual worms against different parameters of their locomotion activity, we found that the locomotion speed at individual days alone (i.e. day 3, 5, 7 or 9) showed either no or weak correlation with lifespan (Figure 2A-D). Nor was a correlation observed between lifespan and the mean or maximal locomotion speed at days 3−9 (Figure 2E and 2F). In contrast, there appeared to be a strong inverse correlation between lifespan and the rate of locomotion speed decay during days 3−9 (r2 = 0.91; r = −0.70; p<0.00001) (Figure 2G). The Spearman rank correlation method also indicates an inverse correlation between lifespan and the rate of locomotion speed decay during days 3−9 (−0.63; p<0.0001). These results suggest that the rate of motor activity decay can be used as a lifespan predictor.
Figure 2. The rate of motor activity decay during the early-to-mid life of a worm inversely correlates with its lifespan.
(A-D) The speed of locomotion of wild-type worms at day 3, 5, 7 or 9 shows no or weak correlation with lifespan. Locomotion speed of all 169 individuals at day 3 (A), day 5 (B), day 7 (C) or day 9 (D) was plotted as a function of their lifespan. The solid line in each plot represents the fitting line.
(E) The mean speed of locomotion of wild-type worms during days 3−9 shows no correlation with lifespan. The speed of locomotion of each worm during days 3−9 was averaged and plotted as a function of its lifespan.
(F) The maximal speed of locomotion of wild-type worms during days 3−9 shows no correlation with lifespan. The highest value of locomotion speed of each worm during days 3−9 was plotted as a function of its lifespan.
(G) The rate of locomotion speed decay of wild-type worms during days 3−9 inversely correlates with lifespan. The speed of locomotion of each worm during days 3−9 was plotted as a function of age, and the rate of locomotion speed decay of each worm during this period was calculated using the equation described in Figure 1D.
To further evaluate this lifespan predictor, we calculated the odds value. If the rate of locomotion activity decay of a worm during days 3−9 is slower and faster than that of a worm with a mean decay rate, the odds value for this worm to live longer and shorter than mean lifespan is 3.4 and 3.1, respectively (p<0.0001); namely, the probability for this worm to live longer and shorter than mean lifespan is 77% and 76%, respectively.
Mutations that extend lifespan reduce the rate of motor activity decay in aging worms
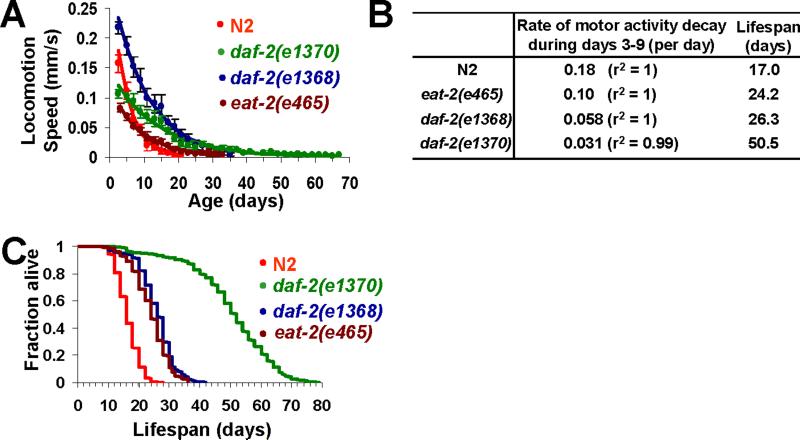
Having identified the rate of motor activity decay as a lifespan predictor, we then asked whether and how this parameter is regulated by genes known to affect lifespan. The insulin/IGF-1 signaling pathway has been found to modulate lifespan [3,7,19,25,33]. In particular, mutations in the daf-2 insulin-like receptor extend lifespan up to one fold [20,21]. daf-2 mutant worms are also more resistant to a variety of stress conditions and less susceptible to tumorigenesis than are wild-type worms [8,26,28,31]. To ascertain whether and how the daf-2 gene affects the rate of motor activity decay, we quantified spontaneous locomotion behavior of daf-2(e1370) and daf-2(e1368) worms throughout their lifesapn. As a control, wild-type worms were assayed under the same conditions. We found that the rate of motor activity decay was greatly reduced in daf-2(e1370) worms (Figure 3A and 3B). A similar phenomenon was also observed in daf-2(e1368) worms (Figure 3A and 3B). These results support the view that daf-2 mutations not only extend worms’ lifespan but also their “healthspan” [7,19,32].
