Abstract
Microglial inflammatory responses affect Parkinson's disease (PD) associated nigrostriatal degeneration. This is triggered, in measure, by misfolded, nitrated alpha-synuclein (N-α-syn) contained within Lewy bodies that are released from dying or dead dopaminergic neurons into the extravascular space. N-α-syn-stimulated microglial immunity is regulated by CD4+ T cells. Indeed, CD4+CD25+regulatory T cells (Treg) induce neuroprotective immune responses. This is seen in rodent models of stroke, amyotrophic lateral sclerosis, human immunodeficiency virus associated dementia, and PD. To elucidate the mechanism for Treg-mediated microglial responses, we used a proteomic platform integrating difference gel electrophoresis and tandem mass spectrometry peptide sequencing. These tests served to determine the consequences of Treg on the N-α-syn stimulated microglia. The data demonstrated that Treg substantially alter the microglial proteome in response to N-α-syn. This is seen through Treg's abilities to suppress microglial neurotoxic proteins linked to cell metabolism, migration, protein transport and degradation, redox biology, cytoskeletal, and bioenergetic activities. We conclude that Treg modulate the N-α-syn microglial proteome and, in this way, can slow the tempo and course of PD.
Keywords: Regulatory T cells, Proteomics, Microglia, Inflammation, Parkinson's disease, Alpha-synuclein
Introduction
Parkinson's disease (PD) is a progressive neurodegenerative disease characterized clinically as gait and motor disturbances such as rigidity, resting tremor, slowness of voluntary movement, and postural instability. In some cases these evolve to frank dementia1-4. A plethora of host and environmental factors affect the onset and progression of PD including genetics, environmental cues, aging, peripheral immunity, impaired energy metabolism, and oxidative stress5-15. Pathologically, PD is characterized by nigrostriatal degeneration precipitated by progressive loss of dopaminergic neuronal cell bodies in the substantia nigra pars compacta (SNpc) and their projections to the dorsal striatum16. This degeneration is associated with alterations in innate, microglial activation17-24 and adaptive T cell immunity5,25-27. Precipitation of immune dysfunction in PD is thought to ensue from the release of cytoplasmic inclusions of fibrillar, misfolded proteins encased in Lewy bodies (LB) and composed principally of aggregated α-synuclein (α-syn)28. Such misfolded proteins can engage innate and adaptive immunity28,29. Indeed, substantive evidence supports the notion that nigrostriatal degeneration is manifest by α-syn mediated microglial activation, oxidative stress and disease inciting adaptive immune responses25-27, 30-33. It is the pathogenic spiral of dopaminergic neuronal death, release of extracellular aggregated α-syn, microglial activation, peripheral immune activation, collateral neuronal injury, sustained α-syn release with ingress into lymphatics, and engagement of specific T cell responses that further damage dopamine neurons.
We previously demonstrated that microglia associated degenerative responses are triggered by nitrated α-syn (N-α-syn)-specific effector T cells (Teff)25; whereas, CD4+CD25+ regulatory T cells (Treg) attenuate microglial activation and promote dopaminergic neuronal survival34. Lacking from our prior works was a mechanism for CD4+ T cell-mediated modulation of microglial function. Based on these observations, we hypothesized that CD4+ T cells have dual roles, and as such, influence microglial responses to evoke biological activities that ultimately effect neuronal survival or loss. In attempts to decipher the mechanisms underlying such responses, we used aggregated N-α-syn as an inducer of microglial activation30-32, then examined the microglial proteome affected by interactions with CD4+ T cell subsets35. Using proteomic approaches, we demonstrate that Treg regulatory activities extend beyond inhibition of cellular activation and include modulation of a broad range of microglial activities involving regulation of phagocytosis and proteasome function, induction of redox-active and bioenergetic proteins, and apoptotic cell processes. Such regulatory events lead to the attenuation of microglial inflammatory neurotoxic responses. Importantly, the data demonstrate that the effects of Treg on N-α-syn-mediated immune activities are multifaceted and of potential therapeutic benefit.
Materials and Methods
Animals
C57BL/6J male mice (7 wks old) were purchased from The Jackson Laboratory (Bar Harbor, ME) and used for CD4+ T cell isolations. C57BL/6J neonates were obtained from breeder colonies housed in the University of Nebraska Medical Center animal facilities. All animal procedures were in accordance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center.
Cell isolates
Microglia were prepared from neonatal mice (1-2 days old) using previously described techniques36. Cultures were consistently >98% CD11b+ microglia37. CD4+ T cell subsets were isolated using previously described techniques34, 38. Treg and Teff isolates were >95% enriched35. CD3-activated T cells were co-cultured with microglia at 1:1 ratio. All analyses of microglia were performed after removal of the T cells from the cultures.
Recombinant α-syn
Purification, nitration and aggregation of recombinant mouse α-syn were performed as previously described30-32. N-α-syn was added to cultures at 100 nmol/L (14.5 ng/ml).
2D Difference Gel Electrophoresis (DIGE) and image analysis
Protein prepared from microglial cell lysates was labeled with the respective CyDyes, followed by separation in the first and second dimension, and the gels were scanned using a Typhoon 9400 Variable Mode Imager. Analyses of Cy3-Cy5 image pairs, adjustment to Cy2 control images and detection of protein spots were performed using DeCyder™ software (GE Healthcare). Statistical significance (P≤ 0.05) was determined with Biological Variance Analysis (BVA).
Mass spectrometry
In gel trypsin digestion were performed as previously described40. The resulting peptides were sequenced using Electrospray Ionization-Liquid Chromatography Mass Spectrometry (ESI-LC MS/MS) (Proteome X System with LCQDecaPlus mass spectrometer, ThermoElectron, Inc.) with a nanospray configuration. The spectra were searched against the NCBI.fasta protein database narrowed to murine proteins using SEQUEST search engine (BioWorks 3.1 SR software from ThermoElectron, Inc.). Validation of select proteins identified by LC-MS/MS was performed using immunocytochemistry or Western blot (Supplementary information)
Cytotoxicity
The Live/Dead Viability/Cytotoxicity kit (Invitrogen) was performed according to manufacturer's protocol. Images were taken using fluorescence microscopy. Cell counts were normalized as the percentage of surviving cells from unstimulated culture controls.
Statistics
For identification of statistically significant proteins, three-to-four analytical gels were analyzed using BVA software by one-way ANOVA for pair-wise comparison between treatment groups. Differences between means ± SEM were analyzed by one-way ANOVA followed by Tukey's post-hoc test for pair-wise comparisons.
Results
Microglial protein profiling techniques following N-α-syn stimulation and Treg co-cultivation
We previously demonstrated that aggregated N-α-syn induces activation of the NF-κB pathway in microglia resulting in a robust inflammatory response characterized by increased production of TNF-α, IFN-γ, IL-6, and IL-1β among others 30, 31. Co-culture of microglia with Treg either pre- or post-stimulation significantly attenuates NF-κB activation as well as inflammatory cytokine and superoxide production in response to N-α-syn, whereas Teff exacerbate these responses 34, 39. Therefore, to uncover putative mechanisms for CD4+ T cell-mediated modulation of the microglial phenotype, 2D DIGE was used to identify differences in protein expression of N-α-syn stimulated of microglia alone and co-cultured with CD4+ T cells. 2D DIGE analysis of microglial cell lysates was repeated three separate times with three independent cell isolations and cultures. Analyses of 2D images from protein lysates of 15 × 106 microglial cells identified an average of approximately 2000 “putative” protein spots. DeCyder™ DIGE Analysis of Cy3-labeled proteins from unstimulated microglia and Cy5-labeled proteins from N-α-syn stimulated microglia obtained from three independent experiments showed an average of 2072 detected spots. Representative analyses revealed 43% differentially expressed protein spots after setting a threshold mode of quantitative differences ≥ 2 standard deviations (SD). Of those uniquely identifiable spots (582), 28% were upregulated and (318) 15% were downregulated in microglial cell lysates in response to 24 h stimulation with N-α-syn. To assess how CD4+ T cells modulate the N-α-syn microglial phenotype, microglia were co-cultured with either Treg or Teff for 24 h either prior to stimulation with N-α-syn (pre-treatment) or following 12 h of stimulation (post-treatment), and comparisons were made using 2D DIGE and nano-LC-MS/MS peptide sequencing (Figure 1). Co-cultivation with Treg prior to stimulation with N-α-syn (pre-treatment) altered the microglial phenotypic response to N-α-syn stimulation. An analysis of Cy3-labeled proteins from N-α-syn stimulated microglia and Cy5-labeled proteins from N-α-syn stimulated microglia pre-treated with Treg obtained from three independent experiments showed an average of 2326 detected spots. Representative analysis revealed 31% differentially expressed protein spots after setting a threshold mode of quantitative differences ≥ 2 SD. Of those uniquely identifiable spots (348), 15% were increased and (365) 16% were decreased in microglial cell lysates in response to Treg treatment prior to N-α-syn stimulation. Pre-treatment with Teff had less robust affects on the microglial phenotype in response to N-α-syn. Of the > 2000 uniquely identifiable spots, approximately 32 (1.8%) were decreased and 22 (1.3%) increased in abundance compared to N-α-syn stimulation alone.
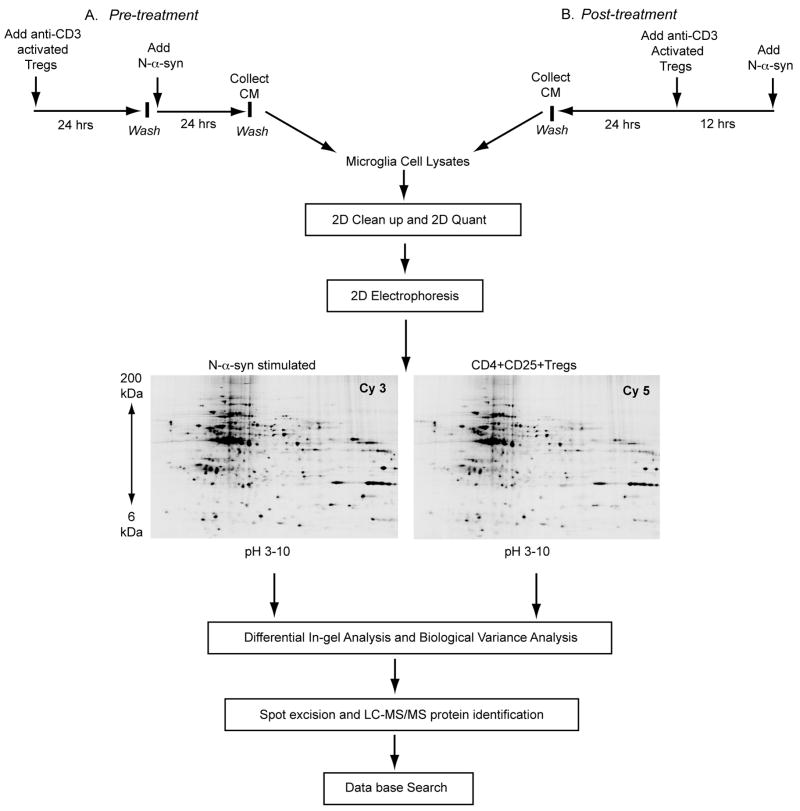
Figure 1.
Experimental design for microglial proteomics protein discovery. Microglia were co-cultured for 24 h with CD4+CD25+ Treg (or Teff) or without as control. Treg (or Teff) were removed from the cultures and the microglia stimulated with aggregated N-α-syn for 24 h [pre-treatment] to represent asymptomatic disease (A). Alternatively, microglia were stimulated with N-α-syn for 12 h prior to the addition of Treg [post-treatment] to represent more overt disease (B). Twenty-four hours later microglial cell protein lysates were prepared and subjected to 2D electrophoresis. Decyder analysis software was used to match spots and identify expression patterns. Selected protein spots were excised, digested with trypsin and identified by nano-LC-MS/MS peptide sequencing. Database searches were performed using SEQUEST with criteria thresholds set to afford greater than 95% confidence level in peptide identification.
To mimic what may occur during overt disease, CD4+ T cells were added to N-α-syn microglial cultures 12 h post-stimulation. Co-cultivation with Treg post-stimulation with N-α-syn (post-treatment) also altered the microglial phenotype. An analysis of Cy3-labeled proteins from N-α-syn stimulated microglia and Cy5-labled proteins from N-α-syn stimulated microglia post-treated with Treg obtained from three independent experiments showed an average of 1905 detected spots. Representative analysis revealed 27% differentially expressed protein spots after setting a threshold mode of quantitative differences ≥ 2 SD. Of those uniquely identifiable spots, (110) 6% were increased and (403) 21% were decreased in microglial cell lysates in response to Treg treatment following N-α-syn stimulation. By comparison, post-treatment with Teff resulted in significant modulation of the microglial proteome in response to N-α-syn stimulation. Of the > 2000 uniquely identifiable spots, approximately 318 (15%) were decreased and 325 (16%) were increased in abundance compared to N-α-syn stimulation alone.
