Abstract
Nucleic acid aptamers are rapidly gaining prominence as diagnostic tools, targeting reagents, and potential therapeutics. To extend the use of aptamers into the biochemical analysis of protein interactions on the surface of live cells, we converted an enzymatically generated RNA aptamer into a photo-cross-linkable affinity tag through the replacement of all uracils with 4-thiouracil. Specifically, we converted a previously selected, inhibitory aptamer that binds the soluble extracellular domains of the ERBB3 receptor into a targeted and highly specific cross-linking reagent in a live cell setting. Since the photo-cross-linkable aptamer has two functionalities, targeted and highly selective as well as unspecific cross-linking capability, the attachment of this inhibitory aptamer converts ERBB3 into a passive and signaling incompetent probe of its immediate receptor environment. This approach detects receptor clustering of endogenous ERBB3 in the breast cancer cell line MCF7 at levels as low as 25000 receptors per cell and at aptamer concentrations as low as 20 nM. Our analysis also indicates that ERBB3 receptors are apparently segregated from ERBB2 receptors in their resting state, and both ligand-activated ERBB3 and ERBB2 do not share the same microenvironment as inactive ERBB3.
In recent years, the selection of nucleic acid aptamers by SELEX has emerged as a powerful route to macromolecules that exhibit high affinities and high specificity comparable to those of antibodies (1, 2). Aptamers are on average one-tenth the size of antibodies and are in their final format usually within range of fully synthetic production. This provides a broad range of options for site-specific chemical modification. Significant efforts, in terms of both time and cost, have to be applied to create aptamers that are chemically stable and sufficiently resistant to nucleases for use as therapeutics or related applications in whole organisms. However, to be used as a diagnostic tool in vitro, both in solution and in cell culture, RNA aptamers can be readily stabilized by the simple addition of RNase inhibitors to experiments. For example, RNA aptamers have been selected for distinguishing cell lines on the basis of the presence or absence of clinically relevant biomarkers (3–5). We wanted to use an existing and inhibitory aptamer against a known cell surface target, the ERBB31 receptor, to create a probe that can evaluate the microenvironment of ERBB3 in a live cell setting. The probe, which ultimately consists of both the targeted and inhibited receptor and the attached photo-cross-linkable aptamer, should preferably remain neutral during signaling to distinguish changes in its microenvironment from changes that occur to the status of the probe itself. In other words, we wanted to convert an ERBB3 receptor into a “passive bystander species”, capable of reporting on the status of its immediate surrounding beyond the well-documented stabilization of receptor heterodimers by ligand.
ERBB receptors are cell surface receptor tyrosine kinases, and the human ERBB family consists of the EGF receptor (EGFR or ERBB1) and its homologues, ERBB2, ERBB3, and ERBB4 (also termed HER2/neu, HER3, and HER4, respectively). The protein interactions in the ERBB system in the presence and absence of ligand are very complex, and many aspects of it are still poorly understood. Additional questions arise when data derived in vitro from soluble segments of the receptors are translated to a live cell membrane setting. One question is the presence and role of higher-order receptor associations of either activated or resting receptors in signaling. We specifically wanted to evaluate whether the previously observed self-association of soluble ERBB3 extracellular domains (ECDs) and overexpressed recombinant cellular ERBB3 can be confirmed for endogenous and nonoverexpressed ERBB3 receptors, whether such cellular preassociation or clustering in the absence of ligand involves coclustered ERBB2, and whether the activation of ERBB3 by ligand results in the recruitment of ERBB2 into such clusters of inactive ERBB3. Alternatively, activated ERBB2 and ERBB3 receptors may be spatially segregated from clusters of inactive ERBB3. We have previously reported the selection of SELEX-derived RNA aptamers that bind the extracellular domains of ERBB3 with high specificity. The most potent aptamer (A30) binds in the low nanomolar range and interferes with ligand-induced signaling, but without competition for the ligand binding site of ERBB3 (6). A30 represented a good candidate aptamer to be targeted to ERBB3 and thereby convert the aptamer-tagged and inactive receptor into a passive probe of its microenvironment.
While little is known about the extent and function of higher-order association states of ERBB receptors, a considerable body of work exists on the control and structure of receptor dimers, which are at the core of activation of ERBB receptors. Within the ligand-activated dimer, tyrosine phosphorylation on the cytoplasmic side of the receptors is critical to the transmission of signal (7). For EGFR, the respective ligands are EGF and EGF-like ligands such as TGFα. ERBB4 and ERBB3 bind isoforms of neuregulin, a large family of EGF-related ligands. Isoforms of neuregulin 1 are also called heregulins. All ERBB3 receptors are capable of forming heterodimers with other ERBB family members, and indeed, ERBB3, the only kinase-deficient member in this family (8), relies exclusively on heterodimerization with ERBB1, ERBB2, or ERBB4 for signaling (9, 10). The preferred heterodimerization partner for ERBB3 is ERBB2, which in turn stands out by being the only member of the family lacking any identified ligand to date. Heterodimers of ERBB3 and ERBB2 form a potent signaling pair (11). Overexpression of ERBB2 can be found in a large number of cancers and results in elevated levels of constitutively activated receptors. The consequences of high ERBB2 levels are further amplified by elevated levels of ERBB3 (12, 13). In this context, the apparent tendency of ERBB3 to cluster in the absence of ligand could potentially represent a significant means of limiting ERBB2/ERBB3 signaling, especially if such clusters occur even at low levels of ERBB3 and exclude ERBB2. So far, the extent and nature of clustering of ERBB3 by methods such as random chemical cross-linking could not be addressed without recombinant overexpression of ERBB3.
Several lines of evidence have pointed toward the existence of higher-order association states of ERBB receptors on the cell surface. These complexes or clusters change in size and composition upon ligand binding. However, compared to dimerization, much less is known about the significance and role of such higher-order and presumably more transient interactions. Preassociated or clustered EGFR has been reported to have reduced kinase activity (14–18), and recent studies have also identified ligand-induced dimer–tetramer transitions of EGFR on live cells (19). However, the role and stability of such clusters are likely to differ between ERBB receptors, and significantly less is known about the role of these clusters during ligand-induced signaling or in the constitutive activation that occurs in many overexpressing cancers. In the context of ERBB interactions, the role of the ERBB4 and ERBB3 ligands of the neuregulin family also appears to be more complex. While neuregulin β1 (NRG1) activates and stabilizes heterodimeric ERBB3–ERBB2 complexes, it destabilizes preexisting and signaling incompetent oligomers of ERBB3 (20). However, the NRG1-induced destabilization of the self-association of overexpressed recombinant ERBB3 or chimeric receptors carrying ERBB3-ECDs is consistently less efficient in a membrane setting when compared to the ligand-dependent destabilization of interactions between soluble ERBB3-ECDs (20, 21). This would suggest that the destabilization of direct ERBB3-ECD interactions by NRG1 may be curtailed by other levels of spatial organization or limits to free receptor diffusion.
Nevertheless, the interactions in oligomers of ERBB3 can, to a large degree, be recapitulated by investigating the interactions of the soluble extracellular domains. This analysis revealed that oligomers are held together by at least two distinct interfaces. Structural data for ERBB receptors, derived from dimers of extracellular domains I–III of EGFR (22, 23), indicate that the ligand does not bind in the dimer interface but at outward-facing binding sites. We previously proposed that oligomers of ERBB3 are held together by the combined interactions of the canonical dimer interface, which is very weak for ERBB3 homodimers (24), and a second outward-projecting interface that results in the formation of oligomers from semistable dimers. This secondary interface is expected to be disrupted by ligand binding (20). Consistent with this model, NRG1 is more efficient at destabilizing oligomers of ERBB3 when its EGF-like receptor-binding domain is combined with naturally occurring Ig-like domains, although the latter do not influence receptor activation and complex stability on the level of dimers. This difference in cluster or oligomer destabilization between NRG1 and Ig-NRG1 correlates with altered signal attenuation properties (25). For soluble ECDs of ERBB3, A30 binding and ligand binding appear to target the same, secondary receptor interface on the “outside” of dimers, as both destabilize oligomers of ERBB3 in solution when used at saturation (shown also in Figure 1) and A30 is capable of binding to a previously characterized variant of the ERBB3-ECD that is constitutively in a locked conformation, a conformation in which access to the canonical dimer interface is blocked (26 and data not shown). However, A30 and NRG1 do not compete for the same binding site. This is evident from earlier competition studies (6) and the formation of a ternary complex containing the ECD and both NRG1 and A30 (shown in this study).
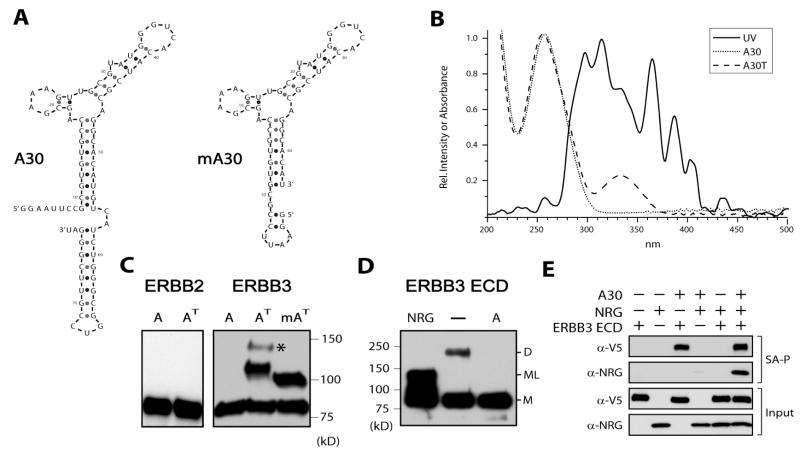
Figure 1.