Figure 3. Mutant strains with deficits in insulin signaling and food intake reduce the rate of motor activity decline.
(A-B) The rate of locomotion speed decay is reduced in daf-2 and eat-2 mutant worms. Locomotion speed of wild-type (N2), daf-2 and eat-2 mutants was plotted as a function of age beginning day 3 (A), and the rate of locomotion speed decay in each strain was calculated and shown in (B). n≥30. Censored worms were not included for analysis. The mean lifespan of each strain was also shown in (B). Error bars represent SD.
(C) Lifespan curve (Kaplan–Meier test) of daf-2(e1370) [mean lifespan: 50.5 ± 0.8 (SEM) days; 75% lifespan: 60 days], daf-2(e1368) [mean lifespan: 26.3 ± 0.4 (SEM) days; 75% lifespan: 30 days], eat-2(e465) worms [mean lifespan: 24.2 ± 0.3 (SEM) days; 75% lifespan: 28 days], and N2 worms [mean lifespan: 17.0 ± 0.2 (SEM) days; 75% lifespan: 20 days] assayed in (A-B).
We also examined whether caloric restriction affects the rate of motor activity decay during aging by quantifying spontaneous locomotion behavior of eat-2 worms. Mutations in eat-2 reduce food intake and thereby extend lifespan, and these worms have been used as a common model for caloric restriction-mediated modulation of lifespan in C. elegans [22,23]. We found that the rate of motor activity decay was also reduced in eat-2 worms (Figure 3A and 3B), supporting the notion that caloric restriction extends both lifespan and healthspan.
The rate of motor activity decay may potentially be used for across-strain comparison of healthspan
Interestingly, while the locomotion speed of daf-2(e1368) worms was higher than that of wild-type worms at the same age, the same phenomenon was not observed in daf-2(e1370) worms (Figure 3A). In fact, the locomotion speed of daf-2(e1370) worms at days 1−5 was lower than that of wild-type worms (Figure 3A). Similarly, though lifespan was extended in eat-2 worms (Figure 3C), their motor activity was lower than those of wild-type worms until day 11 (Figure 3A). Thus, there appears to be no strong correlation between the mean lifespan and the absolute speed of locomotion when comparing different strains, a phenomenon also observed among different individual worms with the same genetic background (Figure 2A-D).
In contrast, there seems to be an inverse correlation between the rate of motor activity decay and the mean lifespan of the strain (Figure 3B). Specifically, the strain with the slowest rate of motor activity decay showed the longest mean lifespan, and vice versa (Figure 3B). This suggests that the rate of motor activity decay might potentially be used for across-strain comparison of healthspan.
Concluding remarks
In summary, using machine vision we have quantified the locomotion behavior of aging worms throughout lifespan. Our method automates and standardizes data acquisition and processing, thus allowing for extraction of behavioral parameters in an objective, quantitative manner. As such, it offers an advantage over previous methods that rely on human observation. Using this method, we have identified the rate of motor activity decay as an endogenous physiological parameter that can predict lifespan at the early-to-mid stage of aging. Interestingly, the absolute motor activity of a worm in early-mid life does not show a strong correlation with lifespan. We have also shown that the rate of motor activity decay is affected by genetic factors, suggesting that it may potentially be used for across-strain comparison of healthspan. As our method assays the rate of motor activity decay but not the absolute motor activity during aging, it may be applied to strains with reduced motor activity. Nevertheless, it should be noted that this method cannot be applied to those severely uncoordinated (Unc) mutant strains that do not possess motor activity throughout lifespan (e.g. unc-13) [27].