To identify differentially expressed proteins (P≤ 0.05), analyses with BVA software were performed on analytical gels from separate lysates comparing microglia cultures stimulated with media alone, N-α-syn or co-cultured with Treg or Teff to facilitate cross-comparisons between treatments by BVA whereby identified spots were compared for area and peak height (3D plots). The 3D peak of each protein spot, comprised of Cy3-labeled and Cy5-labeled cell lysates, was generated based on the pixel intensity versus pixel area, where peak area correlated with the distribution of the protein spot on the gel. 3D images were obtained using 2D Master Imager and were evaluated independently based on their differential fluorescent signal within a constant area for the spot. Their relative peak volumes were normalized to the total volume of the spot (Cy2-labeled). All analytical gels were cross-compared by BVA and matched to a preparative gel consisting of pooled protein from the experimental groups. The proteins identified consisted of structural or cytoskeletal proteins, regulatory proteins, redox-active proteins and enzymes. Figure 2 shows the location of these proteins on the preparative gel selected for LC-MS/MS sequencing.
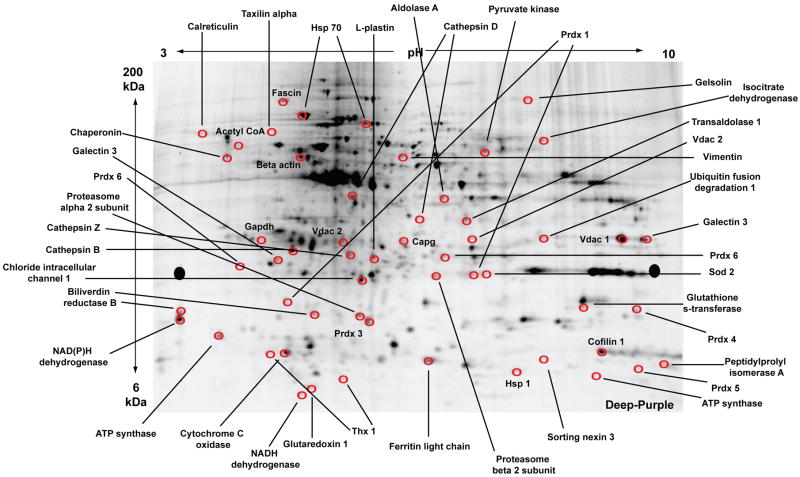
Figure 2.
2D-DIGE of proteins from all experimental groups with matched spots picked for sequencing identifications by nano-LC-MS/MS. A representative preparative gel is shown. Equal amounts of protein were pooled from all experimental groups (unstimulated, N-α-syn-stimulated, pre- and post-treatment with Treg or Teff) and replicates for a total concentration of 450 μg. The pooled sample was applied to a pH gradient strip and separated with isoelectric focusing for the first dimension. For the second dimension, the strip was loaded onto a large format gradient gel and separated based on molecular weight. Following electrophoresis, the gel was fixed and post-stained with Deep Purple for positive detection of protein spots. Circled spots identified by BVA using Decyder analysis software were selected for excision. Proteins with the most peptides positively identified within a specific spot are labeled on the gel. (Abbreviations: Prdx, peroxiredoxin; Thx, thioredoxin, Vdac, voltage-dependent anion channel; Sod, superoxide dismutase; Hsp, heat shock protein; Capg, macrophage capping protein; NAD, nicotinamide adenine dinucleotide).
N-α-syn stimulation and the microglial proteome
In our prior works, we demonstrated that N-α-syn is capable of inducing the temporal activation of a neurotoxic microglial phenotype 30, 31. To extend these works, the time course of activation was extended from 2 h, 4 h, and 8 h to 24 h for the current study. Table 1 shows proteins differentially expressed in microglia that were stimulated in media alone or with N-α-syn. Proteins were considered identified with high confidence with at least two peptides sequenced and met the threshold peptide criteria (Supplementary information). Such threshold criteria have been determined previously to result in a 95% confidence level in peptide identification41, 42. The categories of proteins included regulatory, cytoskeleton/structural, enzymes, mitochondrial, redox-active and others. Figure 3A shows the relative percentages of proteins within each classification based on protein function that were modulated by N-α-syn stimulation and expression trends.
Table 1. N-α-syn stimulated proteome.
| Protein ID by LC/MS/MS* |
SwissProt† | IPI‡ | M.wt.§ (DA) |
pI11 | Subcellular Location¶ | Function# | Peptide**# | DIGE†† Index |
P- value‡‡ |
|---|---|---|---|---|---|---|---|---|---|
| Heterogeneous nuclear ribonucleoprotein A2/B1 isoform 2 | O88569 | IPI00622847 | 37403 | 8.97 | Nucleus | transcription | 2 | -1.69 | |
| Guanine nucleotide-binding protein | P62880 | IPI00162780 | 37331 | 5.60 | Cytoplasm | GTPase activity | 2 | -1.5 | 0.004 |
| Guanine nucleotide-binding protein | Q61011 | IPI00116938 | 37240 | 5.41 | Cytoplasm | GTPase activity | 2 | -1.5 | 0.004 |
| Interleukin-6 receptor subunit beta | Q00560 | IPI00120155 | 102452 | 5.32 | Membrane | signal transduction | 3 | -1.84 | 0.05 |
| Ubiquitin A-52 residue ribosomal protein fusion | B0LAC2 | IPI00138892 | 8038.2 | 6.89 | Ribosome | protein modification | 2 | -1.51 | 0.05 |
| Alpha tubulin | P68369 | IPI00110753 | 50136 | 4.94 | Cytoskeleton | cell motility | 2 | -2.21 | |
| Beta actin | P60710 | IPI00110850 | 269833 | 5.82 | Cytoskeleton | cell motility | 3 | -1.5 | 0.046 |
| Dynein cytoplasmic 1 intermediate chain 2 | O88487 | IPI00131086 | 68394 | 5.16 | Cytoskeleton | cell motility | 2 | -2.21 | |
| Galectin 3 | P16110 | IPI00131259 | 27515 | 8.47 | Cytoplasm/nucleus | protein binding, phagocytosis | 4 | -2.96 | |
| L-plastin | Q61233 | IPI00118892 | 70149 | 5.2 | Cytoskeleton | phagocytosis | 2 | -1.95 | |
| RuvB-like protein 1 | P60122 | IPI00133985 | 50214 | 6.02 | Nucleus | proliferation | 5 | -1.5 | |
| Voltage-dependent anion channel 2 | Q60930 | IPI00122547 | 31733 | 7.44 | Mitochondria | ion transport | 6 | -1.71 | |
| Voltage-dependent anion channel 3 | Q60931 | IPI00876341 | 30753 | 8.96 | Mitochondria | ion transport | 19 | -1.94 | |
| Vacuolar H+ATPase B2 | P62814 | IPI00119113 | 56551 | 5.57 | Membrane | ion transport | 6 | -1.82 | 0.04 |
| Voltage-dependent anion-selective channel protein 1 (VDAC-1) | Q60932 | IPI00230540 | 32351 | 8.55 | Membrane/ Mitochondria | ion transport | 3 | -1.94 | |
| G-protein beta subunit | Q61621 | IPI00120716 | 13533 | 5.50 | Membrane | G-protein signaling | 4 | -1.5 | 0.004 |
| Lamin A isoform C2 | P48678 | IPI00230435 | 74238 | 6.54 | Nucleus | membrane stabilization | 22 | -1.67 | 0.017 |
| Cofilin 1 | P18760 | IPI00890117 | 18559 | 8.22 | Cytoskeleton | actin polymerization | 3 | -2.15 | 0.02 |
| Cofilin 2 | P45591 | IPI00266188 | 18710 | 7.66 | Cytoskeleton | actin polymerization | 2 | -2.15 | 0.02 |
| Vimentin | P20152 | IPI00227299 | 53688 | 5.06 | Cytoskeleton | stabilize cytoskeleton | 5 | -2.35 | 0.05 |
| Peripherin | P15331 | IPI00129527 | 54268 | 5.40 | Cytoskeleton | cytoskeleton organization | 2 | -2.35 | 0.05 |
| Desmin | P31001 | IPI00130102 | 53498 | 5.21 | Cytoskeleton | stabilize cytoskeleton | 2 | -2.35 | 0.05 |
| Adenylyl cyclase-associated protein 1 (CAP 1) | P40124 | IPI00137331 | 51575 | 7.16 | Cytoskeleton | cytoskeleton organization | 3 | -2.39 | 0.05 |
| Fascin | Q61553 | IPI00353563 | 54508 | 6.44 | Filopodium | actin binding | 3 | -1.63 | |
| Annexin A2 | P07356 | IPI00468203 | 38676 | 7.55 | Secreted | matrix | 2 | -1.5 | |
| Annexin A10 | Q9QZ10 | IPI00136659 | 37301 | 5.40 | Mitochondria | matrix | 2 | -1.63 | 0.05 |
| Inner membrane protein, mitochondria | Q8CAQ8 | IPI00228150 | 83900 | 6.18 | Mitochondria | matrix | 7 | -1.58 | |
| Gelsolin | A2AL35 | IPI00117167 | 85942 | 5.83 | Cytoskeleton | apoptosis and inflammation, vesicle transport | 8 | 1.53 | 0.05 |
| Annexin A1 | P10107 | IPI00230395 | 38734 | 6.97 | Cytoplasm | membrane fusion and exocytosis | 2 | -1.5 | |
| Palmitoyl-protein thioesterase 1 | B1B0P8 | IPI00881289 | 19550 | 8.09 | Membrane/ Lysosome | endocytosis/ protein transport | 2 | -1.5 | |
| Rho GDP dissociation inhibitor (GDI) alpha | Q99PT1 | IPI00322312 | 23407 | 5.12 | Cytoplasm/ membrane | protein binding | 9 | -2.08 | |
| Cryptochrome 2 | Q9R194 | IPI00128234 | 66850 | 8.66 | Cytoplasm/nucleus | protein transport | 3 | -1.5 | 0.039 |
| 14-3-3 zeta | P63101 | IPI00116498 | 27111 | 4.73 | Mitochondria | protein targeting | 5 | -2.39 | 0.05 |
| Ferritin light chain 1 | P29391 | IPI00762203 | 20802 | 5.66 | Cytoplasm | iron homeostasis | 5 | -1.75 | 0.05 |
| Ferritin heavy chain 1 | P09528 | IPI00230145 | 21067 | 5.53 | Cytoplasm | iron homeostasis | 3 | -1.61 | 0.02 |
| Acetyl-Coenzyme A acetyltransferase 1 | A8XUS5 | IPI00228253 | 41298 | 7.16 | Cytoplasm | metabolism | 4 | -1.52 | |
| Acetyl-Coenzyme A acyltransferase 2 | A8XUT1 | IPI00881591 | 38147 | 7.63 | Cytoplasm | metabolism | 2 | -1.52 | |
| Aldehyde dehydrogenase, mitochondrial | P47738 | IPI00111218 | 56538 | 7.53 | Mitochondria | metabolism | 23 | -2.79 | |
| Hexosaminidase B | P20060 | IPI00115530 | 61116 | 8.28 | Lysosome | metabolism | 4 | -2.79 | |
| Ugp2 protein | Q8R0M2 | IPI00279474 | 55498 | 6.92 | Cytoplasm | metabolism | 6 | -1.5 | 0.028 |
| Pyrophosphatase | Q9D819 | IPI00110684 | 32667 | 5.37 | Cytoplasm | metabolism | 7 | -1.73 | |
| Aldo-keto reductase family 1, member B8 | Q3UJW9 | IPI00466128 | 36615 | 6.90 | Cytoplasm/ membrane | catabolism | 4 | 1.6 | |
| Catechol O-methyltransferase | O88587 | IPI00759876 | 29496 | 5.52 | Cytoplasm | catabolism | 3 | 1.51 | 0.05 |
| Glutamate oxaloacetate transaminase 2, mitochondrial | P05202 | IPI00117312 | 47411 | 9.13 | Mitochondria | catabolism | 6 | -1.5 | 0.046 |
| Fatty acid-binding protein | P05201 | IPI00230204 | 46232 | 6.68 | Cytoplasm | catabolism | 4 | -3.57 | 0.02 |
| Cathepsin B | P10605 | IPI00113517 | 37280 | 5.57 | Lysosome | thiol protease | 4 | 2.86 | 0.005 |
| Cathepsin D | P18242 | IPI00111013 | 44954 | 6.71 | Lysosome | acid protease | 4 | 1.62 | 0.036 |
| Calreticulin | P14211 | IPI00123639 | 47995 | 4.33 | Membrane/ ER | chaperone | 6 | -4.6 | |
| Calreticulin 3 isoform 1 | Q9D9Q6 | IPI00113023 | 44198 | 5.99 | ER | chaperone | 2 | -2.75 | |
| Chaperonin subunit 6a zeta | Q52KG9 | IPI00116281 | 58076 | 6.46 | Cytoplasm | chaperone | 3 | -1.67 | 0.017 |
| HSP 10 | Q64433 | IPI00263863 | 10963 | 7.91 | Mitochondria | chaperone | 2 | -3.29 | |
| HSP 60 | P63038 | IPI00308885 | 60955 | 5.91 | Mitochondria | chaperone | 17 | -2.35 | 0.05 |