4-Thiouracil-substituted A30 photo-cross-links to ERBB3-ECDs in solution. (A) Sequence and predicted secondary structure of A30 and minimal A30 (mA30) [created using mfold (34)]. (B) Experimentally determined spectrum of the light source used for cross-linking after light passes through a plastic cell culture dish (UV, solid line). The normalized excitation spectrum of the light source is superimposed on the absorption spectrum of A30 (dotted line) and 4-thiouracil-substituted A30 (A30T, dashed line). (C) A30T (AT) cross-links to soluble ECDs of ERBB3 but not ERBB2. Recombinant ECDs were detected by their V5 epitope tags after cross-linking and SDS–PAGE analysis. Cross-linking does not occur for unmodified A30 (A) or with the ECD of ERBB2. Both A30T (AT) and minimal A30T (mAT) cross-link with high efficiency, and A30 but not mA30 cross-links with low abundance in a 1:2 stoichiometry of ECD to aptamer (indicated with an asterisk). (D) The chemical cross-linking of ERBB3-ECDs in solution is disrupted by binding of A30 to the ECD. The homobifunctional cross-linker BS3 was used in the presence and absence of a 5-fold molar excess of either A30 or NRG1 over ERBB3-ECDs (500 nM). The V5 epitope tag of the ECD was visualized by Western blotting. For NRG1, the disruption of ECD interactions leading to cross-linked dimers (D) coincides with the emergence of a chemically cross-linked NRG1-ECD species (ML) in addition to the ECD monomer (M). BS3 targets primary amines and does not cross-link protein and RNA. (E) A30, NRG1, and ERBB3-ECD form a ternary complex. Biotinylated mA30 (mA30B) (1 μM), NRG1 (1 μM), and ERBB3-ECD (300 nM) were incubated in the indicated combinations. mA30B and interacting species were pulled down with agarose-coupled streptavidin and analyzed by SDS–PAGE and Western blotting. The V5 epitope tag on the ERBB3-ECD was visualized, and anti-NRG1 antibody was used to detect NRG1.
A secondary receptor interface or proximity between receptor dimers has also been proposed for EGFR (27). The use of fluorescence resonance energy transfer (FRET) between fluorophore-labeled EGFs revealed strong FRET signals only in a cellular context but not for vesicle-reconstituted EGFR, although reconstituted receptors were fully capable of dimerization and signaling. On the basis of the relevant Foerster distances for this system and crystallographic data on ligand binding sites in the receptor dimer, the FRET signal that was observed in a cellular setting did not arise from the placement of ligands within dimers but from the proximity between dimers. This FRET-inducing spatial organization required a cellular environment. Whether such proximity of EGFR existed or was qualitatively distinct in the absence of ligand could not be addressed by this approach. Recent studies using different size immunogold labels and electron microscopy also indicate that both ERBB2 and ERBB3 preexist in clusters that are distinct and may be “reshuffled” but not dispersed upon ligand binding (28). The use of A30-tagged and signaling incompetent ERBB3 creates a probe that can report on such proximity of neighboring receptors in the ERBB3–ERBB2 system, independent of the activation and dimerization status of ERBB3.
Our study has two major aspects. We first evaluated the functionality and specificity of A30 that has been converted into a targeted live cell photo-cross-linking reagent. To convert A30 into an efficient photo-cross-linking probe, we replaced all uracils with 4-thiouracils during in vitro transcription. Due to a shift in absorption spectra to higher wavelengths in the near UV (Figure 1B), the selective excitation of thio-nucleobases has been used in the past for a broad range of in vitro cross-linking studies involving both RNA–RNA and RNA–protein interactions (refs 29 and 30; reviewed in ref 31). [Note that these thio-nucleobases are not related to 4′-thioribonucleosides which significantly alter duplex stability and ligand binding properties of RNA–protein complexes (32). These 4′-thioribonucleosides are not used as cross-linkers.] While several thio derivatives of bases are photoreactive (4-thiouracil, 4-thiothymine, 6-thioguanine, and 6-mercaptopurine), 4-thiouracil (4-thioU) is the only thio-nucleobase found naturally in select positions of Escherichia coli tRNA (33) and is needed for several photobiological responses of E. coli to near UV (31). Our current study shows, to the best of our knowledge for the first time, that the use of 4-thiouracil-mediated cross-linking can be extended to cross-linking studies on live cells, despite the significantly greater demands in terms of specificity and cross-linking efficiency at low concentrations of reagent. In its current implementation, this approach uses only reagents and methodology that are readily available to most biochemistry laboratories. Cross-linking can also be dissected mechanistically into two distinct steps, a highly specific targeting step and a secondary cross-linking step that detects proteins in the proximity of the primary and aptamer-tagged target. The second step depends only on the length of the tether portion of the aptamer that protrudes from its primary point of targeted attachment. Cross-linked complexes can be enriched for further analysis by the simultaneous incorporation of biotin into the aptamer.
Equipped with this new and highly sensitive method of targeted, live cell photo-cross-linking, we proceeded to evaluate the state of association or clustering of ERBB3 at low endogenous levels in MCF7 cells and in the presence and absence of ligand. We were able to detect the presence of ERBB3 clusters at receptor levels as low as 25000 receptors per cell and at aptamer concentrations as low as 20 nM. Furthermore, aptamers with extended tethers were able to detect neighboring ERBB3 receptors in a diffusion-independent manner and under conditions in which short-range chemical cross-linkers fail to pick up direct association events. Efficient dispersal of ERBB3 clusters beyond the reach of the extended tether of A30 did not occur after the addition of ligand and in the presence of only stoichiometric levels of ERBB2. Instead, more efficient dispersal required the presence of a molar excess of ERBB2. Throughout the process, inactive ERBB3 appears to remain segregated from ERBB2 as well as activated ERBB3. Consistent with recent electron microscopy studies on fixed samples, suggesting preclustering of both receptors and ligand-induced cluster reshuffling, our live cell studies indicate spatial and functionally relevant segregation at a level above dimers and introduce a novel methodology that should be readily adaptable to a broad range of targets.
EXPERIMENTAL PROCEDURES
Plasmid Constructs, Reagents, and Cell Culture
Antibodies, conjugates, and detection reagents were obtained as follows: anti-ERBB2 (Neu C-18), anti-ERBB3 (C-17 and conjugates), goat anti-rabbit HRP, and Protein A/G PLUS-Agarose from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-phosphotyrosine-HRP (4G10-HRP), anti-ERBB2-phosphotyrosine, and anti-myc tag from Upstate Biotechnology, Inc. (Lake Placid, NY); anti-V5-HRP from Invitrogen; anti-ERBB3-phosphotyrosine from Cell Signaling Technology; 4′,6-diamidino-2-phenylindole (DAPI) from Molecular Probes; streptavidin-agarose resin, TEXAS red-conjugated Immunopure Streptavidin, SuperSignal West Pico Chemiluminescent Substrate, and bis(sulfosuccinimidyl) suberate (BS3) from Pierce; and ExGen in vitro transfection reagent from Fermentas. A fusion protein of the EGF-like domain of NRG1-β1 with thioredoxin (Trx–NRG) was generated in a pET-32a vector (Novagen) and purified as described previously (21). Trx–NRG1 has been confirmed to have biological activity comparable to that of human Ig-neuregulin-1-β1 (21, 34) and was used for stimulation and cross-linking studies unless specifically stated otherwise. Recombinant human Ig-neuregulin-1-β1 (Ig-NRG1) and anti-human NRG1-β1 antibody were purchased from R&D Systems Inc. (Minneapolis, MN). CHO cells were obtained from ATCC, and both MCF7 and MCF7/ERBB2 were kindly provided by M. Pegram (University of California, Los Angeles, CA). For the MCF7 lines in our study, ligand responsiveness was confirmed by Western blotting and absolute receptor numbers have been confirmed by quantitative FACS to be comparable to previously published data for cells from the same source (35).
Aptamer Generation
A30 was transcribed as described previously (6) using the RiboMAX Large Scale RNA Production System (Promega). Linearized plasmid (100 ng/μL) or PCR-generated template (50 ng/μL) containing a T7 promotor was used as a template for in vitro transcription. For the generation of aptamers containing 4-thiouracil (AT), 4-thiouridine 5′-triphosphate (TriLink Biotechnology) was substituted for UTP in the reaction mixture. For the generation of biotinylated aptamers (AB), 1 mM biotin-16-amino-allyl-cytidine-5′-triphosphate (TriLink Biotechnology) was added in the presence of all standard trinucleotides (7.5 mM). At this ratio of biotinylated and unbiotinylated CTP, approximately 10% of aptamers incorporate one biotinylated cytosine. Doubly labeled aptamers (ABT) were created by combining the substitution of UTP with 4-thio-UTP and the addition of biotin-CTP without further modifications. The minimal version of A30 (mA30) was generated using a PCR template with a shortened 5′ end. The in vitro transcription was carried out for 5 h at 37 °C, and the reaction mixture was subsequently treated for 15 min at 37 °C with RNase-free DNase (Promega). Unincorporated trinucleotides and DNA degradation products were removed by gel filtration on CentriSep spin columns (Princeton Separations).
ERBB Constructs
Soluble extracellular domains (ECDs) of ERBB3 and ERBB2 were produced in S2 insect cells as described previously (21). In brief, the coding sequence of the ECDs was cloned into the pMT/BiP/V5-His A expression vector (Invitrogen) which carries a metallothionin promoter and a C-terminal hexa-His and V5 epitope tag. Inductions with 500 μM CuSO4 were carried out for 2 days in 500 mL of S2 insect cell medium (Sigma) with 10% FBS at a density of approximately 6 × 106 stably transfected S2 cells/mL. His-tagged ECD was purified on a 5 mL Pharmacia HI-TRAP chelating column. Full-length mammalian expression systems were based on the previously described pIND/TRE vector (20). C-Terminally truncated ERBB3 was expressed in constitutive expression vector pFLAG-MYC-CMV-19 (Sigma). Truncations were carried out at amino acid Thr977. Thus, the last ERBB3-derived amino acids of this construct were “GPEPHGLT”, located at the end of the kinase domain, followed by the vector-derived myc epitope tag.
A30B-Mediated Ternary Complex Pull-Down Assay
ERBB3-ECD (300 nM), Trx–NRG1 (1 μM), and mA30B (1 μM) were incubated for 10 min at room temperature in PBS-BT (PBS containing 0.5% BSA and 0.01% Triton X-100) according to the experimental setup indicated in Figure 1. Immobilized streptavidin (50 μL slurry) was added to each sample and incubated for 2 h at room temperature. Samples were washed five times with 1 mL of PBS-BT for 10 min. Pulled-down materials were eluted from the resin by boiling in SDS–PAGE sample buffer. Following SDS–PAGE separation, pulled-down ERBB3-ECD or Trx–NRG1 was detected by Western blotting using HRP-conjugated anti-V5 antibody (Invitrogen) or anti-NRG1-β1 antibody (R&D Systems).