Progressive motor activity decline also occurs in old animals of other organisms and likely represents a universal phenomenon of aging in the animal kingdom [2,15,30]. Our work raises the possibility that similar strategies may be applied to other organisms to identify endogenous physiological parameters for lifespan prediction and healthspan comparison.
Acknowledgments
We thank John Faulkner for advice, Patrick Hu for critically reading the manuscript, Bin Cao, Alex Ward and Carol Mousigan for technical assistance. Some strains were obtained from the Caenorhabditis Genetics Center. This work was supported by the Ellison Medical Foundation (A.L.H.), Pew Scholarship (X.Z.S.X), and the NIH (X.Z.S.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors declare no actual or potential conflicts of interest.
References
- 1.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41(1):45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 2.Altun M, Bergman E, Edstrom E, Johnson H, Ulfhake B. Behavioral impairments of the aging rat. Physiol Behav. 2007 doi: 10.1016/j.physbeh.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95(2):199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 4.de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 5.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 6.Feng Z, Li W, Ward A, Piggott BJ, Larkspur E, Sternberg PW, Xu XZS. A C. elegans model of nicotine-dependent behavior: regulation by TRP family channels. Cell. 2006;127(3):621–33. doi: 10.1016/j.cell.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finch CE, Ruvkun G. The genetics of aging. Annual review of genomics and human genetics. 2001;2:435–62. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- 8.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300(5627):1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 9.Glenn CF, Chow DK, David L, Cooke CA, Gami MS, Iser WB, Hanselman KB, Goldberg IG, Wolkow CA. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. The journals of gerontology. 2004;59(12):1251–60. doi: 10.1093/gerona/59.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408(6809):255–62. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 11.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1(1):119–28. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419(6909):808–14. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 13.Hosono R, Sato Y, Aizawa SI, Mitsui Y. Age-dependent changes in mobility and separation of the nematode Caenorhabditis elegans. Experimental gerontology. 1980;15(4):285–9. doi: 10.1016/0531-5565(80)90032-7. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101(21):8084–9. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingram DK, Chefer S, Matochik J, Moscrip TD, Weed J, Roth GS, London ED, Lane MA. Aging and caloric restriction in nonhuman primates: behavioral and in vivo brain imaging studies. Ann N Y Acad Sci. 2001;928:316–26. doi: 10.1111/j.1749-6632.2001.tb05661.x. [DOI] [PubMed] [Google Scholar]
- 16.Johnson TE. Aging can be genetically dissected into component processes using long-lived lines of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1987;84(11):3777–81. doi: 10.1073/pnas.84.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson TE. Recent results: biomarkers of aging. Experimental gerontology. 2006;41(12):1243–6. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Johnson TE, Conley WL, Keller ML. Long-lived lines of Caenorhabditis elegans can be used to establish predictive biomarkers of aging. Experimental gerontology. 1988;23(4−5):281–95. doi: 10.1016/0531-5565(88)90031-9. [DOI] [PubMed] [Google Scholar]
- 19.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–60. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 21.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277(5328):942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 22.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95(22):13091–6. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging cell. 2006;5(6):515–24. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Feng Z, Sternberg PW, Xu XZS. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature. 2006;440(7084):684–7. doi: 10.1038/nature04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115(4):489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 26.Lithgow GJ, White TM, Hinerfeld DA, Johnson TE. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. Journal of gerontology. 1994;49(6):B270–6. doi: 10.1093/geronj/49.6.b270. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama IN, Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1991;88(13):5729–33. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313(5789):971–5. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 29.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37(8):894–8. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridgel AL, Ritzmann RE. Insights into age-related locomotor declines from studies of insects. Ageing research reviews. 2005;4(1):23–39. doi: 10.1016/j.arr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Scott BA, Avidan MS, Crowder CM. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science. 2002;296(5577):2388–91. doi: 10.1126/science.1072302. [DOI] [PubMed] [Google Scholar]
- 32.Wolkow CA. Identifying factors that promote functional aging in Caenorhabditis elegans. Experimental gerontology. 2006;41(10):1001–6. doi: 10.1016/j.exger.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290(5489):147–50. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]