| HSP 70 | P63017 | IPI00323357 | 70871 | 5.37 | Cytoplasm | chaperone | 17 | -2.35 | 0.05 |
| Proteasome subunit, alpha type 2 | P49722 | IPI00890001 | 25926 | 8.39 | Cytoplasm | Ubiquitin-Proteasome system | 2 | -1.53 | 0.05 |
| Proteasome subunit, alpha type 3 | O70435 | IPI00331644 | 28405 | 5.29 | Cytoplasm/nucleus | Ubiquitin-Proteasome system | 6 | -2.39 | 0.05 |
| Proteasome subunit, alpha type 6 | Q9QUM9 | IPI00131845 | 27372 | 6.35 | Cytoplasm/nucleus | Ubiquitin-Proteasome system | 4 | -1.54 | |
| Proteasome subunit alpha type 7 | Q9Z2U0 | IPI00131406 | 27855 | 8.59 | Cytoplasm/nucleus | Ubiquitin-Proteasome system | 3 | -1.69 | 0.027 |
| 20S proteasome subunit C2 | Q9JHS5 | IPI00283862 | 4581.4 | 8.07 | Cytoplasm | Ubiquitin-Proteasome system | 2 | -1.54 | 0.05 |
| Ubiquitin-conjugating enzyme E2-25K | P61087 | IPI00322440 | 22407 | 5.33 | Cytoplasm | Ubiquitin-Proteasome system | 8 | -1.51 | 0.05 |
| Superoxide dismutase 1, soluble | P08228 | IPI00130589 | 15943 | 6.02 | Cytoplasm/mitochondria | redox | 9 | -1.54 | 0.05 |
| Thioredoxin reductase 2 | Q9JLT4 | IPI00124699 | 56453 | 8.72 | Mitochondria | redox | 2 | -1.5 | 0.028 |
| Biliverdin reductase B (NADPH) | Q923D2 | IPI00113996 | 22197 | 6.49 | Cytoplasm | redox | 9 | -1.53 | 0.05 |
| Peroxiredoxin 1 | P35700 | IPI00121788 | 22177 | 8.26 | Cytoplasm | redox | 2 | -1.54 | 0.05 |
| Peroxiredoxin 4 | O08807 | IPI00116254 | 31053 | 6.67 | Cytoplasm | redox | 2 | -1.69 | 0.05 |
| Peroxiredoxin 6 | O08709 | IPI00555059 | 24871 | 5.71 | Cytoplasm/lysosome | redox | 6 | -1.59 | 0.05 |
| Isocitrate dehydrogenase [NADP] cytoplasmic | O88844 | IPI00135231 | 46660 | 6.48 | Cytoplasm | redox | 4 | -4.43 | |
| Glutaredoxin 1 | Q9QUH0 | IPI00331528 | 11871 | 8.68 | Cytoplasm | redox | 4 | -1.72 | |
| Glutathione reductase 1 precursor | P47791 | IPI00111359 | 53663 | 8.19 | Cytoplasm/mitochondria | redox | 2 | -1.5 | 0.028 |
| Alpha enolase | P17182 | IPI00462072 | 47141 | 6.37 | Cytoplasm/ membrane | glycolysis | 3 | -2.08 | |
| Enolase 3, beta | P21550 | IPI00228548 | 47025 | 6.73 | Cytoplasm | glycolysis | 2 | 1.56 | |
| Lactate dehydrogenase A | P06151 | IPI00319994 | 36499 | 7.61 | Cytoplasm | glycolysis | 12 | 1.63 | |
| Pyruvate dehydrogenase (lipoamide) beta | Q9D051 | IPI00132042 | 38937 | 6.41 | Mitochondria | glycolysis | 8 | -1.5 | 0.004 |
| Pyruvate dehydrogenase E1 alpha 1 | P35486 | IPI00337893 | 43232 | 8.49 | Mitochondria | glycolysis | 2 | -2.66 | |
| Pyruvate kinase M | P52480 | IPI00407130 | 57845 | 7.17 | Mitochondria | glycolysis | 2 | -2.08 | |
| Triosephosphate isomerase 1 | P17751 | IPI00467833 | 26713 | 6.90 | Cytoplasm | glycolysis | 2 | -1.53 | 0.05 |
| Malate dehydrogenase | P14152 | IPI00336324 | 36511 | 6.16 | Cytoplasm | TCA cycle | 2 | -1.94 | |
| Dihydrolipoamide dehydrogenase | O08749 | IPI00874456 | 54272 | 7.99 | Mitochondria | oxidoreductase | 2 | -1.63 | |
| Nucleoside-diphosphate kinase | Q5NC82 | IPI00127417 | 17363 | 6.97 | Mitochondria | cell survival/apoptosis | 4 | -5.68 | |
| ATP synthase, H+ transporting mitochondrial F1 complex, delta subunit | Q4FK74 | IPI00453777 | 17600 | 5.03 | Mitochondria (Complex V) | oxidative phosphorylation | 3 | -1.84 | |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | B1ASE1 | IPI00230507 | 18749 | 5.52 | Mitochondria (Complex V) | oxidative phosphorylation | 2 | -1.51 | 0.05 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit b | Q5I0W0 | IPI00341282 | 28949 | 9.11 | Mitochondria (Complex V) | oxidative phosphorylation | 3 | -2.66 | |
| ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit e | Q5EBI8 | IPI00111770 | 8236.5 | 7.99 | Mitochondria (Complex V) | oxidative phosphorylation | 20 | -1.88 | 0.011 |
| Electron transferring flavoprotein, alpha polypeptide | B1B1B4 | IPI00116753 | 35009 | 8.62 | Mitochondria | electron transport | 2 | -1.71 |
The CID spectra were compared against those of the EMBL nonredundant protein database by using SEQUEST (ThermoElectron, San Jose, CA). After filtering the results based on cross correlation Xcorr (cutoffs of 2.0 for [M+ H]1+, 2.5 for [M + 2H]2+, and 3.0 for [M+ 3H]3+), peptides with scores greater than 3000 and meeting delta cross-correlation scores (ΔCn) > 0.3, and fragment ion numbers > 60% were deemed valid by these SEQUEST criteria thresholds, which have been determined to afford greater than 95% confidence level in peptide identification.
SwissProt accession number (accessible at http://ca.expasy.org/sprot/).
International Protein Index (IPI) (accessible at http://www.ebi.ac.uk/IPI/).
Theoretical molecular mass for the primary translation product calculated from protein DNA sequences.
Theoretical isoelectric point.
Postulated subcellular location (accessible at http://locate.imb.ug.edu.au).
Postulated cellular function (accessible at http://ca.expasy.org/sprot/).
Number of different peptides identified for each protein.
Fold changes of proteins in N-α-syn stimulated microglial lysates versus unstimulated microglial lysates. Negative DIGE index indicates decreased expression in N-α-syn stimulated microglia relative to controls.
P-values as determined by Biological Variation Analysis by one-way ANOVA for pair-wise comparison between treatments.
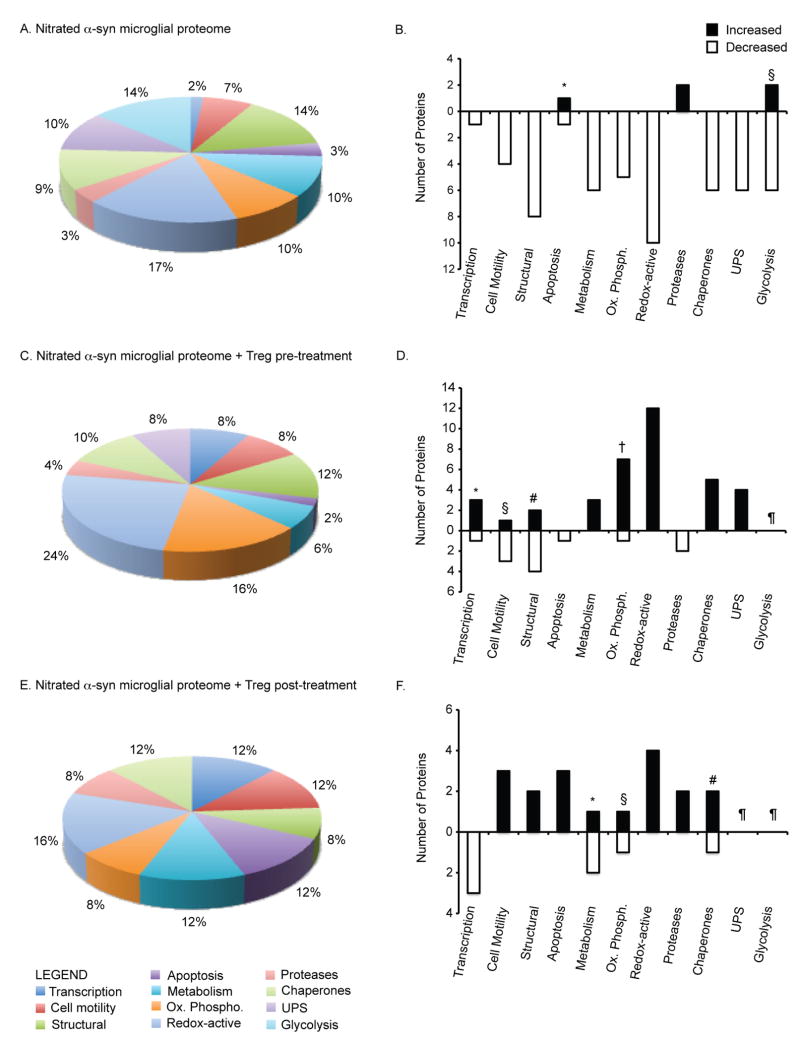
Figure 3.
Classification of proteins modulated by N-α-syn stimulation and Treg treatment. Pie-chart diagrams represent the proportion (%) of proteins within specific categories based on classification and function identified by mass spectrometry. (A) Classification of proteins differentially expressed by microglia in response to N-α-syn stimulation alone. (B) Relative expression of proteins in response to N-α-syn stimulation compared to unstimulated controls. Several proteins within each category showing both increased and decreased proteins were identified including those for apoptosis (*gelsolin increased; nucleoside-diphosphate kinase decreased) and glycolysis (§enolase 3 and lactate dehydrogenase increased; alpha enolase, pyruvated dehydrogenase, pyruvate kinase, and triosphosphate isomerase 1 decreased) (Table 1). (C) Proportion of microglial proteins differentially expressed in response to N-α-syn following Treg pre-treatment and the relative expression trends shown in D. Categories associated with transcription (*VIP-receptor gene repressor protein, TAR DNA binding protein, and Ubiquitin conjugating enzyme E2N increased; MRG-binding protein decreased), cell motility (§microtubule associated protein increased; laminin B2, beta actin, and alpha tubulin decreased), structural (#Capg and guanine nucleotide exchange factor increased; vimentin, cofilin 1 and 2 decreased), and oxidative phosphorylation (†NADH dehydrogenase Fe-S, ATP synthase O subunit, H+-ATP synthase e subunit, and cytochrome c oxidase increased; ATP synthase F0 complex decreased) consisted of both increased and decreased expression of proteins (Table 2). (E) Proportion of microglial proteins differentially expressed in response to N-α-syn following Treg post-treatment and the relative expression trends shown in F. Categories associated with metabolism (#phosphoglycerate mutase 1 increased; aldolase 1 and aldehyde dehydrogenase 2 decreased), oxidative phosphorylation (§ATP synthase D increased; H+-transporting two-sector ATPase alpha chain decreased), and chaperones (#cyclophilin A increased; protein disulfide isomerase decreased) consisted of both increased and decreased expression of proteins (Table 3). (¶Proteins within this category were not identified as differentially expressed).
A majority of the proteins positively identified by mass spectrometry were decreased in expression. A large percentage of the proteins that were decreased in response to N-α-syn stimulation following 24 h were cytoskeletal associated including vimentin, cofilin 1, beta-actin and alpha-tubulin (Table 1). N-α-syn stimulation also resulted in decreased expression of proteins involved in protein processing, transport, and folding. These included cryptochrome 2, 14-3-3 zeta, and annexin A1, as well as several molecular chaperones including heat shock protein (Hsp) 10, Hsp 60, and Hsp 70. Moreover, stimulation with N-α-syn decreased expression of proteins associated with the ubiquitin-proteasome system (UPS) greater than 1.5-fold compared to unstimulated microglia (Table 1). Several proteins associated with mitochondrial function and redox biology were also decreased as a result of stimulation with N-α-syn. Of interest, proteins of the electron transport chain (ETC), specifically complex V involved in adenosine triphosphate (ATP) synthesis, were decreased in expression. Redox-active proteins were also decreased following 24 h of exposure to N-α-syn including superoxide dismutase (Sod)1, biliverdin reductase B, peroxiredoxin (Prdx) 1 and glutaredoxin 1 (Table 1). Other proteins decreased following stimulation with N-α-syn stimulation were metabolic proteins such as acetyl-coenzyme A and aldehyde dehydrogenase, and proteins involved in glycolysis such as alpha enolase, pyruvate dehydrogenase, and pyruvate kinase (Table 1). Despite the even-distribution of up- and down-regulated proteins identified in the initial analysis, many of the proteins that were increased in expression did not reach the confidence interval threshold for adequate identification by mass spectrometry. Nonetheless, those identified included lysosomal proteases cathepsins B and D, gelsolin implicated in inflammation and proteins involved in catabolism including aldo-keto reductase family 1 member B8 and catechol o-methyltransferase (Table 1).