UV Cross-Linking of A30 to ERBB3-ECDs in Solution
UV cross-linking of V5 and His6 epitope-tagged ERBB3-ECDs and ERBB2-ECDs to aptamers was evaluated in PBS (pH 7.4) and in the presence of 40 units/mL recombinant RNasin ribonuclease inhibitor (Promega). The indicated aptamer species (A30T or mA30T, each at 300 nM) and 30 nM ERBB3-ECD or ERBB2-ECD were preincubated prior to UV irradiation at room temperature for 15 min. Samples were irradiated on a benchtop UV trans-illuminator (UVP M15-E, UVP) for 15 min. Samples were placed on a Petri dish, approximately 1 cm above the illuminator screen surrounded by ice during irradiation. After irradiation, each sample was mixed with 3× SDS–PAGE loading buffer [24 mM Tris-HCl (pH 6.8), 30% (w/v) glycerol, 6% (w/v) SDS, 50 mM DTT, and 0.1% (w/v) bromophenol blue] and then heated to 95 °C for 5 min. Samples were then loaded onto a 5% polyacrylamide SDS–PAGE gel for separation of the cross-linked and uncross-linked ECDs. ECDs were detected by Western blotting, using its C-terminal V5 epitope tag.
Photo-Cross-Linking of A30 on Live Cells
Semiconfluent, attached MCF7 parental cells or transfected CHO cells (approximately 350000 cells per sample, seeded in 2.5 cm wells) were treated with A30BT or mA30BT for 40 min at 37 °C. A30BT was added directly to serum-free medium which was pretreated with 400 units/mL Rnasin (Promega). The medium was RPMI for MCF7 cells and αMEM for CHO cells. Cells were cooled on ice before UV-cross-linking, and for one-step cross-linking, A30BT-containing medium was not removed prior to UV irradiation. Samples were irradiated for a designated amount of time as described above. For two-step cross-linking, cells were incubated with A30BT and initially irradiated for 5 min under temperature-controlled conditions followed by a washing step prior to irradiation for an additional 10 min. When ligand stimulation was included, NRG was added after the washing step, and cells were stimulated for 10 min at 37 °C followed by 10 min on ice and a subsequent second round of irradiation for 10 min in the continued presence of NRG1. Temperature control during cross-linking was achieved by placing the cell culture dish into a larger transparent plastic dish with the same UV filter properties. The system was temperature-equilibrated to the larger water reservoir of the outer container (ice-cold or 37 °C) prior to being cross-linked for 5 min. After cross-linking, cells were washed with ice-cold PBS containing 1 mM phenylmethanesulfinyl fluoride (PMSF) and lysed in mild lysis buffer [20 mM Tris (pH 8.0), 137 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA, 1 mM sodium orthovanadate, 1 mM PMSF, 1 μg/mL leupeptin, and 1 μg/mL aprotinin]. The cell lysate was further homogenized by being passed five times through a 26-gauge needle, followed by incubation at 37 °C for 10 min to disrupt Triton-resistant lipid fractions. Insoluble material was removed by centrifugation for 10 min at 25000g, and the supernatant was further purified by being filtered through an Ultrafree-MC 0.45 μm filter (Millipore) to ensure that insoluble material did not enter affinity purification procedures. The supernatant was used for the subsequent pull-down assay using immobilized streptavidin.
Chemical Cross-Linking
For the chemical cross-linking of membrane-localized ERBB3, medium on ERBB3-transfected CHO/Tet(off) cells was replaced with fresh medium containing 400 units/mL RNasin. After 5 min, NRG1 or A30 was added at either 10 or 100 nM as indicated in the legend of Figure 4. After 5 min, samples were cooled to 4 °C. After an additional 10 min at 4 °C, medium was replaced with PBS containing 10 mM MgCl2 and the bifunctional and cell impermeable cross-linker BS3 (2 mM). BS3 (Pierce Biotechnology) is a homobifunctional, water soluble, noncleavable, and membrane impermeable cross-linker. It contains an amine-reactive N-hydroxysulfosuccinimide (NHS) ester at each end of an eight-carbon spacer arm. At pH 7–9, it forms stable amides with primary amines and its effective cross-linking distance is 11.4 Å. After 1 h at 4 °C, cells were washed twice with PBS containing 50 mM glycine. Cells were lysed in hot SDS–PAGE sample buffer [75 mM Tris-HCl, 10% (w/v) glycerol, 3% (w/v) SDS, 2.4 mM bromophenol blue, and 52 mM dithiothreitol] containing fresh DTT. Samples were analyzed by Western blot analysis with antibodies (C17) against the C-terminus of ERBB3. Chemical cross-linking of ECDs was carried out at 500 nM ECD in PBS in the presence of a 100-fold molar excess of BS3 and a 5-fold molar excess of either NRG1 or A30. After 1 h, the reaction was stopped with 50 mM glycine. Samples were boiled in SDS–PAGE buffer and analyzed by Western blot analysis using the C-terminal V5 epitope tag of recombinant ECDs.
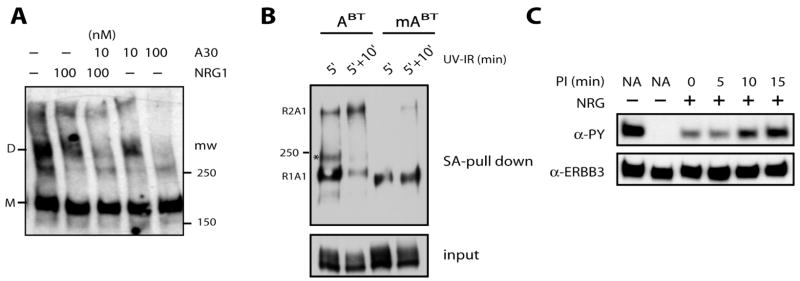
Figure 4.
A30 destabilizes direct ECD interactions and cross-links proximal receptors by a two-step mechanism. (A) Random chemical cross-linking of ERBB3 receptors with a shorter-range cross-linking reagent is suppressed by high concentrations of A30 (100 nM) and to a lesser degree by NRG1 or low concentrations of A30 (10 nM). Overexpressed recombinant ERBB3 receptors in CHO cells were cross-linked with BS3 in the presence of the indicated concentrations of NRG1 or A30, and cross-linked species were detected by Western blotting for ERBB3. Monomers (M) and dimers (D) of ERBB3 are indicated at the left. (B) Two-step cross-linking to adjacent ERBB3 receptors is enhanced by the non-ERBB3 binding tail region of A30. A30BT (ABT) or minimal A30BT (mABT) was cross-linked for 5 min to live CHO cells transfected previously with ERBB3. Cells were processed directly, or free aptamers were washed off and secondary cross-links initiated with cross-linking for an additional 10 min (labeled 5′ + 10′). Both minimal and full-length A30 show secondary cross-linking, but the tail region of A30 significantly enhances secondary cross-linking to neighboring receptors, evaluated by Western blotting after streptavidin enrichment and SDS–PAGE analysis. (C) The ability of preirradiated A30BT to inhibit ERBB2 and ERBB3 signaling was assessed as the readout for its ability to bind its cellular target after UV treatment. Preirradiation times (PI) are indicated as is the addition or absence of ligand stimulation. A decline in inhibitory properties occurs after preirradiation in solution for 10 and 15 min.
Cross-Linking and Fluorescent Imaging of A30BT and ERBB3 in Live Cells
CHO cells were seeded into 12-well culture dishes and were transiently transfected with N-terminally FLAG-tagged ERBB3 (pFLAG-ERBB3). Cells were treated with 300 nM A30BT in standard medium containing 400 units/mL RNasin (Promega) for 30 min at 37 °C. UV cross-linking was performed for 10 min as described above. A30BT-cross-linked cells were visualized by incubation with ALEXA-488-conjugated anti-FLAG antibody and 1.5 μg/mL TEXAS red-conjugated streptavidin (SA-TR, Pierce). After being incubated for 1 h at 37 °C in a CO2 incubator, cells were washed twice with PBS/Ca/Mg (PBS with 1 mM MgCl2 and 1 mM CaCl2). Images were acquired in PBS/Ca/Mg on a Zeiss Axiovert 200M fluorescence microscope equipped with an ORCA-ER digital camera. For the detection of ALEXA-488, a filter cube with HQ480/40 excitation, HQ535/50 emission, and a Q505lp beam splitter configuration was used. TEXAS red was visualized using HQ560/55 excitation, HQ645/75 emission, and a Q595lp beam splitter configuration (both from Chroma Technology).
Streptavidin Pull-Down Assay
Immobilized streptavidin (Pierce) was washed thoroughly with mild lysis buffer prior to addition to the prepared lysate. Cell lysates were incubated with streptavidin-agarose resin on a rotator for 2 h at room temperature. The streptavidin-agarose resin was washed three times with urea wash buffer [4 M urea, 10 nM Tris (pH 7.3), 5 mM EDTA, 137 mM NaCl, and 1% Triton X-100], and bound material was eluted by boiling with 1× SDS sample buffer at 95 °C for 15 min, followed by SDS–PAGE and Western blot analysis.
Immunoprecipitations
Stable CHO/Tet(off) cells were transfected or cotransfected with pIND-TRE-based full-length ERBB3 (B3C17), pIND-TRE-based full-length ERBB2 (B2C17), and pFLAG-based truncated and myc-tagged ERBB3 (B3myc) as indicated in the figures. For co-immunoprecipitation studies, MCF7 and MCF/ERBB2 cells were used. Lysate preparation for immunoprecipitation was carried out as described above for streptavidin-agarose resin, but instead, 3 μg of anti-myc antibody (Upstate) and 30 μL of a slurry of protein A/G PLUS-Agarose or 30 μL of C17 AC-agarose were applied to the lysates. Immunoprecipitations were incubated for 2 h at room temperature. The immunoprecipitated material was washed three times with 10 mL of cold mild lysis buffer for anti-myc pull downs. For co-immunoprecipitations, lysates were immunoprecipitated with resin-conjugated anti-ERBB3 (C17), and samples were washed three times with 1 mL of cold mild lysis buffer. Immunoprecipitates were eluted from the resin with SDS–PAGE sample buffer. Following SDS–PAGE, immunoblot analysis was performed with anti-ERBB2 and anti-ERBB3 as the primary antibodies and goat anti-rabbit IgG-HRP as the secondary antibody.
Aptamer Competition Assay
V5 epitope-tagged ERBB3-ECD (20 nM) was incubated for 10 min at room temperature with 100 nM A30BT in the presence of increasing concentration of A30, as indicated in the figures. Cross-linking to the ECD was conducted as described above, followed by enrichment of cross-linked species on streptavidin-agarose resin and Western blot analysis. ECDs were detected by immunoblot analysis with HRP-conjugated anti-V5 antibody (Invitrogen).