Treg-microglial co-cultivation followed by N-α-syn stimulation (pre-treatment)
To simulate preclinical disease and assess putative mechanisms for early affects of CD4+ T cells on the microglial phenotype in response to N-α-syn, microglial cells were co-cultured with CD3-activated CD4+ T cells for 24 h prior to exposure to N-α-syn. Table 2 shows those proteins differentially expressed in microglia stimulated with N-α-syn alone or pre-treated with Treg. The relative percentages of proteins within each classification based on protein function that were modulated by Treg pre-treatment together with N-α-syn stimulation and expression trends are shown in Figure 3B. Among the proteomic changes induced by pre-treatment of microglia with Treg prior to N-α-syn stimulation were decreased expression in several cytoskeletal proteins such as β-actin, vimentin, cofilin 1, and gelsolin, involved in regulation of cell motility and vesicle transport. Treatment with Treg also resulted in increased expression of microglial proteins involved in exocytosis such as annexin A1 and annexin A4, and phagocytosis such as L-plastin (Table 2). In addition, pre-treatment with Treg increased expression of UPS-related proteins including proteasome subunit alpha type-2, proteasome subunit beta type-2, ubiquitin specific protease 19 and ubiquitin fusion degradation. Treatment with Treg also increased the expression of molecular chaperones including HSPs and calreticulin. Whereas lysosomal proteases cathepsins B and D were increased by N-α-syn stimulation alone, microglia pre-treated with Treg showed decreased abundance of the same proteins. Regulatory proteins involved in cellular metabolism (transaldolase 1) and catabolism (α-mannosidase) were increased in Treg pre-treated cultures (Table 2).
Table 2. Modulation of the N-α-syn microglial proteome by Treg pretreatment.
| LC/MS/MS* Protein ID by |
SwissProt† | IPI‡ | M.wt.§ (DA) |
pI11 | Subcellular Location¶ | Function# | Peptide#** | DIGE†† Index |
P-value‡‡ |
|---|---|---|---|---|---|---|---|---|---|
| VIP-receptor-gene repressor protein | O88461 | IPI00209665 | 72972 | 9.57 | Nucleus | transcription | 2 | 1.5 | 0.0011 |
| TAR DNA binding protein | Q921F2 | IPI00121758 | 44548 | 6.26 | Nucleus | transcription | 2 | 1.52 | 0.003 |
| MRG-binding protein | Q9DAT2 | IPI00119018 | 23888 | 4.87 | Nucleus | transcription | 2 | -3.79 | <0.0001 |
| Ubiquitin conjugating enzyme E2N | P61089 | IPI00165854 | 17138 | 6.13 | Nucleus | transcription | 3 | 2.15 | 0.0076 |
| Eukaryotic translation initiation factor 3, subunit H | Q91WK2 | IPI00128202 | 39832 | 6.19 | Nucleus | translation | 2 | -1.52 | 0.0037 |
| Laminin B2 | Q61292 | IPI00119065 | 196352 | 6.28 | Secreted | cell motility | 2 | -1.51 | 0.0038 |
| Beta actin | P60710 | IPI00110850 | 269833 | 5.82 | Cytoskeleton | cell motility | 5 | -2.09 | 0.0008 |
| Alpha-tubulin | P68369 | IPI00110753 | 50136 | 4.94 | Cytoskeleton | cell motility | 2 | -1.51 | 0.0038 |
| Microtubule-associated protein, RP/EB family, member 1 | Q7TN34 | IPI00117896 | 29885 | 5.12 | Cytoskeleton | cell motility | 9 | 1.5 | 0.0011 |
| Chloride intracellular channel 1 | Q9Z1Q5 | IPI00130344 | 27013 | 5.09 | Cytoplasm | ion channel | 6 | 1.55 | 0.003 |
| Voltage-dependent anion channel 2 | Q60930 | IPI00122547 | 31733 | 7.44 | Mitochondria | ion channel | 3 | 1.53 | 0.005 |
| Voltage-dependent anion channel 1 | Q60932 | IPI00122549 | 32351 | 8.55 | Mitochondria | ion channel | 5 | 1.88 | <0.0001 |
| Vimentin | P20152 | IPI00227299 | 53688 | 5.06 | Cytoskeleton | stabilize cytoskeleton | 6 | -1.63 | 0.0002 |
| Cofilin 1 | P18760 | IPI00890117 | 18559 | 8.22 | Cytoskeleton | actin polymerization | 3 | -1.61 | 0.0022 |
| Cofilin 2 | P45591 | IPI00266188 | 18710 | 7.66 | Cytoskeleton | actin polymerization | 2 | -1.5 | 0.05 |
| Macrophage capping protein (CAPG) | P24452 | IPI00136906 | 39240 | 6.73 | Cytoplasm | inhibits actin polymerization | 9 | 1.82 | 0.036 |
| Guanine nucleotide exchange factor GEFT | Q9CWR0 | IPI00109434 | 68262 | 5.19 | Cytoplasm | actin reorganization | 4 | 2.7 | <0.0001 |
| Gelsolin | A2AL35 | IPI00117167 | 85942 | 5.83 | Cytoskeleton | apoptosis and inflammation, vesicle transport | 5 | -2.48 | 0.0002 |
| Galectin 3 | P16110 | IPI00131259 | 27515 | 8.47 | Cytoplasm/nucleus | protein binding, phagocytosis | 26 | -1.68 | 0.0016 |
| Early endosome antigen 1 | Q8BL66 | IPI00453776 | 160915 | 5.99 | Cytoplasm | endosomal trafficking | 8 | -2.41 | <0.0001 |
| Annexin A1 | P10107 | IPI00230395 | 38734 | 6.97 | Cytoplasm | membrane fusion and exocytosis | 3 | 1.53 | 0.0047 |
| Annexin A4 | P97429 | IPI00353727 | 35990 | 5.43 | Cytoplasm | membrane fusion and exocytosis | 18 | 1.5 | 0.0011 |
| Nestin | Q6P5H2 | IPI00453692 | 207124 | 4.3 | Cytoplasm | protein trafficking | 2 | 1.51 | 0.023 |
| cAMP-dependent protein kinase | P05132 | IPI00227900 | 40571 | 8.84 | Cytoplasm | protein trafficking | 5 | -1.59 | 0.05 |
| Non-specific lipid transfer protein | P32020 | IPI00134131 | 59126 | 7.16 | Cytoplasm | lipid protein transfer | 3 | 4.42 | <0.0001 |
| Glycolipid transfer protein | Q9JL62 | IPI00229718 | 23690 | 6.9 | Cytoplasm | lipid protein transfer | 2 | 1.55 | 0.003 |
| Peptide chain release factor 1 | Q8BWY3 | IPI00312468 | 49031 | 5.51 | Cytoplasm | termination of peptide synthesis | 3 | 1.71 | 0.034 |
| L-Plastin | Q61233 | IPI00118892 | 70149 | 5.2 | Cytoskeleton | phagocytosis | 25 | 2.1 | 0.0029 |
| Ferritin light chain 1 | P29391 | IPI00762203 | 20802 | 5.66 | Cytoplasm | iron homeostasis | 2 | 1.51 | 0.023 |
| Ferritin heavy chain | P09528 | IPI00230145 | 21067 | 5.53 | Cytoplasm | iron homeostasis | 8 | -1.54 | 0.0033 |
| Transaldolase 1 | Q93092 | IPI00124692 | 37387 | 6.57 | Cytoplasm | metabolism | 7 | 1.71 | 0.034 |
| Hypoxanthine guanine phosphoribosyl transferase 1 | P00493 | IPI00284806 | 24570 | 6.21 | Cytoplasm | metabolism | 4 | 2.33 | <0.0001 |
| Sterol carrier protein 2 | A2APS3 | IPI00134131 | 59126 | 7.16 | Mitochondria | metabolism | 9 | 4.42 | <0.0001 |
| Aconitate hydratase | Q99KI0 | IPI00116074 | 85464 | 8.08 | Mitochondria | enzyme | 2 | 1.51 | 0.023 |
| Lysosomal alpha-mannosidase precursor | O09159 | IPI00381303 | 114604 | 8.3 | Lysosome | catabolism | 3 | 2.59 | <0.0001 |
| Contrapsin | P07759 | IPI00131830 | 46880 | 5.05 | Secreted | protease inhibitor | 3 | 6.56 | <0.0001 |
| Calpastatin | P51125 | IPI00409176 | 84922 | 5.37 | Cytoplasm | protease inhibitor | 2 | -1.86 | 0.0004 |
| Cathepsin B | P10605 | IPI00113517 | 37280 | 5.57 | Lysosome | thiol protease | 15 | -3.34 | <0.0001 |
| Cathepsin D | P18242 | IPI00111013 | 44954 | 6.71 | Lysosome | acid protease | 5 | -2.09 | 0.0008 |
| Cathepsin Z | Q9R1T3 | IPI00207663 | 34194 | 6.74 | Cytoplasm/Secreted | peptidase | 2 | 1.5 | 0.0011 |
| SDF2 like protein 1 | Q9ESP1 | IPI00227657 | 23648.34 | 6.92 | ER | stress response | 3 | 2.59 | 0.0067 |
| Calreticulin | P14211 | IPI00123639 | 47995 | 4.33 | Membrane/ ER | chaperone | 20 | 2.32 | <0.0001 |
| HSP 10 | Q64433 | IPI00263863 | 10962.7 | 7.91 | Mitochondria | chaperone | 5 | 1.52 | 0.0025 |
| HSP 70 | P63017 | IPI00323357 | 70871 | 5.37 | Cytoplasm | chaperone | 3 | 1.59 | 0.0097 |
| HSP 90 | Q80Y52 | IPI00330804 | 84788 | 4.93 | Cytoplasm | chaperone | 2 | 2.09 | 0.0004 |
| Proteasome subunit beta type-2 | Q9R1P3 | IPI00128945 | 22906 | 6.52 | Cytoplasm | Ubiquitin-Proteasome system | 4 | 2.59 | <0.0001 |
| Proteasome (prosome, macropain) subunit, alpha type 2 | P49722 | IPI00890001 | 25926 | 8.39 | Cytoplasm | Ubiquitin-Proteasome system | 7 | 6.56 | <0.0001 |
| Ubiquitin specific protease 19 | Q3UJD6 | IPI00420483 | 150549 | 5.99 | Cytoplasm | Ubiquitin-Proteasome system | 2 | 4.42 | <0.0001 |
| Ubiquitin fusion degradation | P70362 | IPI00656165 | 34484 | 6.97 | Cytoplasm/ ER | Ubiquitin-Proteasome system | 2 | 1.52 | 0.0037 |
| Immune costimulatory protein B7-H4 | Q7TSP5 | IPI00169522 | 30875 | 5.69 | Membrane | immune response | 2 | -1.59 | 0.05 |
| Interferon-alpha/beta receptor alpha chain precursor | P33896 | IPI00115420 | 65777 | 5.37 | Membrane | immune response | 2 | 3.29 | <0.0001 |
| Interferon-induced GTP-binding protein | Q01514 | IPI00124675 | 67712 | 5.41 | Membrane | immune response | 2 | -1.83 | 0.0011 |
| Peroxiredoxin 1 | P35700 | IPI00121788 | 22177 | 8.26 | Cytoplasm | redox | 7 | 2.59 | <0.0001 |
| Peroxiredoxin 3 | Q9Z0V6 | IPI00208215 | 28295 | 7.14 | Mitochondria | redox | 7 | 2.7 | <0.0001 |
| Peroxiredoxin 4 | O08807 | IPI00116254 | 31053 | 6.67 | Cytoplasm | redox | 8 | 2.33 | <0.0001 |
| Peroxiredoxin 5 | P99029 | IPI00129517 | 21897 | 9.1 | Mitochondria | redox | 3 | 2.43 | <0.0001 |
| Peroxiredoxin 6 | O08709 | IPI00555059 | 24871 | 5.71 | Cytoplasm/lysosome | redox | 4 | 1.7 | 0.05 |
| Superoxide dismutase 1 [Cu-Zn] | P08228 | IPI00130589 | 15943 | 6.02 | Cytoplasm/mitochondria | redox | 2 | 1.6 | 0.05 |
| Superoxide dismutase 2 [Mn] | P09671 | IPI00109109 | 24603 | 8.8 | Mitochondria | redox | 2 | 1.71 | <0.0001 |
| Glutaredoxin 1 | Q9QUH0 | IPI00331528 | 11871 | 8.68 | Cytoplasm | redox | 2 | 1.82 | <0.0001 |
| Biliverdin reductase B (NADPH) | Q923D2 | IPI00113996 | 22197 | 6.49 | Cytoplasm | redox | 13 | 6.56 | <0.0001 |
| Oxidation resistance 1 | Q4KMM3 | IPI00277552 | 83016 | 4.9 | Mitochondria | redox | 2 | 1.51 | 0.05 |
| Thioredoxin 1 | P10639 | IPI00226993 | 11675 | 4.8 | Mitochondria | redox | 3 | 3.36 | <0.0001 |
| Catalase | P24270 | IPI00312058 | 59765 | 7.72 | Mitochondria | redox | 2 | 2.13 | 0.014 |
| Prohibitin | P67778 | IPI00133440 | 29820 | 5.57 | Mitochondria | respiration activity | 2 | 2.7 | <0.0001 |
| NADH dehydrogenase (ubiquinone) Fe-S protein-2 | Q923F9 | IPI00229008 | 18518 | 9.9 | Mitochondria (Complex I) | oxidative phosphorylation | 3 | 2.94 | 0.0012 |
| Mitochondrial ATP synthase, O subunit | Q9DB20 | IPI00118986 | 23364 | 10 | Mitochondria (Complex V) | oxidative phosphorylation | 3 | 2.59 | 0.0067 |
| H(+)-ATP synthase subunit e | P56382 | IPI00230241 | 5838 | 10.01 | Mitochondria (Complex V) | oxidative phosphorylation | 2 | 1.52 | 0.0025 |
| ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit e | Q5EBI8 | IPI00111770 | 8237 | 7.99 | Mitochondria (Complex V) | oxidative phosphorylation | 4 | 1.52 | 0.0025 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | B1ASE1 | IPI00230507 | 18749 | 5.52 | Mitochondria (Complex V) | oxidative phosphorylation | 21 | -1.83 | 0.0011 |
| Cytochrome c oxidase, subunit Va | P12787 | IPI00120719 | 16101 | 6.08 | Mitochondria (Complex IV) | oxidative phosphorylation | 4 | 2.65 | 0.014 |
| Cytochrome c oxidase, subunit VIb polypeptide 1 | P56391 | IPI00225390 | 10071 | 8.96 | Mitochondria (Complex IV) | oxidative phosphorylation | 2 | 2 | 0.0012 |
The CID spectra were compared against those of the EMBL nonredundant protein database by using SEQUEST (ThermoElectron, San Jose, CA). After filtering the results based on cross correlation Xcorr (cutoffs of 2.0 for [M+ H]1+, 2.5 for [M + 2H]2+, and 3.0 for [M+ 3H]3+), peptides with scores greater than 3000 and meeting delta cross-correlation scores (ΔCn) > 0.3, and fragment ion numbers > 60% were deemed valid by these SEQUEST criteria thresholds, which have been determined to afford greater than 95% confidence level in peptide identification.
SwissProt accession number (accessible at http://ca.expasy.org/sprot/).
International Protein Index (IPI) (accessible at http://www.ebi.ac.uk/IPI/).
Theoretical molecular mass for the primary translation product calculated from protein DNA sequences.
Theoretical isoelectric point.
Postulated subcellular location (accessible at http://locate.imb.ug.edu.au).
Postulated cellular function (accessible at http://ca.expasy.org/sprot/).
Number of different peptides identified for each protein.
Fold changes of proteins in Treg pre-treated microglia versus N-α-syn alone stimulated microglial lysates. Negative DIGE index indicates decreased expression in N-α-syn stimulated microglia relative to controls.
P-values as determined by Biological Variation Analysis by one-way ANOVA for pair-wise comparison between treatments.
ETC proteins such as nicotinamide adenine dinucleotide (NADH) dehydrogenase (ubiquinone) Fe-S protein-2 of complex I, cytochrome c oxidase of complex III and the subunits that comprise the components of ATP synthase were increased by microglia in response to N-α-syn stimulation following Treg pre-treatment. Changes in the mitochondrial response to Treg were not limited to proteins involved in cellular energetic, but included redox proteins, chaperones, and structural proteins. Other proteins increased as a result of pre-treatment with Treg were mitochondrial redox-active proteins including peroxiredoxins, Sod 1, Sod 2, thioredoxin (Thrx) 1 and catalase. In addition, cytoplasmic redox-active proteins were also increased including Prdx 1, biliverdin reductase B and glutaredoxin 1 (Table 2).
Cross-comparison of Teff pre-treatments was facilitated by the BVA module to compare protein expression trends. In contrast to pre-treatment with Treg, pre-treatment with Teff did not alter the expression of structural proteins including cofilin 1 and 2, taxilin alpha or beta actin in response to N-α-syn stimulation. Expression of lysomal proteases including cathepsin B and D were also not changed. In addition, pre-treatment with Teff did not affect expression of redox-active proteins such as Prdx 5, cytochrome c reductase, Thrx 1, or biliverdin reductase B. However, enzymatic proteins that were involved in glycolysis and metabolism were decreased in expression following Teff pre-treatment included pyruvate kinase M, phosphoglycerate kinase and aldolase A. Proteins of the ETC were also decreased including ATP synthase (Complex V). Compared to N-α-syn stimulation alone, Teff pre-treatment resulted in greater than 1.5 fold increased expression of voltage-dependent anion channel-1 (Vdac-1), the interferon α/β receptor, and Prdx 1, whereas Hsp 90, chaperonin, galectin 3 and gelsolin were decreased greater than 1.5 fold in expression (data not shown).
N-α-syn stimulation followed by Treg-microglial co-cultivation (post-treatment)
For comparison of the microglial phenotype after commitment to activation by N-α-syn stimulation and modulation by CD3-activated CD4+ T cells, microglia were first stimulated with N-α-syn for 12 h prior to the addition of Treg or Teff for an additional 24 h and the T cells removed prior to microglial cell lysis. Table 3 shows those proteins differentially expressed in microglia stimulated with N-α-syn alone or post-treated with Treg. The relative percentages of proteins within each classification based on protein function that were modulated by Treg post-treatment together with N-α-syn stimulation and expression trends is shown in Figure 3C.
Table 3. Modulation of the N-α-syn microglial proteome by Treg post-treatment.
| LC/MS/MS* Protein ID by |
SwissProt† | IPI‡ | M.wt.§ (DA) |
p11 | Subcellular Location¶ | Function# | Peptide#** | DIGE†† Index |
P-value‡‡ |
|---|---|---|---|---|---|---|---|---|---|
| Histone H4 | P62806 | IPI00407339 | 11367 | 11.21 | Nucleus | nucleosome component | 3 | -1.56 | 0.021 |
| Histone H2B | Q64475 | IPI00554853 | 13592 | 10.31 | Nucleus | nucleosome component | 2 | -2.28 | |
| Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3) | Q8BG05 | IPI00269661 | 39652 | 8.46 | Nucleus | cytoplasmic trafficking of RNA | 5 | -1.62 | |
| GTP-binding nuclear protein Ran | P62827 | IPI00134621 | 24423 | 7.19 | Nucleus/Cytoplasm | GTPase activity | 5 | 1.35 | 0.033 |
| Rho GTPase-activating protein 1 | Q5FWK3 | IPI00404970 | 50411 | 5.97 | Membrane | GTPase activity | 2 | -1.75 | 0.05 |
| Rho GDP-dissociation inhibitor | Q99PT1 | IPI00322312 | 23407 | 5.12 | Cytoplasm | GTPase activity | 2 | 1.48 | 0.046 |
| Guanine nucleotide-binding protein subunit beta-2 | P62880 | IPI00162780 | 37331 | 7.06 | Membrane | signaling | 14 | 1.32 | 0.018 |
| Stathmin | P54227 | IPI00551236 | 17274 | 5.77 | Cytoplasm | cell motility | 3 | 1.52 | 0.0024 |
| Beta-actin | P60710 | IPI00110850 | 41737 | 5.78 | Cytoplasm | cell motility | 7 | 1.53 | 0.015 |
| Gamma-actin | P63260 | IPI00874482 | 41793 | 5.56 | Cytoplasm/Cytoskeleton | cell motility | 9 | 1.54 | |
| Cofilin-1 | P18760 | IPI00890117 | 18560 | 8.22 | Cytoplasm | actin polymerization | 2 | 1.63 | 0.0065 |
| Brain acid soluble protein 1 | Q91XV3 | IPI00129519 | 22087 | 4.5 | Membrane | nurite outgrowth | 9 | 1.55 | |
| Gelsolin | A2AL35 | IPI00117167 | 85942 | 5.83 | Cytoskeleton | apoptosis and inflammation, vesicle transport | 7 | 1.73 | 0.004 |
| Galectin-3 | P16110 | IPI00131259 | 27515 | 8.5 | Nucleus | protein binding, phagocytosis | 7 | 1.65 | 0.067 |
| Cyclophilin A | P17742 | IPI00554989 | 17971 | 7.74 | Cytoplasm | protein folding | 6 | 1.48 | 0.05 |
| Protein disulfide isomerase | Q8BXZ1 | IPI00453798 | 51848 | 5.02 | ER | protein folding | 18 | -1.53 | |
| L-Plastin | Q61233 | IPI00118892 | 70149 | 5.21 | Cytoplasm | Phagocytosis | 17 | -1.53 | |
| Ferritin heavy chain | P09528 | IPI00230145 | 21067 | 5.53 | Cytoplasm | iron homeostasis | 2 | 1.50 | 0.0072 |
| Ferritin Light Chain 1 | P29391 | IPI00762203 | 20802 | 5.66 | Cytoplasm | iron homeostasis | 2 | 1.5 | 0.0072 |
| Leupaxin | Q8R355 | IPI00387515 | 43460 | 5.88 | Cytoplasm | zinc ion binding | 2 | 1.4 | 0.0021 |
| Aldolase I | P05064 | IPI00221402 | 39356 | 8.31 | Cytoplasm | metabolism | 4 | -2.16 | |
| Aldehyde dehydrogenase 2 | Q3TVM2 | IPI00111218 | 56596 | 7.03 | Mitochondria | metabolism | 4 | -1.75 | 0.05 |
| Phosphoglycerate mutase 1 | Q9DBJ1 | IPI00457898 | 28832 | 6.75 | Cytoplasm | metabolism | 8 | 1.37 | 0.044 |
| Transmembrane glycoprotein NMB (Dendritic cell-associated transmembrane protein) | Q99P91 | IPI00311808 | 63675 | 7.88 | Membrane | enzyme | 2 | -3.50 | 0.0089 |
| Alpha-enolase | P17182 | IPI00462072 | 47141 | 6.36 | Cytoplasm | enzyme | 9 | 1.40 | 0.0021 |
| Beta enolase | P21550 | IPI00228548 | 47025 | 6.73 | Cytoplasm | enzyme | 3 | 1.4 | 0.0021 |
| S-formylglutathione hydrolase | Q9R0P3 | IPI00109142 | 31320 | 6.70 | Cytoplasm | enzyme | 5 | 1.49 | 0.0068 |
| Peptidylprolyl isomerase A | Q8CEC6 | IPI00229025 | 73431 | 6.58 | Cytoplasm | enzyme | 6 | 1.48 | 0.05 |
| Malate dehydrogenase, cytosolic | P14152 | IPI00336324 | 36511 | 6.16 | Cytoplasm | enzyme | 2 | 1.49 | 0.0068 |
| Nucleoside diphosphate kinase | Q9WV84 | IPI00125448 | 20549 | 9.21 | Mitochondria | enzyme | 5 | 1.48 | 0.05 |
| Phosphoglycerate kinase 1 | P09411 | IPI00555069 | 44540 | 8.02 | Cytoplasm | enzyme | 3 | 1.63 | 0.0065 |
| Adenylosuccinate synthase | P28650 | IPI00123190 | 50254 | 8.57 | Cytoplasm/Membrane | enzyme | 2 | -1.74 | |
| Cathepsin B precursor | P10605 | IPI00113517 | 37280 | 5.57 | Lysosome | thiol protease | 7 | 1.6 | 0.022 |
| Cathepsin D precursor | P18242 | IPI00111013 | 44954 | 6.71 | Lysosome | acid protease | 2 | 1.63 | 0.0065 |
| Vacuolar proton pump subunit E 1 | P50518 | IPI00119115 | 26157 | 8.44 | Cytoplasm | proton pump for acidification of intracellular compartments | 8 | -2.08 | |
| Beta-N-acetylhexosaminidase | P29416 | IPI00125522 | 60599 | 6.09 | Lysosome | protein degradation | 2 | -1.75 | 0.05 |
| Peroxiredoxin-1 | P35700 | IPI00121788 | 22176 | 8.26 | Cytoplasm | redox | 6 | 1.38 | 0.022 |
| Peroxiredoxin-5 | P99029 | IPI00129517 | 21897 | 9.1 | Mitochondria/Cytoplasm | redox | 6 | 1.46 | 0.003 |
| Superoxide dismutase [Cu-Zn] | P08228 | IPI00130589 | 15943 | 6.03 | Cytoplasm | redox | 4 | 1.51 | 0.048 |
| Vat1 | Q62465 | IPI00126072 | 43097 | 5.95 | Membrane | redox | 15 | 1.4 | 0.0021 |
| H+ transporting two-sector ATPase alpha chain | Q03265 | IPI00130280 | 59753 | 9.22 | Mitochondria (Complex V) | oxidative phosphorylation | 18 | -1.75 | 0.048 |
| ATP synthase D chain, mitochondrial | Q9DCX2 | IPI00230507 | 18250 | 5.52 | Mitochondria (Complex V) | oxidative phosphorylation | 2 | 1.5 | 0.0072 |
| Translation elongation factor 1 | Q9D1M4 | IPI00133928 | 19859 | 8.6 | Nucleus/Cytoplasm | DNA damage response | 2 | -1.74 | |
| Apoptosis-associated speck-like protein containing a CARD | Q9EPB4 | IPI00109709 | 21459 | 5.03 | Cytoplasm | caspase-mediated apoptosis | 5 | 1.51 |
The CID spectra were compared against those of the EMBL nonredundant protein database by using SEQUEST (ThermoElectron, San Jose, CA). After filtering the results based on cross correlation Xcorr (cutoffs of 2.0 for [M+ H]1+, 2.5 for [M + 2H]2+, and 3.0 for [M+ 3H]3+), peptides with scores greater than 3000 and meeting delta cross-correlation scores (ΔCn) > 0.3, and fragment ion numbers > 60% were deemed valid by these SEQUEST criteria thresholds, which have been determined to afford greater than 95% confidence level in peptide identification.
SwissProt accession number (accessible at http://ca.expasy.org/sprot/).
International Protein Index (IPI) (accessible at http://www.ebi.ac.uk/IPI/).