Inhibition of Tyrosine Phosphorylation
MCF7 cells were seeded at a density of 500000 cells/well in a six-well plate. The 4-thioU-substituted aptamer (A30T) was preirradiated under standard cross-linking conditions for 0, 5, 10, and 15 min. Preirradiated aptamer was subsequently added to cells for 30 min at 37 °C before Trx–NRG1 was added for 10 min at 37 °C and cells were kept on ice for additional 10 min to simulate cross-linking conditions. Following stimulation, cells were washed with phosphate-buffered saline (PBS) and lysed in SDS lysis buffer [1% SDS, 20 mM Tris (pH 8.0), 137 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA, 1 mM sodium orthovanadate, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mM phenylmethanesulfonyl fluoride]. Lysates were evaluated directly for tyrosine phosphorylation by SDS–PAGE and Western blot analysis.
RESULTS
4-Thiouracil Substitution Converts A30 into an Efficient and Targeted Cross-Linking Reagent
Full-length A30 is composed of a core region, responsible for binding to ERBB3, and a nonbinding tail region. The latter is largely derived from nonrandomized flanking sequences used during the initial selection (6) (Figure 1A). These flanking regions can be omitted, yielding minimal A30 (mA30), which retains full ERBB3 binding affinity. To convert either variant of A30 into a highly effective cross-linking reagent without prior mapping of specific protein aptamer interfaces, we substituted all uracils with 4-thiouracil, yielding A30T and mA30T, respectively. While the complete replacement of multiple uracils with 4-thiouracil in synthetic aptamers is challenging at best, even for short sequences, efficient complete substitution is feasible by enzymatic incorporation (Figure 1A of the Supporting Information). On the basis of gel mobility shift assays and the ability of the modified and unmodified aptamers to inhibit NRG1-induced signaling, we confirmed that the complete substitution of all uracils with 4-thiouracil did not alter the binding affinity of A30 significantly (Figure 1B–D of the Supporting Information).
We first evaluated the efficiency and specificity of photo-cross-linking in solution. For in vitro cross-linking, samples were irradiated for 15 min on a standard transilluminator and low-range UV was blocked out by the use of a plastic cell culture dish, exposing samples to irradiation in the range between 280 and 410 nm (Figure 1B). Consistent with the established binding specificity of A30 (6), this analysis demonstrated that A30T cross-links with high efficiency and specificity to the ECD of ERBB3 but not to the ECD of the highly homologous ERBB2 receptor (Figure 1C). Cross-linking was strictly dependent on the incorporation of 4-thiouracil which exhibits a characteristic absorption between 320 and 360 nm. No cross-linking was observed with control aptamers (A) containing unmodified uracil instead. This initial test confirms that photo-cross-linking by A30T is both efficient and specific in solution.
For the cross-linking of A30T to ERBB3-ECD, the primary cross-linked species corresponds in molecular weight to a 1:1 complex of ECD and A30. A second minor band was observed with an apparent molecular weight matching one ECD and two aptamers, consistent with the ability of 4-thioU to efficiently cross-link to both nucleic acids and proteins. The removal of the 3′ region flanking the randomized and selected core region does not alter the binding properties of the aptamer but eliminates the second cross-linking product. This suggests that the 1:2 ECD–aptamer complex does not reflect specific binding of the aptamer to two binding sites but is more likely to reflect additional cross-linking of refolded aptamers to each other through weak interactions in their 3′ tail regions. The more pronounced tendency of the larger aptamer to assume alternate secondary structures, which may include unstable dimers, is evident in the direct ethidium bromide staining of aptamer preparations after refolding. This alternate folding or association is less pronounced for the fully 4-thiouracil-substituted aptamer compared to unmodified A30 (Figure 1A of the Supporting Information). However, while shortening of the 3′ termini minimizes alternate folding and aptamer association events, it also reduces the extent to which additional cross-linkers can be introduced and decreases the spatial reach of the probe for the analysis of proteins in the proximity of ERBB3 receptors on live cells. The implications of this difference in the reach of the probe will be discussed in more detail for cell-based cross-linking data.
We at no point observed a photo-cross-linked dimer of ERBB3-ECDs. This is consistent with earlier observations in cross-linking and gel mobility shift studies that indicated that A30 disrupts direct ECD interactions even more efficiently than NRG1. This aspect is mechanistically relevant for the interpretation of subsequent studies in a live cell setting and also provides additional support for the assumption that A30 does not bring receptors together by binding two ECDs through intrinsic bivalency or the formation of functionally bivalent dimers of the aptamer.
To confirm the disruption of direct ECD interactions by both ligand and A30, we carried out chemical cross-linking of ERBB3-ECDs with the homobifunctional cross-linker BS3 in the presence and absence of A30, but without photo-cross-linking (Figure 1D). BS3 has two amino-reactive succinimido groups that target primary amines and that are spaced 11.4 Å apart. Due to its lack of membrane permeability, it is widely used for the evaluation of direct protein–protein complexes on the cell surface. On the basis of BS3 cross-linking, direct interactions of ERBB3-ECDs are efficiently disrupted by both NRG1 and A30. However, despite comparable effects of A30 and NRG1 on direct interactions of ERBB3-ECDs, both macromolecules do not target the same binding site on the ECD. Earlier competition studies with NRG1 and A30 had suggested this, and the level of binding of A30 to soluble ECDs even increases in the presence of NRG1 (6). We confirmed the ability of simultaneous binding of NRG1 and A30 to the ECD through a streptavidin enrichment of an ECD-mediated ternary complex containing NRG1 and biotinylated A30 (Figure 1E). Thus, A30 binding creates a signaling incompetent species of ERBB3, while ligand binding and aptamer binding independently destabilize direct ERBB3-ECD interactions and disrupt ECD oligomers in solution. This disruption of direct ECD interactions and lack of cross-linking of multiple ECDs by A30 are in direct contrast to later observations in a live cell setting, and this contrast provides insights into the limits of diffusion imposed on cellular ERBB receptors.
A30BT Cross-Linking Is Specific on Live Cells
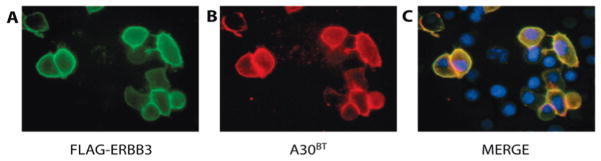
A significant extension of the use of cross-linkable aptamers would be the ability to target receptors in live cells. Our previous functional evaluation of unmodified A30 had shown that A30 does not inhibit the closely related EGF receptor in ligand stimulation assays. However, to probe the potentially complex microenvironment of ERBB3 in a cellular setting, it is crucial that photo-cross-linking does not covalently capture a broad range of weak and unspecific interactions on the cell surface that cannot readily be excluded by testing functional inhibition. We therefore visualized A30BT by fluorescence microscopy after photo-cross-linking to CHO cells (Figure 2). Specifically, A30BT (200 nM) was added to live hamster (CHO) cells that were transiently transfected with FLAG-tagged human ERBB3. The cross-linking was carried out without prior fixation or additional blocking reagents, and the biotin-tagged aptamer was visualized with TEXAS red-conjugated streptavidin and compared with ERBB3 expression (visualized by FITC-conjugated anti-FLAG antibodies). Both transfected and untransfected cells were localized by staining their nuclei with DAPI. Under those conditions, A30BT cross-linking was specific to cells displaying recombinant ERBB3 extracellular domains, confirming that A30BT binding and the conditions chosen for cross-linking had sufficient specificity for the use of A30BT as a targeted cross-linking reagent in a live cell setting.
Figure 2.
A30BT cross-linking is specific on live cells. ERBB3, carrying an N-terminal FLAG epitope tag, was transfected transiently into CHO cells, and A30BT was added and cross-linked for 10 min in serum-free cell culture medium. ERBB3 was subsequently visualized on live cells, without fixation, using anti-FLAG-ALEXA-488. A30BT was labeled with streptavidin-labeled with TEXAS red. Nuclei were stained with DAPI. Both ERBB3 and A30BT show overlapping staining, and untransfected cells (stained with DAPI) do not carry detectable cross-linked A30BT.
A30-Mediated Photo-Cross-Linking Detects ERBB3 Clustering at Low Endogenous Receptor Levels
ERBB3 is not found to be overexpressed in established cancer cell lines and is indeed mostly present in low abundance. The interactions of ERBB3 receptors at these low endogenous levels have therefore been difficult to analyze by established biophysical and biochemical methods. The comparable fluorescence-based detection of FLAG-ERBB3 by either anti-FLAG antibodies or A30 coupling suggested a high coupling efficiency for A30BT photo-cross-linking. Combined with the high specificity of cross-linking, targeted photo-cross-linking of A30 promised to provide a route to the study of putative ERBB receptor interactions at low endogenous expression levels. We chose MCF7 cells as our model system. MCF7 cells carry low endogenous levels of both ERBB2 and ERBB3 and represent a widely used system for NRG1-induced ERBB2 and ERBB3 signaling. A stable derivative line (MCF7/ERBB2) exists and was used for later studies. MCF7/ERBB2 carries highly elevated levels of ERBB2 and represents a model system for cancers with a more aggressive phenotype that is driven by ERBB2 overexpression. As previously reported (35) and confirmed by quantitative FACS for our line of MCF7 cells (data not shown), the endogenous ERBB3 receptor levels in MCF7 parental cells are very low. In addition, the low ERBB3 receptor levels of approximately 15000–25000 receptors per cell are approximately stoichiometric to the levels of ERBB2 (35).