Theoretical molecular mass for the primary translation product calculated from protein DNA sequences.
Theoretical isoelectric point.
Postulated subcellular location (accessible at http://locate.imb.ug.edu.au).
Postulated cellular function (accessible at http://ca.expasy.org/sprot/).
Number of different peptides identified for each protein.
Fold changes of proteins in Treg-post-treated microglia versus N-α-syn alone stimulated microglial lysates. Negative DIGE index indicates decreased expression in N-α-syn stimulated microglia relative to controls.
P-values as determined by Biological Variation Analysis by one-way ANOVA for pair-wise comparison between treatments.
Similar proteins were affected by post-treatment with Treg as with pre-treatment; interestingly, some exhibited opposite expression patterns observed after pre-treatment with Treg. Akin to pre-treatment, post-treatment with Treg yielded increased redox-active protein expression by activated microglia including Sod1 and Prdx1 and 5. Several proteins differentially expressed in the pre-treatment analysis were also identified in post-treatment analysis, but were expressed in opposite directions, including increased expression of structural proteins involved in cell motility, such as β-actin and γ-actin, decreased expression of mitochondrial proteins including ETC complex V, and decreased expression of L-plastin (Table 3). Induction of pro-apoptotic protein expression was observed and included increased expression of apoptosis-associated speck-like protein containing a caspase recruitment domain, gelsolin, eukaryotic translation elongation factor 1, and cathepsins B and D. Decreased expression of proteins involved in cellular metabolism such as aldolase I and aldehyde dehydrogenase 2 was also observed in response to Treg post-treatment (Table 3).
Cross-comparison of protein expression trends following post-treatment with Teff revealed that in contrast to pre-treatment, post-treatment with Teff increased expression of redox-active proteins including Prdx 1, Thrx 1, and cytochrome c oxidase in N-α-syn stimulated microglia compared to N-α-syn stimulation alone. Ferritin light chain, Hsp 70, and transaldolase 1 were also increased. Similar to pre-treatment, expression of cathepsins B and D were not affected. Moreover, expression of pro-apoptotic proteins was not affected with Teff post-treatment (data not shown).
Validation of protein identification and biological significance
Immunocytochemistry and Western blot analyses were used to validate protein expression trends identified in our proteomic analyses. Immunoflourescent cytochemistry revealed that stimulation with N-α-syn significantly reduced Prx1 expression in microglial cells compared to unstimulated microglia. In contrast, Treg pre-treatment protected against a decrease in Prx1 expression (Fig. 4A). In comparison, post-treatment with Treg rescued microglial Prx1 expression and restored expression levels to near 100% of the unstimulated control. The effect of Teff was more variable and depended on the temporal engagement of Teff with stimulated microglia. Pre-treatment with Teff did not effectively alter Prx1 expression in response to N-α-syn stimulation, however Prx1 expression appeared to be partially rescued following post-treatment with Teff although this did not reach statistical significance.
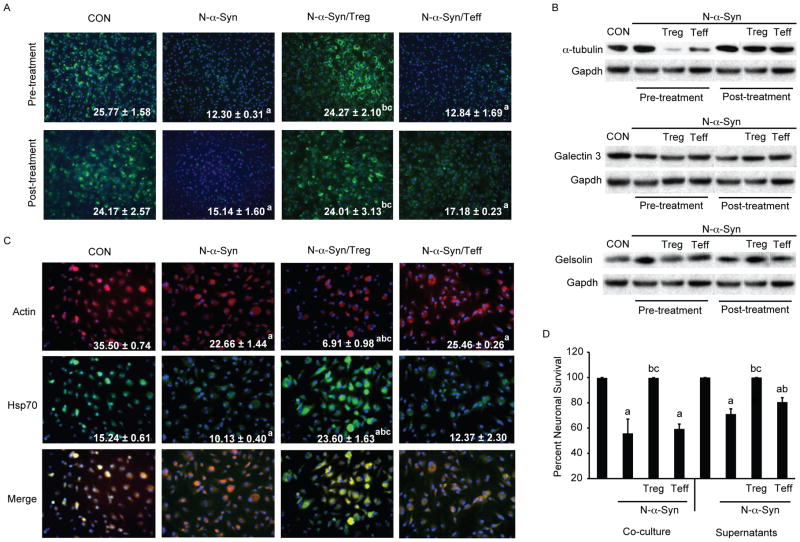
Figure 4.
Treg modulate microglial inflammation to attenuate the neurotoxic phenotype of N-α-syn stimulated microglia. (A) Photomicrographs (20× magnification) of Prx1 expression (green) in microglia treated with media alone (CON), N-α-syn, or cultured with CD4+ T cell subsets following pre-and post-treatment. Values shown are the mean fluorescence intensity (MFI) per field ± SEM. (B) Western blot analysis for α-tubulin, galectin 3 and gelsolin in response to treatment normalized to Gapdh expression within the same blot for comparisons. (C) Photomicrographs (20× magnification) of actin expression (red) or Hsp70 (green) in microglia treated with media alone (CON), N-α-syn, or cultured with CD4+ T cell subsets following pre-and post-treatment. Values shown are the MFI per field ± SEM. (D) Survival of MES23.5 cells after co-culture with N-α-syn stimulated microglia with and without Treg or Teff or after culture with condition media (supernatants) of N-α-syn stimulated microglia treated with either Treg or Teff. Values ± SEM (P< 0.01 vs. aCON, bN-α-syn alone, cN-α-syn/Teff).
Western blot validation for cytoskeletal and inflammatory proteins that were involved both in cell mobilization as well as survival, confirmed expression trends of select proteins following different culture conditions (Fig. 4B). Expression of alpha-tubulin was decreased nearly 6-fold following Treg pre-treatment, and compared to a 1.5 fold increase by N-α-syn stimulation alone. In comparison, alpha tubulin expression in N-α-syn–stimulated microglia following Teff pre-treatment was reduced by 2-fold. Post-treatment with Treg or Teff failed to alter alpha-tubulin expression levels in N-α-syn stimulated microglia. Analysis of gelsolin confirmed the increased expression in N-α-syn stimulated microglial lysates compared to control (1.5 fold). Pre-treatment with Treg reduced gelsolin expression to control levels, while, post-treatment increased gelsolin expression compared to N-α-syn stimulation alone. Albeit pre-treatment with Treg had no effect on galectin 3 expression, post-treatment with Treg resulted in a 1.4 fold increase compared to N-α-syn stimulation alone. No change in expression of gelsolin or galectin 3 was detected in response to Teff treatment by Western blot.
Immunofluorescence cytochemistry for actin and Hsp70 also confirmed differential expression of these proteins following N-α-syn stimulation and pre-treatment with CD4+ T cells. Whereas pre-treatment with Treg significantly decreased fluorescence intensity of beta-actin expression in response to N-α-syn stimulation, expression of Hsp70 was increased compared with N-α-syn stimulation alone to levels and exceeded those observed in unstimulated controls. By comparison, pre-treatment with Teff had no observed affect on either actin or Hsp70 expression compared with N-α-syn stimulation alone (Fig 4C).
Deleterious microglial activation is postulated to affect a neurodegenerative process in PD. For this reason, suppression of microglial activation by Treg may be responsible for the profound protection observed in vivo34. To investigate whether phenotypic modulation of microglia by Treg co-culture affected neuronal survival, an in vitro model of microglia-mediated cytotoxicity was established using N-α-syn-activated microglia and the dopaminergic cell line MES23.5. We observed a 56% loss of MES23.5 cells after co-culture for 24 h with N-α-syn stimulated microglia compared to control co-cultures of MES23.5 with unstimulated microglia (Fig. 4D). In contrast, co-culture of N-α-syn stimulated microglia with Treg inhibited microglial-mediated MES23.5 cytotoxicity, while activated Teff afforded no cytotoxic protection. These data suggested that Treg modulation of microglia attenuates the neurocytotoxic responses mediated by activated microglia. In addition, supernatants from microglia stimulated with N-α-syn alone or N-α-syn and cultured in the presence of Teff were cytotoxic to MES23.5 cells, whereas neurocytotoxicity was abrogated in supernatants from stimulated microglia co-cultured with Treg. Surprisingly, there was less cytotoxicity induced from culture supernatants from N-α-syn microglia treated with Teff than seen in supernatants from N-α-syn microglia alone. How this occurred awaits further study. These data demonstrate the potential of Tregs to suppress cytotoxicity afforded by N-α-syn-activated microglia, and suggest that direct modulation of microglial responses provides a primary mechanism for Treg-mediated neuronal protection.
Discussion
The events that lead to microglial activation in PD and its effects on neuronal survival can be attributed to the formation of aggregated α-syn in the cytosol or in LB, the death of dopaminergic neurons, and the release of these modified aggregates to activate microglia and induce a lethal cascade of neuroinflammation and neuronal destruction33, 43. Oxidation of α-syn leads to formation of aggregates and filaments found to be a major component of LB44, 45. α-Syn released from dying dopaminergic neurons activates microglia, causing release of reactive oxygen species (ROS) and neurotoxicity30-34. Indeed, oxidized and aggregated α-syn, when released from dying neurons, may stimulate scavenger receptors on microglia resulting in their sustained activation and dopaminergic neurodegeneration29, 33, 43. Alternatively, microglia may internalize α-syn through the formation of clathrin pits and secondarily activate microglia46. Activated microglia generate nitric oxide and superoxide that rapidly react to form peroxynitrite47, which can then traverse cell membranes resulting in nitrotyrosine formation and further nitration of α-syn, DNA damage, mitochondrial inhibition, and lipid peroxidation48. The mechanisms by which α-syn activates microglia have been extensively studied and include endocytosis of α-syn by microglia with subsequent cell stimulation resulting in NF-κB activation and secretion of pro-inflammatory cytokines and chemokines as well as production of ROS30, 33, 46. Moreover, α-syn alters the microglial genome, proteome, and secretome leading to the temporal conversion from a neuroregulatory phenotype to an activated phenotype; the latter characterized by differential expression of regulatory, structural, and redox-active proteins, and enzymes together with altered biochemical functions including protein processing, trafficking and degradation30, 31.
Microglia serve as the first line of defense and protects the host against pathogenic microbes through phagocytosis, antigen presentation, and secretion of biologically active factors, as well as mediation of pathological processes. During homeostatic conditions, microglial cells are in a resting state, their cell bodies barely visible and few ramified processes. Neuroprotective functions of homeostatic microglia are suggested by their abilities to produce neurotrophins and eliminate excitotoxins present in the extracellular spaces49 and may also promote neuronal survival following injury50, 51. Underlying these cellular functions is inflammation. Altogether, the inflammatory process serves as a sensor against invasion of bacteria, viruses, and parasites, as well as wound healing following acute tissue infection and injury. However, inflammation is closely linked to neurodegenerative processes. In the central nervous system (CNS), neuroinflammation perpetrated through activation of microglia and other glial elements act in concert as a central pathway in a multitude of neurodegenerative disorders including PD. These initial responders of innate immunity set up a cascade, and later involve the activation and recruitment of adaptive immunity and ultimately neurodegeneration.
A role for adaptive immunity in the pathogenesis of PD has been proposed as a result of several independent lines of investigation that demonstrated a robust adaptive immune response to the CNS consisting of T cell and B cell infiltration, and immunoglobulin deposition within the SNpc to a greater extent than could be attributed to normal immune survelience5, 25-27. Importantly, an intact adaptive immune system with functional CD4+ T cells are required for 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced Parkinsonism in rodents26. As microglial activation is a key pathological feature of PD, we hypothesized that microglial interactions with specific CD4+ T cell subsets may affect the microglial activation phenotype and thus the tempo of disease. Specifically, we posited that interactions with Teff would exacerbate neurotoxic responses whereas Treg would attenuate microglial activation. This is in contrast to current hypotheses that a synergistic immune response between Th1 and Th2 modulate the microglial neurotoxic phenotype to neuroprotective52, 53. According to this hypothesis, synergy between IFN-γ and IL-4 is necessary to modulate neuroprotective innate immunity, in that protection cannot be afforded without the innate immune cells first being activated. However, during neurodegenerative disease, microglia and astrocytes are already in the activated state and induce Th1 responses that exacerbate the microglial neurotoxic phenotype, whereas the induction of Th2/Treg responses would effectively attenuate disease pathogenesis 39, 54.
Genomic and proteomic methodologies are widely used to evaluate changes in gene transcripts and protein expression linked to PD pathogenesis. Analyses of SN of PD patients revealed dysregulation in gene expression, including substantial down-regulation of genes involved in synapse function, dopaminergic phenotype, cytoskeletal maintenance60,and components of the proteasome and ETC complexes61. The results of which provide support for the impairment of multiple ETC complexes and the UPS in PD. Of these, genes involved in the ETC62, as well as genes encoding components of the UPS were decreased. These analyses also revealed an upregulation of genes that participate in protein disposal and degradation60, induction of HSPs, anti-apoptotic gene groups, and genes involved in mitogenic pathways62. Analysis of protein expression of the SN from PD patients implicates an inflammatory process in disease pathogenesis. Higher expression of redox-active proteins64, along with reduced complex I, II, and III activity was also identified to support this contention. Studies have thus far revealed relatively comprehensive quantitative changes in gene expression and protein expression, as well as post-translational modifications (mostly oxidative damage) of high abundance proteins, thus confirming deficits in energy production, protein degradation, antioxidant protein function, and cytoskeletal regulation associated with neurodegenerative diseases such as PD65-72.
Interaction between Treg and microglia affect microglial processes including inflammation, cell function, and specific enzymatic activities ultimately resulting in the conversion of microglia from a neurotoxic to neuroprotective phenotype. This change is multifunctional as the microglial response to stimuli can induce reversion to its original function in maintenance of homeostasis and prevention of neuronal damage. We show that the cellular proteome of microglia in response to N-α-syn stimulation is modulated by pre-treatment with Treg and consists of increased expression of redox-active proteins, altered expression of cytoskeletal proteins involved in phagocytosis and migration, increased expression of HSPs and proteins of the UPS, increased expression of proteins of the ETC, and decreased expression of lysosomal proteases. Taken together, our data suggest that Treg are able to facilitate microglial homeostatic functions to cope with oxidative stress and accumulation of aberrant proteins. Indeed, HSPs have been shown to protect cells from toxicity associated with inhibition of proteasomal function and form excess levels of normal or abnormal proteins55, 56. As inhibition of the proteasomal system has been implicated in PD pathogenesis, stimulation of UPS-mediated proteolysis could serve as a potential therapeutic avenue induced by Treg to reduce protein aggregation and pathology linked to PD. As disease progresses, the effects of Treg on the microglial phenotype may be subverted as a result of reduced numbers or function of Treg, reduced microglial susceptibility to Treg regulation or robust effector T cell responses that overwhelm regulatory functions. The result is a compromise of microglial function and homeostasis and induction of an inflammatory phenotype that mediates neurodegenerative processes.
Proteomic changes observed by addition of Treg post-treatment were less robust than with pre-treatment. In keeping with prior studies, the proteomic profile of the stimulated microglia following co-culture with Treg revealed increased expression of apoptotic proteins, which parallel decreased expression in proteins related to ATP synthesis and cellular metabolism. N-α-syn induces ROS production and NF-κB activation by microglia30-32, and oxidative modification of several proteins may result in altered structure and function or targeted degradation following treatment with Treg that were not targeted with pre-treatment. Less robust changes observed with post-treatment may also be attributed to increased caspase activation and apoptosis of microglia35 resulting in altered protein synthesis and processing. Increased expression of lysosomal proteases including cathepsins B and D suggest that post-stimulatory Treg interactions induce autolysis57. Indeed, we have shown that the pro-apoptotic effect of Tregs on activated microglia is mediated, in part, by the Fas-FasL pathway and is contingent on cathepsin B expression35.
A novel hypothesis for Treg modulation of microglial function during the asymptomatic and overt disease stages in PD is proposed. This hypothesis is based on the activation of innate immune responses by aggregated and oxidized neural proteins, particularly α-syn. We posit that during the asymptomatic stage, adaptive immune responses are operative on microglia that attenuate microglial activation and neuroinflammatory responses including ROS that parallel nigral neuronal damage and subsequent release of α-syn from LB. Treg at this stage of disease modulate microglia to be actively phagocytic and produce a spectrum of regulatory factors that maintain CNS homeostasis. This limits accumulation of α-syn in the extravascular space. Such biochemical events preclude the development of potent neurodegenerative immune responses and the widespread, often adverse affects of oxidized and misfolded proteins. During overt disease, regulation of adaptive immunity breaks down and significantly influences control of the neural environment25, 26. Indeed, the effects of aging on microglial function have been proposed to result in chronic microgliosis or cellular senescence leading to the production of pro-inflammatory and neurotoxic mediators58, 59. Treg may also be, in part, reduced in numbers and function as a result of age, decreased T cell receptor repertoire and N-α-syn immunity. However, clinical analyses of T cell subsets yielded conflicting results in regards to CD4+CD25+ Treg numbers and function5,11 for aging and PD. Nonetheless, the neuroinflammatory events seen in disease are known to result in more widespread nigrostriatal damage, recruitment of immunocytes into the brain and a spiral of pathogenic activities facilitating accelerated neuronal damage and loss. Profound oxidative-associated neuronal damage and death of nigral neurons lead to increased release of α-syn and drives subsequent oxidation and folding. With increased exposure to N-α-syn, microglia are activated yielding a phenotype with reduced phagocytic capacity and homeostatic secretory processes. During this phase, Treg likely engages activated microglia for apoptosis or affect neurotrophic activities while showing limited, in part, pro-inflammatory activity. Our results, taken together, demonstrate the importance of adaptive immune responses in the tempo, progression and control of PD. How such immunomodulators can be controlled for the benefit of the patient will continue to be a target area for future research.
Conclusion
These studies corroborate observations of others73-75 that uncover important differences in the mechanism of Treg-mediated suppression of inflammation. While pre-treatment with Treg alter the microglial activation phenotype to stimulation, Treg interactions following stimuli-mediated activation induce apoptosis. The ability of Treg to regulate microglial inflammation, cell function, and specific enzymatic activities provide novel tools to manipulate ongoing microglial inflammatory responses. In light of these and previously published findings, we now propose a model for disease with regards to a role for Treg in both the pathogenesis and therapeutic intervention of PD. As such, these data support the use of therapeutics that manipulates Treg responses within the brain or that target specific protein changes linked to reversion of a neurotoxic microglial phenotype to neurotrophic.
Supplementary Material
Acknowledgments
The authors thank Drs. Tong Wang, Eric Anderson, Pawel Ciborowski, Joshua Schlautman, Dr. Ronald Cerny for technical assistance and data analysis, and Robin Taylor for critical reading of the manuscript. This work was funded by the Frances and Louie Blumkin Foundation, the Community Neuroscience Pride of Nebraska Research Initiative, the Alan Baer Charitable Trust (to H.E.G.), the UNMC Patterson Fellowship (to A.D.R.), and NIH grants 5P01NS31492, 2R37 NS36126, 2R01 NS034239, P20RR15635, U54NS43011, P01MH64570 and P01 NS43985 (to H.E.G.).
Footnotes
Supplementary Information. An extended material and methods is available in the supplementary information. This information is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Ashley D. Reynolds, Email: areynold@unmc.edu.
David K. Stone, Email: dstone@unmc.edu.
R. Lee Mosley, Email: rlmosley@unmc.edu.
Howard E. Gendelman, Email: hegendel@unmc.edu.
References
- 1.Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39(6):889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 2.Fahn S, Przedborski S. Parkinsonism. In: Rowland LP, editor. Merritt's Neurology. Lippincott Williams & Wilkins; New York: 2000. pp. 679–693. [Google Scholar]
- 3.Fahn S, Sulzer D. Neurodegeneration and neuroprotection in Parkinson disease. NeuroRx. 2004;1(1):139–54. doi: 10.1602/neurorx.1.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayeux R. Epidemiology of neurodegeneration. Annu Rev Neurosci. 2003;26:81–104. doi: 10.1146/annurev.neuro.26.043002.094919. [DOI] [PubMed] [Google Scholar]
- 5.Baba Y, Kuroiwa A, Uitti RJ, Wszolek ZK, Yamada T. Alterations of T-lymphocyte populations in Parkinson disease. Parkinsonism Relat Disord. 2005;11(8):493–8. doi: 10.1016/j.parkreldis.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Klockgether T. Parkinson's disease: clinical aspects. Cell Tissue Res. 2004;318(1):115–20. doi: 10.1007/s00441-004-0975-6. [DOI] [PubMed] [Google Scholar]
- 7.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5(2):133–9. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 8.Naylor K, Li G, Vallejo AN, Lee WW, Koetz K, Bryl E, Witkowski J, Fulbright J, Weyand CM, Goronzy JJ. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174(11):7446–52. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 9.Orr CF, Rowe DB, Mizuno Y, Mori H, Halliday GM. A possible role for humoral immunity in the pathogenesis of Parkinson's disease. Brain. 2005;128(Pt 11):2665–74. doi: 10.1093/brain/awh625. [DOI] [PubMed] [Google Scholar]
- 10.Reale M, Iarlori C, Thomas A, Gambi D, Perfetti B, Di Nicola M, Onofrj M. Peripheral cytokines profile in Parkinson's disease. Brain Behav Immun. 2008 doi: 10.1016/j.bbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Rosenkranz D, Weyer S, Tolosa E, Gaenslen A, Berg D, Leyhe T, Gasser T, Stoltze L. Higher frequency of regulatory T cells in the elderly and increased suppressive activity in neurodegeneration. J Neuroimmunol. 2007;188(12):117–27. doi: 10.1016/j.jneuroim.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Sian J, Dexter DT, Lees AJ, Daniel S, Agid Y, Javoy-Agid F, Jenner P, Marsden CD. Alterations in glutathione levels in Parkinson's disease and other neurodegenerative disorders affecting basal ganglia. Ann Neurol. 1994;36(3):348–55. doi: 10.1002/ana.410360305. [DOI] [PubMed] [Google Scholar]
- 13.Taki J, Nakajima K, Hwang EH, Matsunari I, Komai K, Yoshita M, Sakajiri K, Tonami N. Peripheral sympathetic dysfunction in patients with Parkinson's disease without autonomic failure is heart selective and disease specific. taki@med.kanazawa-u.ac.jpEur J Nucl Med. 2000;27(5):566–73. doi: 10.1007/s002590050544. [DOI] [PubMed] [Google Scholar]
- 14.Tanner CM. Occupational and environmental causes of parkinsonism. Occup Med. 1992;7(3):503–13. [PubMed] [Google Scholar]
- 15.Tanner CM. Epidemiology of Parkinson's disease. Neurol Clin. 1992;10(2):317–29. doi: 10.1016/S0733-8619(18)30212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornykiewicz O, Kish SJ. Biochemical pathophysiology of Parkinson's disease. Adv Neurol. 1987;45:19–34. [PubMed] [Google Scholar]
- 17.Banati RB, Daniel SE, Blunt SB. Glial pathology but absence of apoptotic nigral neurons in long-standing Parkinson's disease. Mov Disord. 1998;13(2):221–7. doi: 10.1002/mds.870130205. [DOI] [PubMed] [Google Scholar]
- 18.Block ML, Wu X, Pei Z, Li G, Wang T, Qin L, Wilson B, Yang J, Hong JS, Veronesi B. Nanometer size diesel exhaust particles are selectively toxic to dopaminergic neurons: the role of microglia, phagocytosis, and NADPH oxidase. Faseb J. 2004;18(13):1618–20. doi: 10.1096/fj.04-1945fje. [DOI] [PubMed] [Google Scholar]
- 19.Cicchetti F, Brownell AL, Williams K, Chen YI, Livni E, Isacson O. Neuroinflammation of the nigrostriatal pathway during progressive 6-OHDA dopamine degeneration in rats monitored by immunohistochemistry and PET imaging. Eur J Neurosci. 2002;15(6):991–8. doi: 10.1046/j.1460-9568.2002.01938.x. [DOI] [PubMed] [Google Scholar]
- 20.Forno LS, DeLanney LE, Irwin I, Di Monte D, Langston JW. Astrocytes and Parkinson's disease. Prog Brain Res. 1992;94:429–436. doi: 10.1016/s0079-6123(08)61770-7. [DOI] [PubMed] [Google Scholar]
- 21.Hong JS. Role of inflammation in the pathogenesis of Parkinson's disease: models, mechanisms, and therapeutic interventions. Ann N Y Acad Sci. 2005;1053:151–2. doi: 10.1196/annals.1344.054. [DOI] [PubMed] [Google Scholar]
- 22.McGeer PL, McGeer EG. Glial cell reactions in neurodegenerative diseases: pathophysiology and therapeutic interventions. Alzheimer Dis Assoc Disord. 1998;12 2:S1–6. [PubMed] [Google Scholar]