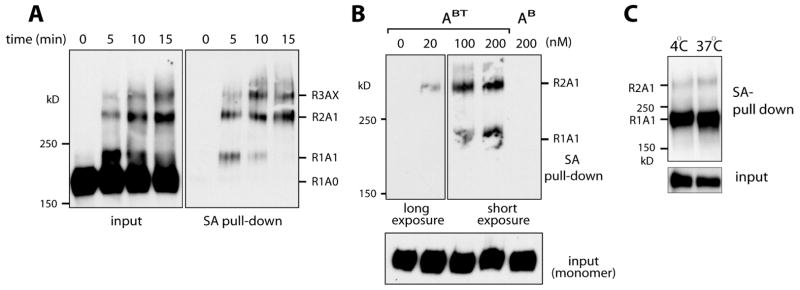
Western blot analysis following live cell photo-cross-linking of A30BT readily indicated photo-cross-linking of A30 to ERBB3 (Figure 3A) as judged by a shift in the apparent molecular weight of ERBB3. For this study, A30BT was added directly to the medium of MCF7 cells prior to cross-linking. Subsequent enrichment with immobilized streptavidin identified aptamer-tagged ERBB3 in samples treated with A30BT but not in samples treated with biotinylated A30B. Hence, the affinity purification is dependent on 4-thiouracil-mediated cross-linking (Figure 3B). No unspecific cross-linking of the nucleic acids was detected at the wavelengths used for these studies. Affinity purification and the apparent molecular weight of enriched samples also confirm that the primary cross-linked species after short durations of photo-cross-linking represents a covalently linked complex of A30BT and ERBB3 (band labeled R1A1 in Figure 3A,B). The primary band at extended irradiation times has an apparent molecular mass that is consistent with A30-mediated cross-linking of two ERBB3 receptors. Confirmation that A30, in contrast to its interaction with soluble ECDs, indeed cross-links two cellular ERBB3 receptors is later provided in experiments using recombinant and differentially epitope-tagged receptors (Figure 5). Additional bands, consistent with complexes containing more than two receptors, are present and become more prominent with longer irradiation times. These prominent higher-order products are also present in later studies using recombinant myc-tagged ERBB3s in CHO cells (Figure 5B). Given that canonical dimers of ERBB3 have been shown to be unstable and considering that direct ERBB3-ECD interactions are further destabilized by A30 (Figure 1D and confirmed later for full-length ERBB3 in Figure 4A), the relative prominence of these higher-order products already suggested spatial proximity of multiple receptors rather than the cross-linking of canonical dimers. While we could demonstrate the molecular composition of the R2A1 dimer band experimentally, the presence of more than two copies of the receptors in larger cross-linked species can be inferred only by molecular mass. We therefore focus in the following analysis on the transition of R1A1 complexes to R2A1 complexes. We will use this transition as our primary readout for the presence of other ERBB receptors in the proximity of aptamer-tagged and inactive ERBB3.
Figure 3.
A30BT detects clusters of endogenous ERBB3 in MCF7 cells. (A) Cross-linked species transition from a 1:1 (R1A1) to a 2:1 receptor:aptamer stoichiometry (R2A1) with an increase in the time of irradiation. A30BT (100 nM) cross-linking to endogenous ERBB3 in MCF7 cells was analyzed by SDS–PAGE and Western blotting after different times of irradiation (indicated above lanes) with (right) and without (left) subsequent enrichment of cross-linked species on streptavidin beads. The estimated stoichiometry of receptors (R) and aptamer (A) is indicated, with an X denoting species for which only the number of receptors is predicted with reasonable accuracy. (B) Cellular A30 cross-linking occurs at concentrations as low as 20 nM but shows a more pronounced 1:1 species at elevated concentrations. Cross-links (15 min) in the presence of varying concentrations of A30BT (0, 20, 100, and 200 nM) were analyzed by streptavidin enrichment and ERBB3 Western blotting. No cellular cross-linking was observed for biotinylated A30 carrying standard uracil (AB). (C) The secondary cross-link of two ERBB3 receptors by A30 is the result of the intrinsic proximity of the receptors rather than diffusion. A30BT (100 nM) was cross-linked to endogenous ERBB3 in MCF7 cells for 5 min at 37 or 4 °C. A30-cross-linked species were enriched by a streptavidin pull down, separated by SDS–PAGE, and probed for ERBB3 by Western blotting.
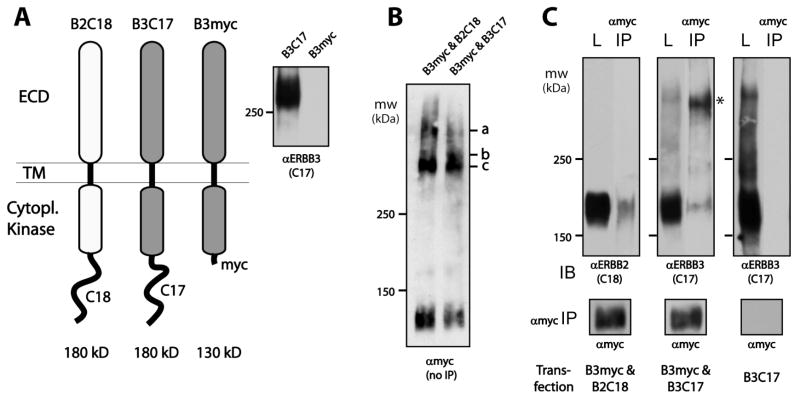
Figure 5.
A30 cross-links two ERBB3 receptors. (A) Cartoon representation of differentially tagged ERBB constructs showing the location of the naturally occurring epitopes of the C18 and C17 antibody in ERBB2 (B2C18) and ERBB3 (B3C17), respectively, as well as the location of the engineered myc epitope tag in ERBB3 (B3myc) lacking the C-terminal tail region and C17 epitope. The absence of a C17 signal for B3myc was confirmed by Western blotting. (B) Direct analysis of A30BT-mediated B3myc cross-linking in transiently transfected CHO cells in the presence of full-length ERBB2 or ERBB3 as indicated above each lane. Cross-linking was evaluated directly by anti-myc Western blotting. The apparent molecular weight of bands c and a is consistent with two and three B3myc species, respectively. In the presence of full-length ERBB3, band b appears at a molecular weight consistent with a mixed dimer of B3C17 and B3myc. (C) Using anti-myc immunoprecipitation instead of streptavidin enrichment, only ERBB3 shows a mixed dimer complex of B3myc and B3C17 (asterisk). The input load control (L) and myc immunoprecipitate (αmyc-IP) were probed for ERBB2 and ERBB3 as indicated for each panel in cells that were transfected as indicated below the panels. Exposures were matched by the relative intensity of the input signal for ERBB2 and ERBB3, and the comparable efficiency of the anti-myc IP was confirmed by Western blotting (dominant, non-cross-linked monomer band shown for the anti-myc blot as a reference). No pull down was obtained in cells transfected with only ERBB3 (B3C17).
For the cross-linking of endogenous receptors, we evaluated the extent to which A30-mediated cross-linking of two receptors requires diffusion or represents the intrinsic proximity of receptors. Toward this end, cross-linking times were limited to 5 min and cross-linking was carried out at controlled temperatures of either 4 °C, thereby restricting receptor diffusion, or 37 °C. Following enrichment of cross-linked species on streptavidin beads, the primary cross-linked product represents the 1:1 complex of ERBB3 and A30. The transition of the R1A1 species to a R2A1 species, although overall inefficient at this shorter cross-linking time, was comparable at both cross-linking temperatures. This indicates that the diffusion of aptamer-tagged receptors did not significantly contribute to the A30-mediated cross-linking of two ERBB3 receptors. Therefore, the formation of higher-order cross-links of ERBB3 indicates the presence of clusters of ERBB3 with restricted receptor diffusion, even at an endogenous receptor level as low as 15000–25000 receptors per cell.
The findings described above indicate the presence of ERBB3 clusters, as defined by the reach of the photo-cross-linking probe. This raises the question of whether the detection of proximal ERBB3 receptors is a reflection of strengthened direct interactions of the ECDs in a membrane setting compared to solution studies (Figure 1D). Alternatively, we may be detecting spatial proximity that is being maintained by other means. Within the limitations of our methodology, we approached this question through two additional tests. First, we asked whether the disruption of direct interactions of ERBB3-ECDs by NRG1 and A30 could be confirmed for the full-length receptors in a live cell setting using the same short-range (11.4 Å) and non-cell permeable cross-linker BS3. Second, we asked if the ability of A30-tagged ERBB3 to reach a neighboring ERBB3 receptor was limited by the dimensions of A30. The latter would be expected if the observed cross-linking was mainly the consequence of spatial proximity rather than direct receptor complexes. The low yield of chemical cross-linking, limited by the availability of suitable lysine residues, does makes this analysis difficult for endogenous receptors at low expression levels. We instead evaluated the disruption of chemical cross-linking with recombinant and overexpressed ERBB3 receptors in CHO cells (Figure 4A). Random chemical cross-linking is partially reduced by 100 nM NRG1 and fully disrupted by 100 nM A30. At lower concentrations of aptamer (10 nM), A30 synergistically enhances NRG1-induced disruption but is insufficient to cause efficient disruption of ECD interactions in isolation.
A minor cross-linked species that is insensitive to disruption can be observed under all conditions. On the basis of its molecular mass (approximately 250 kDa) and antibody reactivity, this species is likely to represent a cross-linked receptor species that includes an N-terminal degradation product of ERBB3 lacking significant portions of the extracellular domains and hence binding sites for NRG1 and A30. The need for higher concentrations of A30 to achieve disruption of short-range cross-linking in a cellular setting is in contrast to findings with soluble ECDs. In solution, the disruption of chemical ECD cross-linking occurs even at low A30 concentrations, and A30-mediated cross-linking of ECDs was likewise not observed, even at lower concentrations of the aptamer (data not shown). On the other hand, cross-linking of full-length cellular ERBB3 receptors occurs even at short irradiation times and low concentrations of A30 (Figure 3) or extended radiation times and elevated aptamer concentrations that are sufficient to block cellular BS3 cross-linking. These findings suggest that, compared to those of soluble ECDs, direct ERBB3 interactions are stabilized in the context of full-length and membrane-localized receptors. However, beyond this stabilization of direct interactions, A30 is also capable of detecting neighboring ERBB3 receptors that may not be engaged in direct ECD interactions but remain within the proximity of each other.
Given that A30 appears to detect ERBB3 receptors that are limited in diffusion but out of reach of short-range chemical cross-linkers, we questioned whether this reach of the photo-cross-linking probe could be altered. Conceivably, changes in the reach of the probe may be systematically exploited in the future in order to explore the environment of the targeted receptor within a defined radius. For distance measurements in a live cell setting to be reliable, photo-cross-linking has to be able to be dissected into two distinct components, rapid and highly specific cross-linking to the targeted receptor and cross-linking to adjacent molecules in a manner that is both strictly dependent on the first, targeted coupling event and dependent on the length of the nontargeting component of the aptamer. To evaluate the dependency on the reach of the aptamer, we made use of the already existing and functionally confirmed variants described in Figure 1, full-length A30 and a minimal variant of A30 (mA30), which retains full binding and cross-linking specificity and a total of 11 cross-linking capable thiouracils. We combined the availability of both variants with the observation of a pronounced two-step nature in photo-cross-linking. Short irradiation times yield 1:1 complexes, which subsequently proceed to higher-order complexes that are dominated by species containing two receptors by molecular mass (Figure 3A). Brief live cell cross-linking of ERBB3-transfected CHO cells with A30BT or minimal A30BT (mA30BT) results in mostly 1:1 cross-linked species for both aptamers. As we saw for soluble ECDs and A30BT, a minor species (denoted with an asterisk) is visible at the anticipated molecular mass equivalent to one ERBB3 receptor and two A30BT aptamers. This double-aptamer species did not exhibit functional bivalency for ECD studies and does not make the transition preferentially into higher-order complexes for full-length receptors. The efficient formation of 1:1 complexes by both aptamers confirms that the minimal version of A30 retains its ability to efficiently target ERBB3 and to “lock in” the resulting complexes through cross-linking events in its receptor-binding region. However, when nonbound aptamers were removed via washes, and previously created covalent 1:1 ERBB3–aptamer complexes were further irradiated for an additional 10 min, complexes based on full-length A30BT made the transition efficiently to 2:1 receptor–aptamer complexes while this reaction occurred at a much lower rate for mA30BT. This length-dependent difference in the transition to higher-order cross-linking products suggests that the length of the tether region, which is not needed for specific targeting and cross-linking in proximity, does indeed determine the extent to which proximal receptors are detected by secondary cross-links.
For the secondary cross-linking reaction to report on the proximity of the ERBB3 receptor, this second cross-linking reaction must be purely based on proximity. In other words, the secondary cross-linking event should be intrinsically unspecific but highly dependent on the aptamer being selectively tethered to its primary target. To evaluate this premise, we compared the progression of cross-linking over time with the extent to which aptamers remain competent to target ERBB3. We used the ability of A30 to inhibit NRG1-induced signaling as a surrogate for the ERBB3-specific binding competency of the aptamer (Figure 4C). Nonirradiated A30BT has potency comparable to that of unmodified A30 with respect to the inhibition of NRG1-induced activation of ERBB2 and ERBB3 signaling (see the Supporting Information). However, over the same time course of irradiation during which the transition from 1:1 to higher-order complexes is the most pronounced (Figure 3A), the inhibitory properties of A30BT are lost when an equivalent dosage of irradiation is delivered to the aptamer prior to inhibition studies. This suggests that the primary binding event but not the secondary cross-linking event requires a structurally and functionally intact aptamer. This finding also provides further support against the functional bivalency of aptamers as a driving force in higher-order cross-links and demonstrates instead that the secondary cross-link is driven by enforced spatial proximity, not by the formation of a second and specific aptamer–ERBB3 interface. The ability to detect proximity is instead only dependent on the length of the nonbinding tether and the timing of irradiation.
The Higher-Order A30-Cross-Linked Species Consist of Dimers of ERBB3
The apparent molecular weight of the primary higher-order complex (Figure 3) suggests two ERBB3 receptors. To prove that A30BT cross-links two ERBB3 receptors, we had to rely on differentially epitope-tagged and hence recombinant ERBB3 receptors (Figure 5A). Full-length ERBB3 harbors the epitope for the C17 antibody in its C-terminal tail region and is therefore called B3C17. We created a second ERBB3 construct (B3myc) that is truncated at the C-terminus of the kinase domain and carries a myc epitope tag but lacks the natural epitope for the C17 antibody (Figure 5A, right). Assuming that the higher-order cross-linked species is composed of two ERBB3 receptors, A30-mediated photo-cross-linking of doubly transfected CHO cells should result in homodimers of both ERBB3 species as well as mixed dimers.
The direct evaluation of aptamer-mediated photo-cross-linking of the myc-tagged species by anti-myc Western blotting indeed shows the formation of a primary myc-tagged species (c) with an apparent molecular weight consistent with a dimer of the truncated B3myc receptor as well as a minor band consistent with the projected molecular weight of a trimer of B3myc (a). In the case of the cotransfection of full-length ERBB3 (B3C17) and B3myc, a faint intermediate band (b) is visible at an apparent molecular weight that is consistent with the formation of a mixed dimer of B3C17 and B3myc. The dominance of myc dimers and trimers over mixed complexes is likely a reflection of the stronger promoter of the B3myc construct. Immunoprecipitation with anti-myc antibodies followed by Western blotting for C17 or C18 (Figure 5C) demonstrates the selective formation of a mixed dimer containing B3myc and B3C17. For the cotransfection of B3myc and B3C17 (center panel), a strong signal (asterisk) matching the anticipated molecular weight of the mixed dimer was obtained for both the input material (L) and the immunoprecipitate (IP). Except for a weak signal corresponding to residual monomeric input, seen in both cotransfections, no signal matching a mixed dimer was seen with the ERBB2-specific antibody C18 in the corresponding cotransfection of B2C18 and B3myc (left panel). The absence of a signal for ERBB2 occurred under conditions in which the input generated comparable Western blot signals for ERBB2 and ERBB3 and comparable amounts of immunoprecipitated myc-tagged receptor was recovered (monomer band shown at the bottom after anti-myc Western blotting of IP). The observed heterodimeric band in the C17 Western blot is also strictly dependent on the presence of B3myc (see the right panel of “B3C17 only” transfection). Therefore, A30BT does efficiently cross-link two ERBB3 receptors but not complexes of ERBB2 and ERBB3.
Inactive ERBB3 Receptors Are Spatially Separated from Activated ERBB3 and ERBB2
Utilizing the ability of A30BT to probe the immediate surroundings of ERBB3 by cross-linking, we evaluated the use of A30BT as a probe for ligand-induced changes in receptor interactions on the cell surface (Figure 6). Although A30 and ligand are not in direct competition, A30 binding converts ERBB3 into a signaling incompetent species while NRG1 does the opposite. Hence, both molecules individually alter the nature of receptor states that the second molecule encounters. To evaluate changes in the proximity of ERBB3 receptors, other than the well-established ligand-induced stabilization of ERBB2 or ERBB3 dimers, we focused our analysis on preformed ERBB3–A30 complexes. To this end, A30BT is first covalently attached to ERBB3 in a predominantly 1:1 receptor:aptamer stoichiometry through brief irradiation. Excess free aptamer is subsequently removed before ligand is added. Since bound A30 destabilizes direct interactions of ERBB3-ECDs and bound aptamer is inhibitory to NRG1-induced signaling, this approach does not probe for the dimerization of the primary tagged species. Instead, it creates an isolated and “passive bystander” species of ERBB3 and queries the extent to which nontagged ERBB3 receptors remain in its proximity, whether those nontagged and signaling competent ERBB3 receptors recruit ERBB2 within cross-linking range of the tagged ERBB3 receptor, and whether these neighboring receptors become tyrosine-phosphorylated.
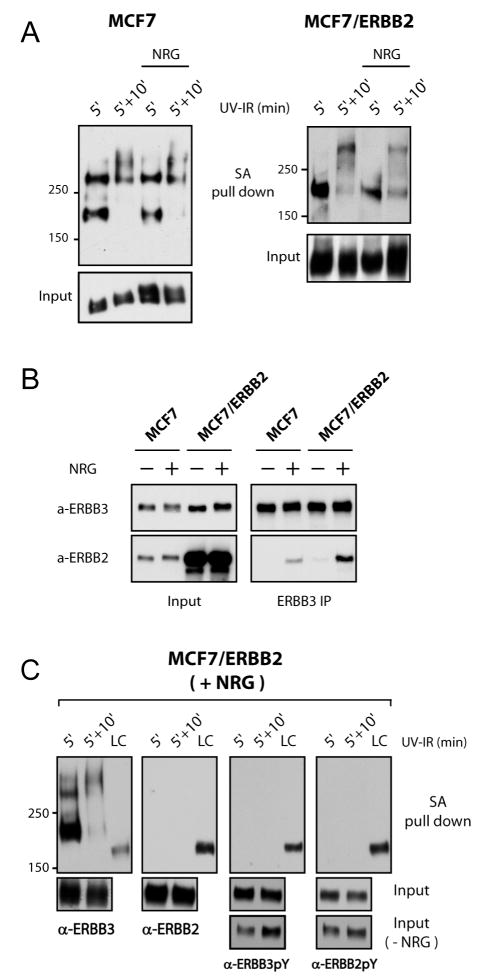
Figure 6.
In the presence of ligand, most of the inactive ERBB3 remains sequestered together with nonactivated ERBB3. (A) A30 was cross-linked in two stages to MCF7 parental and MCF7/ERBB2 cells. ERBB3 was “tagged” with A30 for 5 min in the absence of NRG1. Secondary cross-linking for 10 min was initiated after removal of free aptamers and in the presence or absence of 500 nM NRG1. Cross-linking was evaluated by Western blotting for ERBB3 after enrichment with streptavidin. (B) Efficient heterodimerization of ERBB2 and ERBB3 requires a molar excess of ERBB2. Anti-ERBB3 immunoprecipitation in MCF7 cells and MCF7/ERBB2 cells was conducted after the stimulation with NRG1 (10 nM) for 10 min. Samples were analyzed by Western blotting to detect co-immunoprecipitation of ERBB2 at comparable loads of immunoprecipitated ERBB3. (C) After photo-cross-linking in MCF7/ERBB2 cells, the recovered cross-linked species does not contain ERBB2 or activated receptors. Except for the unstimulated load controls (Input, –NRG1) in Western blots probed for ERBB2-pY and ERBB3-pY, all samples were treated with 500 nM NRG1 in the presence or absence of a second wave of cross-linking. The streptavidin-enriched samples were probed by Western blotting as indicated and compared at exposure settings at which load controls (LC), derived from pooled input, were comparable in intensity for the respective detection system.
To evaluate the ligand-induced changes to the surrounding of A30-tagged ERBB3, we chose two cell lines with different ERBB receptor expression levels, MCF7 and MCF7/ERBB2. While MCF7 cells express low and approximately matched levels of both ERBB2 and ERBB3 (approximately 15000–25000), MCF7/ERBB2 cells contain highly elevated levels of ERBB2 (approximately 1.5 million) and a molar excess of ERBB2 over ERBB3 receptors (35). On the basis of published numbers, the molar excess of ERBB2 over ERBB3 is expected to be 10-fold due to a compensatory 10-fold increase in ERBB3 levels that occur in response to 100-fold overexpressed ERBB2. However, on the basis of Western blot analysis, we find ERBB3 levels to be <10-fold elevated in our line of MCF7/ERBB2 cells, thereby further increasing the molar excess of ERBB2 (see the ERBB3 panel in Figure 6B).