- 23.Mirza B, Hadberg H, Thomsen P, Moos T. The absence of reactive astrocytosis is indicative of a unique inflammatory process in Parkinson's disease. Neuroscience. 2000;95:425–432. doi: 10.1016/s0306-4522(99)00455-8. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Chen S, Ma G, Ye M, Lu G. Involvement of proinflammatory factors, apoptosis, caspase-3 activation and Ca2+ disturbance in microglia activation-mediated dopaminergic cell degeneration. Mech Ageing Dev. 2005;126(12):1241–54. doi: 10.1016/j.mad.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Benner EJ, Banerjee R, Reynolds AD, Sherman S, Pisarev VM, Tsiperson V, Nemachek C, Ciborowski P, Przedborski S, Mosley RL, Gendelman HE. Nitrated alpha-synuclein immunity accelerates degeneration of nigral dopaminergic neurons. PLoS ONE. 2008;3(1):e1376. doi: 10.1371/journal.pone.0001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brochard V, Combadiere B, Prigent A, Laouar Y, Perrin A, Beray-Berthat V, Bonduelle O, Alvarez-Fischer D, Callebert J, Launay JM, Duyckaerts C, Flavell RA, Hirsch EC, Hunot S. Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J Clin Invest. 2009;119(1):182–92. doi: 10.1172/JCI36470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theodore S, Cao S, McLean PJ, Standaert DG. Targeted overexpression of human alpha-synuclein triggers microglial activation and an adaptive immune response in a mouse model of Parkinson disease. J Neuropathol Exp Neurol. 2008;67(12):1149–58. doi: 10.1097/NEN.0b013e31818e5e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 29.Croisier E, Moran LB, Dexter DT, Pearce RK, Graeber MB. Microglial inflammation in the parkinsonian substantia nigra: relationship to alpha-synuclein deposition. J Neuroinflammation. 2005;2:14. doi: 10.1186/1742-2094-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds AD, Glanzer JG, Kadiu I, Ricardo-Dukelow M, Chaudhuri A, Ciborowski P, Cerny R, Gelman B, Thomas MP, Mosley RL, Gendelman HE. Nitrated alpha-synuclein-activated microglial profiling for Parkinson's disease. J Neurochem. 2008;104(6):1504–25. doi: 10.1111/j.1471-4159.2007.05087.x. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds AD, Kadiu I, Garg SK, Glanzer JG, Nordgren T, Ciborowski P, Banerjee R, Gendelman HE. Nitrated alpha-synuclein and microglial neuroregulatory activities. J Neuroimmune Pharmacol. 2008;3(2):59–74. doi: 10.1007/s11481-008-9100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas MP, Chartrand K, Reynolds A, Vitvitsky V, Banerjee R, Gendelman HE. Ion channel blockade attenuates aggregated alpha synuclein induction of microglial reactive oxygen species: relevance for the pathogenesis of Parkinson's disease. J Neurochem. 2007;100(2):503–19. doi: 10.1111/j.1471-4159.2006.04315.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Wang T, Pei Z, Miller DS, Wu X, Block ML, Wilson B, Zhou Y, Hong JS, Zhang J. Aggregated alpha-synuclein activates microglia: a process leading to disease progression in Parkinson's disease. Faseb J. 2005;19(6):533–42. doi: 10.1096/fj.04-2751com. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds AD, Banerjee R, Liu J, Gendelman HE, Mosley RL. Neuroprotective activities of CD4+CD25+ regulatory T cells in an animal model of Parkinson's disease. J Leukoc Biol. 2007;82(5):1083–94. doi: 10.1189/jlb.0507296. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds AD, Stone DK, Mosley RL, Gendelman HE. Nitrated alpha-synuclein induced alteration in microglial immunity is regulated by CD4+ T cell subsets. J Immunol. 2009;182(7):4137–49. doi: 10.4049/jimmunol.0803982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobrenis K. Microglia in cell culture and in transplantation therapy for central nervous system disease. Methods. 1998;16(3):320–44. doi: 10.1006/meth.1998.0688. [DOI] [PubMed] [Google Scholar]
- 37.Enose Y, Destache CJ, Mack AL, Anderson JR, Ullrich F, Ciborowski PS, Gendelman HE. Proteomic fingerprints distinguish microglia, bone marrow, and spleen macrophage populations. Glia. 2005;51(3):161–72. doi: 10.1002/glia.20193. [DOI] [PubMed] [Google Scholar]
- 38.Banerjee R, Mosley RL, Reynolds AD, Dhar A, Jackson-Lewis V, Gordon PH, Przedborski S, Gendelman HE. Adaptive immune neuroprotection in G93A-SOD1 amyotrophic lateral sclerosis mice. PLoS ONE. 2008;3(7):e2740. doi: 10.1371/journal.pone.0002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds AD, Stone DK, Mosley RL, Gendelman HE. Nitrated {alpha}-synuclein-induced alterations in microglial immunity are regulated by CD4+ T cell subsets. J Immunol. 2009;182(7):4137–49. doi: 10.4049/jimmunol.0803982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciborowski P, Enose Y, Mack A, Fladseth M, Gendelman HE. Diminished matrix metalloproteinase 9 secretion in human immunodeficiency virus-infected mononuclear phagocytes: modulation of innate immunity and implications for neurological disease. J Neuroimmunol. 2004;157(12):11–6. doi: 10.1016/j.jneuroim.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 41.Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363(1):198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricardo-Dukelow M, Kadiu I, Rozek W, Schlautman J, Persidsky Y, Ciborowski P, Kanmogne GD, Gendelman HE. HIV-1 infected monocyte-derived macrophages affect the human brain microvascular endothelial cell proteome: new insights into blood-brain barrier dysfunction for HIV-1-associated dementia. J Neuroimmunol. 2007;185(12):37–46. doi: 10.1016/j.jneuroim.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wersinger C, Sidhu A. An inflammatory pathomechanism for Parkinson's disease? Curr Med Chem. 2006;13(5):591–602. doi: 10.2174/092986706776055760. [DOI] [PubMed] [Google Scholar]
- 44.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290(5493):985–9. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 45.Souza JM, Giasson BI, Chen Q, Lee VM, Ischiropoulos H. Dityrosine cross-linking promotes formation of stable alpha -synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J Biol Chem. 2000;275(24):18344–9. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Zhou Y, Wang Y, Fong H, Murray TM, Zhang J. Identification of proteins involved in microglial endocytosis of alpha-synuclein. J Proteome Res. 2007;6(9):3614–27. doi: 10.1021/pr0701512. [DOI] [PubMed] [Google Scholar]
- 47.Dringen R. Oxidative and antioxidative potential of brain microglial cells. Antioxid Redox Signal. 2005;7(910):1223–33. doi: 10.1089/ars.2005.7.1223. [DOI] [PubMed] [Google Scholar]
- 48.Ischiropoulos H. Oxidative modifications of alpha-synuclein. Ann N Y Acad Sci. 2003;991:93–100. doi: 10.1111/j.1749-6632.2003.tb07466.x. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz M. Macrophages and microglia in central nervous system injury: are they helpful or harmful? J Cereb Blood Flow Metab. 2003;23(4):385–94. doi: 10.1097/01.WCB.0000061881.75234.5E. [DOI] [PubMed] [Google Scholar]
- 50.Batchelor PE, Liberatore GT, Wong JY, Porritt MJ, Frerichs F, Donnan GA, Howells DW. Activated macrophages and microglia induce dopaminergic sprouting in the injured striatum and express brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor. J Neurosci. 1999;19(5):1708–16. doi: 10.1523/JNEUROSCI.19-05-01708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batchelor PE, Porritt MJ, Martinello P, Parish CL, Liberatore GT, Donnan GA, Howells DW. Macrophages and Microglia Produce Local Trophic Gradients That Stimulate Axonal Sprouting Toward but Not beyond the Wound Edge. Mol Cell Neurosci. 2002;21(3):436–53. doi: 10.1006/mcne.2002.1185. [DOI] [PubMed] [Google Scholar]
- 52.Garg SK, Kipnis J, Banerjee R. IFN-gamma and IL-4 differentially shape metabolic responses and neuroprotective phenotype of astrocytes. J Neurochem. 2009;108(5):1155–66. doi: 10.1111/j.1471-4159.2009.05872.x. [DOI] [PubMed] [Google Scholar]
- 53.Butovsky O, Talpalar AE, Ben-Yaakov K, Schwartz M. Activation of microglia by aggregated beta-amyloid or lipopolysaccharide impairs MHC-II expression and renders them cytotoxic whereas IFN-gamma and IL-4 render them protective. Mol Cell Neurosci. 2005;29(3):381–93. doi: 10.1016/j.mcn.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto M, Kiyota T, Walsh SM, Liu J, Kipnis J, Ikezu T. Cytokine-mediated inhibition of fibrillar amyloid-beta peptide degradation by human mononuclear phagocytes. J Immunol. 2008;181(6):3877–86. doi: 10.4049/jimmunol.181.6.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan GH, Zhou HY, Yang H, Chen SD. Heat shock proteins reduce alpha-synuclein aggregation induced by MPP+ in SK-N-SH cells. FEBS Lett. 2006;580(13):3091–8. doi: 10.1016/j.febslet.2006.04.057. [DOI] [PubMed] [Google Scholar]
- 56.Uryu K, Richter-Landsberg C, Welch W, Sun E, Goldbaum O, Norris EH, Pham CT, Yazawa I, Hilburger K, Micsenyi M, Giasson BI, Bonini NM, Lee VM, Trojanowski JQ. Convergence of heat shock protein 90 with ubiquitin in filamentous alpha-synuclein inclusions of alpha-synucleinopathies. Am J Pathol. 2006;168(3):947–61. doi: 10.2353/ajpath.2006.050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bird PI. Regulation of pro-apoptotic leucocyte granule serine proteinases by intracellular serpins. Immunol Cell Biol. 1999;77(1):47–57. doi: 10.1046/j.1440-1711.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 58.Choi DY, Zhang J, Bing G. Aging enhances the neuroinflammatory response and alpha-synuclein nitration in rats. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.09.010. in press. [DOI] [PubMed] [Google Scholar]
- 59.Miller KR, Streit WJ. The effects of aging, injury and disease on microglial function: a case for cellular senescence. Neuron Glia Biol. 2007;3(3):245–53. doi: 10.1017/S1740925X08000136. [DOI] [PubMed] [Google Scholar]
- 60.Wang YM, Pu P, Le WD. ATP depletion is the major cause of MPP+ induced dopamine neuronal death and worm lethality in alpha-synuclein transgenic C. elegans. Neurosci Bull. 2007;23(6):329–35. doi: 10.1007/s12264-007-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stack EC, Ferro JL, Kim J, Del Signore SJ, Goodrich S, Matson S, Hunt BB, Cormier K, Smith K, Matson WR, Ryu H, Ferrante RJ. Therapeutic attenuation of mitochondrial dysfunction and oxidative stress in neurotoxin models of Parkinson's disease. Biochim Biophys Acta. 2008;1782(3):151–62. doi: 10.1016/j.bbadis.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Cole NB, Dieuliis D, Leo P, Mitchell DC, Nussbaum RL. Mitochondrial translocation of alpha-synuclein is promoted by intracellular acidification. Exp Cell Res. 2008;314(10):2076–2089. doi: 10.1016/j.yexcr.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gan L, Ye S, Chu A, Anton K, Yi S, Vincent VA, von Schack D, Chin D, Murray J, Lohr S, Patthy L, Gonzalez-Zulueta M, Nikolich K, Urfer R. Identification of cathepsin B as a mediator of neuronal death induced by Abeta-activated microglial cells using a functional genomics approach. J Biol Chem. 2004;279(7):5565–72. doi: 10.1074/jbc.M306183200. [DOI] [PubMed] [Google Scholar]
- 64.Reinheckel T, Deussing J, Roth W, Peters C. Towards specific functions of lysosomal cysteine peptidases: phenotypes of mice deficient for cathepsin B or cathepsin L. Biol Chem. 2001;382(5):735–41. doi: 10.1515/BC.2001.089. [DOI] [PubMed] [Google Scholar]
- 65.Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, Nixon R, Nutt J, Chung K, Zabetian C, Samii A, Lin M, Hattan S, Pan C, Wang Y, Jin J, Zhu D, Li GJ, Liu Y, Waichunas D, Montine TJ, Zhang J. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9(3):293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 66.Basso M, Giraudo S, Corpillo D, Bergamasco B, Lopiano L, Fasano M. Proteome analysis of human substantia nigra in Parkinson's disease. Proteomics. 2004;4(12):3943–52. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 67.Chin MH, Qian WJ, Wang H, Petyuk VA, Bloom JS, Sforza DM, Lacan G, Liu D, Khan AH, Cantor RM, Bigelow DJ, Melega WP, Camp DG, 2nd, Smith RD, Smith DJ. Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson's disease. J Proteome Res. 2008;7(2):666–77. doi: 10.1021/pr070546l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J. Identification of novel proteins associated with both alpha-synuclein and DJ-1. Mol Cell Proteomics. 2007;6(5):845–59. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Jin J, Meredith GE, Chen L, Zhou Y, Xu J, Shie FS, Lockhart P, Zhang J. Quantitative proteomic analysis of mitochondrial proteins: relevance to Lewy body formation and Parkinson's disease. Brain Res Mol Brain Res. 2005;134(1):119–38. doi: 10.1016/j.molbrainres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 70.McLaughlin P, Zhou Y, Ma T, Liu J, Zhang W, Hong JS, Kovacs M, Zhang J. Proteomic analysis of microglial contribution to mouse strain-dependent dopaminergic neurotoxicity. Glia. 2006;53(6):567–82. doi: 10.1002/glia.20294. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, James M, Middleton FA, Davis RL. Transcriptional analysis of multiple brain regions in Parkinson's disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2005;137(1):5–16. doi: 10.1002/ajmg.b.30195. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Y, Wang Y, Kovacs M, Jin J, Zhang J. Microglial activation induced by neurodegeneration: a proteomic analysis. Mol Cell Proteomics. 2005;4(10):1471–9. doi: 10.1074/mcp.M500114-MCP200. [DOI] [PubMed] [Google Scholar]
- 73.Taams LS, van Amelsfort JM, Tiemessen MM, Jacobs KM, de Jong EC, Akbar AN, Bijlsma JW, Lafeber FP. Modulation of monocyte/macrophage function by human CD4+CD25+ regulatory T cells. Hum Immunol. 2005;66(3):222–30. doi: 10.1016/j.humimm.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tiemessen MM, Jagger AL, Evans HG, van Herwijnen MJ, John S, Taams LS. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc Natl Acad Sci U S A. 2007;104(49):19446–51. doi: 10.1073/pnas.0706832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venet F, Pachot A, Debard AL, Bohe J, Bienvenu J, Lepape A, Powell WS, Monneret G. Human CD4+CD25+ regulatory T lymphocytes inhibit lipopolysaccharide-induced monocyte survival through a Fas/Fas ligand-dependent mechanism. J Immunol. 2006;177(9):6540–7. doi: 10.4049/jimmunol.177.9.6540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.