Previous gel mobility shift studies and current chemical cross-linking experiments on soluble ECDs indicate that both the ligand and A30 destabilize the direct interactions of ERBB3-ECDs that favor receptor oligomerization. For the biologically relevant ligand-induced reorganization of ERBB3 interactions, this had previously been confirmed by chemical cross-linking of overexpressed full-length receptors, albeit with reduced potency of ligand-induced disruption (20). In Figure 4A, we confirmed the disruption of direct ECD interactions by both, NRG1 and A30, as well as the synergistic nature of disruption by both, yet when we photo-cross-linked cellular ERBB3 in MCF7 cells with A30BT, the transition of preformed 1:1 complexes of ERBB3 and A30BT to 2:1 receptor–aptamer complexes was not suppressed efficiently by NRG1 (Figure 6A). Ligand-induced suppression of this transition did occur in MCF7/ERBB2 cells, which feature a large stoichiometric excess of ERBB2 over ERBB3. Combined, this relative failure of ERBB3 receptors to be dispersed, and the requirement for a molar excess for ERBB2 to achieve dispersal of ERBB3, suggests that heterodimerization was relatively inefficient at low but matched receptor levels in MCF7 cells when evaluated relative to ERBB3 levels.
This need for excess ERBB2 to achieve efficient heterodimerization of ERBB3 was confirmed by co-immunoprecipitation of both receptors normalized for the amount of precipitated ERBB3 (Figure 6B). Both observations indicate that at low stoichiometric concentrations of ERBB3 and ERBB2, heterodimerization is inefficient and most ERBB3 remains in clusters of ERBB3, despite the high thermodynamic stability of ligand-bound heterodimers of ERBB2 and ERBB3.
Previous studies had suggested that the heterodimerization of ERBB3 and ERBB2 occurs under conditions at which direct ERBB3 interactions are destabilized by ligand binding, yet our current study indicates that A30 cross-linking is detecting the proximity of ERBB3 receptors beyond direct ECD interactions and in the presence of ligand. This also creates different scenarios with respect to ligand-induced interactions of ERBB3 and ERBB2. While direct interactions of ERBB3-ECDs are destabilized, ERBB2 may be recruited into more loosely defined clusters of ERBB3. Alternatively, the localization to clusters of ERBB3 or heterodimerization with ERBB2 may be mutually exclusive for individual ERBB3 receptors. If ERBB2 were to be recruited into loosely defined clusters of ERBB3, we would expect A30-tagged ERBB3, though not signaling competent itself, to detect ERBB2 that is recruited into its proximity by nontagged ERBB3 receptors and these proximal ERBB3 receptors to become phosphorylated. We therefore “pretagged” ERBB3 with A30BT, removed free aptamers, and subsequently stimulated samples with NRG1 prior to a second round of cross-linking (Figure 6C). Streptavidin-enriched fractions from these lysates did not contain any signal for cross-linked ERBB2 under conditions of normalized input signals and strong cross-linking of A30 to ERBB3. In addition, all cross-linked and streptavidin-enriched aptamer–ERBB3 species fail to show any signs of tyrosine-phosphorylated ERBB3 or ERBB2. MCF7/ERBB2 cells show elevated basal activation of ERBB2 that increases only modestly after addition of ligand, but the level of ERBB3 does show a significant increase in ligand-induced tyrosine phosphorylation. This is evident from the analysis of input material in the presence and absence of ligand (Figure 6C, bottom panel) and demonstrates that the cells were responsive to NRG1, even following prior UV irradiation. However, the difference between the ligand-dependent and independent tyrosine phosphorylation decreases with longer UV irradiation (5 + 10 min), mainly due to an elevated level of UV-induced activation in the absence of ligand. Therefore, in a ligand responsive setting, aptamer-tagged and signaling incompetent ERBB3 appears to remain surrounded by inactive ERBB3 receptors, and ERBB2 is not recruited into its proximity and into clusters of inactive ERBB3 receptors.
DISCUSSION
In the context of translating findings derived from soluble ERBB3 receptor ECDs to membrane-localized receptors, several key questions are difficult to address with currently available technology. Specifically, the extent to which spatial clustering of cellular ERBB3 manifests itself without receptor overexpression is not clear, and neither is the extent to which clustering may be driven by spatial confinement, direct ECD interactions, or both. We therefore carried out two sequential studies. We first evaluated the extent to which the previously characterized anti-ERBB3 aptamer A30 may be suitable as a targeted photo-cross-linking reagent that converts aptamer-tagged ERBB3 into a highly sensitive probe of its cellular microenvironment. This would extend our ability to probe ERBB3 receptor interactions beyond the levels accessible with conventional assays. Second, we applied this novel methodology to the analysis of higher-order ERBB3 association states in a live cell setting.
The fact that A30 itself is not neutral in its binding to ERBB3 but destabilizes direct interactions of receptor ECDs is both an advantage and a disadvantage. On the one hand, A30 is limited as a probe for direct ERBB3-ECD interactions since higher concentrations of A30 destabilize such direct protein–protein interactions. In addition, A30 has been selected as an inhibitor of ERBB2 and ERBB3 signaling. Although the aptamer and ligand do not compete for binding to ERBB3 (confirmed in Figure 1E), the ability of A30 to query activation-related changes in heterodimerization or conformation within conventional dimers is intrinsically limited. However, transitions on the level of receptor heterodimers are accessible to analysis by other methods. In contrast, significant questions remain regarding the larger spatial organization of ERBB3 clusters that cannot be readily addressed by conventional cross-linking, in terms of both specificity and the reach and efficiency of probes. We reasoned that the anticipated properties of A30-bound ERBB3, including its inability to form signaling competent heterodimers with ERBB2, would effectively convert aptamer-tagged ERBB3 receptors into a targeted but functionally passive probe of the microenvironment of ERBB3 receptors.
We first confirmed that the complete substitution of uracil with 4-thiouracil has a minimal impact on A30 binding (Figure 1 of the Supporting Information). An analysis of the specificity of binding in a cellular context also confirmed that the substituted aptamer binds and cross-links with high specificity in a live cell setting and regular serum-free growth medium (Figure 2). Moreover, photo-cross-linking of substituted A30, even in the presence of excess A30 in the medium, does not result in unspecific cross-linking to a broad range of low-affinity targets, as one might expect for UV-based cross-linking schemes of nucleic acids in a complex setting. Although not a primary objective of our study, we note that this high specificity in a cellular setting makes 4-thioU substitution well suited to the conversion of randomly selected aptamers into cross-linking reagents without the need for prior mapping of tolerated incorporation sites. This is particularly important when the nature of the aptamer target is uncertain. The approach we describe does not require any specialized equipment or custom-synthesized reagents and should be able to be readily adapted by most biochemistry laboratories.
Equipped with an efficient and ERBB3-targeted photo-cross-linking reagent, we proceeded to evaluate ERBB3 clustering. An independent evaluation of direct interactions of ERBB3 receptors with the short-range chemical cross-linker BS3 confirms both the disruption of direct ERBB3 interactions by NRG1 and A30 and the synergistic nature of disruption by both reagents (Figure 4A). This approach also demonstrates the weakened ability of A30 to destabilize direct ERBB3 interactions at low aptamer concentrations and is consistent with the observed preferential photo-cross-linking of two ERBB3 receptors by A30 at the lowest aptamer concentration we tested [20 nM (Figure 3B)]. Also consistent with these individual observations, the transition at which the 1:1 complex becomes the preferred photo-cross-linking product at short irradiation times coincides with aptamer concentrations at which chemical cross-linking indicates a more potent disruption of direct ERBB3 interactions. It is at this elevated aptamer concentration that A30 (but not BS3 and to a lesser extent mA30) detects neighboring ERBB3 receptors at a second stage of photo-cross-linking. Our study therefore indicates that ERBB3 does indeed form clusters in live cells, even at low endogenous levels that pose a challenge for conventional methods of interaction studies. Furthermore, it suggests that despite the destabilization of direct ERBB3-ECD interactions by A30 itself, ERBB3 receptors are restricted in their diffusion such that they remain within reach for cross-linking by the extended and nonbinding tether of A30 while being out of reach for shorter chemical cross-linkers or shorter forms of A30.
A more detailed analysis of the underlying kinetics of the A30-mediated cross-linking reaction showed that at higher aptamer concentrations it proceeds in two stages. First, a covalent 1:1 complex of ERBB3 and A30 forms. This reaction is very efficient, presumably due to the presence of multiple cross-linking moieties in the binding region of A30 that are seamlessly integrated into the binding interfaces. The subsequent conversion into cross-linked complexes containing two ERBB3 receptors occurs in a manner that does not require binding competent A30 (Figure 4C). In other words, once covalently attached, the entire aptamer becomes a neutral tether during subsequent rounds of irradiation. These distinct stages of A30 cross-linking, specific tagging followed by binding-independent probing of receptor proximity, may explain the high overall efficiency of photo-cross-linking. In addition, they may also provide the foundation for future applications in which a systematic variation of the tether length may provide probes for live cell distance measurements.
However, while it is biochemically compatible with a live cell setting, one clear limitation of our current methodology is the potential damage to cells induced by UV irradiation. We opted for an experimental design that relied on readily available equipment, including a standard trans-illuminator light source combined with a simple filter, provided by the plastic Petri dish, to eliminate the most hazardous wavelength UV. This approach was suitable for the question at hand, and cells irradiated in this manner were still capable of responding globally by ligand-induced tyrosine phosphorylation of ERBB3 (Figure 6C), especially at short irradiation times. However, after prolonged irradiation, the level of ligand-induced activation of total ERBB3 was diminished and the level of constitutive, presumably UV-induced, phosphorylation increased. After irradiation for 15 min, we also noticed a decrease in the levels of full-length ERBB3 that was proportional to the emergence of clipped receptor species (data not shown). This cellular UV response would therefore limit the extent to which questions beyond the initial receptor interactions, such as the correlation of clustering behavior with downstream signaling events, could be addressed in live cells with this simplified setup. More narrowly targeted excitation conditions that optimize the available excitation range of 4-thioU would have to be evaluated for more advanced types of analysis.
Having demonstrated the basic tendency of ERBB3 to cluster and remain restricted in diffusion, even in the absence of direct ECD contacts and at low receptor levels, we asked if our methodology could be applied to evaluate the dynamic nature of clustering in the absence and presence of ligand. Specifically, we wanted to know whether the addition of the ligand induced a gradual scattering of ERBB3 receptors, which should ultimately favor heterodimerization with ERBB2, or whether ERBB2 would be recruited into preexisting clusters of ERBB3.
Given that our approach to creating aptamer-tagged ERBB3 results in a signaling incompetent receptor species, we have limited information about the ligand responsiveness of the tagged receptor itself. While ligand and aptamer are not in direct competition for binding to the ECD, the aptamer may interfere with or destabilize heterodimerization or may alternatively bind heterodimers in a way that places ERBB2 out of reach for cross-linking to the aptamer but enforces an inactive state of the dimer. In either case, aptamer binding does ensure that A30-tagged ERBB3 becomes a passive bystander in signaling events, and hence a selective probe of ligand-induced changes to its broader defined environment. NRG1 and A30 stabilize alternative states of the receptor, and hence, each will alter the equilibrium of states that the other compound encounters. Furthermore, a model that assumes for ERBB3 not to be distributed as dispersed monomers effectively stipulates potentially distinct classes of receptors that may differ in their accessibility for A30. We therefore made use of the pronounced two-step nature of A30 cross-linking and preformed a population of tagged ERBB3 receptors prior to the removal of non-cross-linked aptamers and the optional addition of ligand.
Under the same conditions of ligand stimulation and global tyrosine phosphorylation in which no aptamer-cross-linked ERBB2 is detected, cross-linking to proximal ERBB3 receptors continues, yet neither 1:1 complexes of aptamer and ERBB3 nor complexes containing a second cross-linked ERBB3 contain phosphotyrosine. This suggests that ERBB2 recruitment and activation do apparently not occur inside ERBB3 clusters and that the ligand-induced dispersal of ERBB3 receptors and subsequent heterodimerization is inefficient with the bulk of ERBB3 receptors not engaging in heterodimerization. We confirmed by co-immunoprecipitation that indeed excess ERBB2 is needed to drive efficient heterodimerization. This would imply that regardless of the anticipated thermodynamic stability of heterodimers of ERBB2 and ERBB3, cellular barriers exist that prevent the free mixing of both receptor pools.
Traditional models of ERBB receptor activation have largely been built on the assumption of isolated and inactive receptors and signaling from clearly defined and likewise isolated dimers. However, distinct spatial sequestration of ERBB receptors into functional units may significantly alter signaling properties by spatially restricting access of ERBB receptors to each other or by limiting the free diffusion of activated complexes after activation. Such spatial sequestration may not merely rely on direct receptor interactions but may include externally enforced spatial segregation. In this context, it may be of interest that the cross-linking of ERBB3 to higher-order species in the absence of ligand and at short cross-linking times is significantly higher in MCF7 cells than in MCF7/ERBB2 cells (Figure 6A). This would suggest a decreased level of spatial sequestration of ERBB3 in MCF7/ERBB2 cells. Combined with the excess of ERBB2 in MCF7/ERBB2 cells, this may favor heterodimerization. Constitutively activated ERBB3 provides a PI3K/Akt-mediated pro-survival signal in this cancer cell line that is characterized by significantly more aggressive growth and metastatic potential compared to MCF7 cells. Decreased or incomplete sequestration of ERBB3 may further enhance this constitutive signaling event.
Earlier fluorescence-based studies (36) had already suggested that ERBB receptors are not randomly distributed but cluster in a way that at least in part involves raft microdomains as one critical component of organization. More recent studies, using electron microscopy combined with immunogold particles with a distinct diameter, provided further evidence of spatial clustering of ERBB2 and ERBB3 but suggested that ERBB2 and ERBB3 clusters are largely distinct and that ligand activation results in the formation of spatially structured clusters (28). Our ability to improve our understanding of these additional layers of control that are imposed by spatial segregation is largely driven by the emergence of suitable techniques. Our study demonstrates that the use of photo-cross-linkable aptamers represents a valuable addition to our repertoire of methods that are capable at penetrating this layer of cellular organization. The relative ease with which conventional RNA aptamers can be converted into photo-cross-linking probes, using reagents and equipment available to most biochemistry laboratories, is likely to make this a valuable method for a broad range of investigations.
Supplementary Material
Acknowledgments
We thank Dr. Feng Guo for helpful advice during the search for suitable cross-linking options and during the creation of the manuscript.
Footnotes
This work was supported by the National Institutes of Health (Grant CA098881-01A1 to R.L.) and the Susan G. Komen Foundation (Grant BCTR0504291 to R.L.).
Abbreviations: ERBB3, erythroblastic leukemia viral oncogene homologue 3 (also HER3); EGFR, epidermal growth factor receptor; NRG1, neuregulin 1; ECD, extracellular domain; A30, aptamer 30 (from a random screen against ERBB3-ECD); mA30, minimal ERBB3-ECD binding region of A30; A30BT, biotinylated and 4-thiouracil-substituted A30; SA, streptavidin.
SUPPORTING INFORMATION AVAILABLE
Figure 1 shows ethidium bromide-stained samples of preparations of A30 and A30BT, a gel mobility shift analysis of the binding of 32P-labeled A30 and A30BT to purified ERBB3-ECDs, and the competition between A30 and A30BT for binding to purified ERBB3-ECD. The comparable ability to inhibit ERBB3 and ERBB2 signaling is demonstrated by Western blot analysis of ligand-induced tyrosine phosphorylation in MCF7 cells, treated with either A30 or A30BT. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Labeling tumor cells with fluorescent nanocrystal-aptamer bioconjugates. Biosens Bio-electron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu T, Ebright J, Ellington AD. Using aptamers to identify and enter cells. Curr Opin Mol Ther. 2007;9:137–144. [PubMed] [Google Scholar]
- 6.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci USA. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 8.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., III Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin I, Yarden Y. The basic biology of HER2. Ann Oncol. 2001;12(Suppl 1):S3–S8. doi: 10.1093/annonc/12.suppl_1.s3. [DOI] [PubMed] [Google Scholar]
- 10.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 11.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 12.Naidu R, Yadav M, Nair S, Kutty MK. Expression of c-erbB3 protein in primary breast carcinomas. Br J Cancer. 1998;78:1385–1390. doi: 10.1038/bjc.1998.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Yeung TK, Wang Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem Biophys Res Commun. 2000;277:757–763. doi: 10.1006/bbrc.2000.3731. [DOI] [PubMed] [Google Scholar]
- 14.Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem. 1995;270:3068–3073. [PubMed] [Google Scholar]
- 15.Sherrill JM, Kyte J. Activation of epidermal growth factor receptor by epidermal growth factor. Biochemistry. 1996;35:5705–5718. doi: 10.1021/bi9602268. [DOI] [PubMed] [Google Scholar]
- 16.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E. Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell. 2002;13:2547–2557. doi: 10.1091/mbc.01-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker F, Orchard SG, Jorissen RN, Hall NE, Zhang HH, Hoyne PA, Adams TE, Johns TG, Ward C, Garrett TPJ, Zhu HJ, Nerrie M, Scott AM, Nice EC, Burgess AW. CR1/CR2 Interactions Modulate the Functions of the Cell Surface Epidermal Growth Factor Receptor. J Biol Chem. 2004;279:22387–22398. doi: 10.1074/jbc.M401244200. [DOI] [PubMed] [Google Scholar]
- 19.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor: A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 20.Kani K, Warren CM, Kaddis CS, Loo JA, Landgraf R. Oligomers of ERBB3 have two distinct interfaces that differ in their sensitivity to disruption by heregulin. J Biol Chem. 2005;280:8238–8247. doi: 10.1074/jbc.M410944200. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf R, Eisenberg D. Heregulin reverses the oligomerization of HER3. Biochemistry. 2000;39:8503–8511. doi: 10.1021/bi000953+. [DOI] [PubMed] [Google Scholar]
- 22.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 23.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 24.Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 2004;569:332–336. doi: 10.1016/j.febslet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Warren CM, Kani K, Landgraf R. The N-terminal Domains of Neuregulin 1 Confer Signal Attenuation. J Biol Chem. 2006;281:27306–27316. doi: 10.1074/jbc.M512887200. [DOI] [PubMed] [Google Scholar]
- 26.Kani K, Park E, Landgraf R. The extracellular domains of ErbB3 retain high ligand binding affinity at endosome pH and in the locked conformation. Biochemistry. 2005;44:15842–15857. doi: 10.1021/bi0515220. [DOI] [PubMed] [Google Scholar]
- 27.Whitson KB, Beechem JM, Beth AH, Staros JV. Preparation and characterization of Alexa Fluor 594-labeled epidermal growth factor for fluorescence resonance energy transfer studies: Application to the epidermal growth factor receptor. Anal Biochem. 2004;324:227–236. doi: 10.1016/j.ab.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J Cell Sci. 2007;120:2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 29.Podar M, Perlman PS. Photocrosslinking of 4-thio uracil-containing RNAs supports a side-by-side arrangement of domains 5 and 6 of a group II intron. RNA. 1999;5:318–329. doi: 10.1017/s1355838299981724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moor NA, Ankilova VN, Lavrik OI, Favre A. Determination of tRNA(Phe) nucleotides contacting the subunits of Thermus thermophilus phenylalanyl-tRNA synthetase by photoaffinity crosslinking. Biochim Biophys Acta. 2001;1518:226–236. doi: 10.1016/s0167-4781(00)00294-3. [DOI] [PubMed] [Google Scholar]
- 31.Favre A, Saintome C, Fourrey JL, Clivio P, Laugaa P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J Photochem Photobiol, B. 1998;42:109–124. doi: 10.1016/s1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- 32.Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: Application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipsett MN. The isolation of 4-thiouridylic acid from the soluble ribonucleic acid of Escherichia coli. J Biol Chem. 1965;240:3975–3978. [PubMed] [Google Scholar]
- 34.Jones JT, Ballinger MD, Pisacane PI, Lofgren JA, Fitzpatrick VD, Fairbrother WJ, Wells JA, Sliwkowski MX. Binding interaction of the heregulinbeta egf domain with ErbB3 and ErbB4 receptors assessed by alanine scanning mutagenesis. J Biol Chem. 1998;273:11667–11674. doi: 10.1074/jbc.273.19.11667. [DOI] [PubMed] [Google Scholar]
- 35.Aguilar Z, Akita RW, Finn RS, Ramos BL, Pegram MD, Kabbinavar FF, Pietras RJ, Pisacane P, Sliwkowski MX, Slamon DJ. Biologic effects of heregulin/neu differentiation factor on normal and malignant human breast and ovarian epithelial cells. Oncogene. 1999;18:6050–6062. doi: 10.1038/sj.onc.1202993. [DOI] [PubMed] [Google Scholar]
- 36.Nagy P, Vereb G, Sebestyen Z, Horvath G, Lockett SJ, Damjanovich S, Park JW, Jovin TM, Szollosi J. Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2. J Cell Sci. 2002;115:4251–4262. doi: 10.1242/jcs.00118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.