Abstract
A putative DNA glycosylase encoded by the Rv3297 gene (MtuNei2) has been identified in Mycobacterium tuberculosis. Our efforts to express this gene in Escherichia coli either by supplementing tRNAs for rare codons or optimizing the gene with preferred codons for E. coli resulted in little or no expression. On the other hand, high-level expression was observed using a bicistronic expression vector in which the target gene was translationally coupled to an upstream leader sequence. Further comparison of the predicted mRNA secondary structures supported the hypothesis that mRNA secondary structure(s) surrounding the translation initiation region (TIR), rather than codon usage, played the dominant role in influencing translation efficiency, although manipulation of codon usage or tRNA supplementation did further enhance expression in the bicistronic vector. Addition of a cleavable N-terminal tag also facilitated gene expression in E. coli, possibly through a similar mechanism. However, since cleavage of N-terminal tags is determined by the amino acid at the P1′ position downstream of the protease recognition sequence and results in the addition of an extra amino acid in front of the N-terminus of the protein, this strategy is not particularly amenable to Fpg/Nei family DNA glycosylases which carry the catalytic proline residue at the P1′ position and require a free N-terminus. On the other hand, the bicistronic vector constructed here is potentially valuable particularly when expressing proteins from G/C rich organisms and when the proteins carry proline residues at the N-terminus in their native form. Thus the bicistronic expression system can be used to improve translation efficiency of mRNAs and achieve high-level expression of mycobacterial genes in E. coli.
Keywords: Codon usage, mRNA secondary structure, Translation initiation, Bicistronic vector
Introduction
Escherichia coli remains a common host for high-level expression of heterologous genes, however, this often depends on the source of the target genes. A number of factors that significantly influence gene expression at the translation level have been identified with codon usage and mRNA secondary structures being the major concerns.
There are marked differences in codon usage from one organism to another. Significant variation in codon usage patterns among genes in one organism appears to be associated with their expression levels. Genes with a high proportion of optimal codons are highly expressed, whereas those with rare codons are poorly expressed [1,2]. Moreover, the presence of rare codons can cause ribosome stalling, slow translation, pre-mature translation termination and translation errors and therefore inhibit proper protein synthesis and even cell growth [3–7]. To avoid the potential expression problems resulting from rare codons, one can either optimize codon usage in the target gene by silent mutations for expression in E. coli or expand the intracellular tRNA pool of rare codons by introducing a plasmid which encodes these tRNAs [3]. Both strategies have been successfully used to enhance the expression of heterologous genes with rare codons in E. coli [2,8]. However, negative results have been reported indicating that factors other than codon usage can affect protein expression [8,9].
It has been widely accepted that secondary structure of mRNA in the translation initiation region (TIR)2 plays a crucial role in controlling translation efficiency. A quantitative analysis revealed a strict correlation between the translational efficiency and the stability of the local secondary structure [10]. Since the 30S ribosomal subunit most likely binds to single-stranded regions of mRNA and slides into place while unfolding the TIR, the secondary structure(s) by sequestering either the ribosome binding site (RBS) and/or the initiation codon from ribosome binding will block translation initiation thereby inhibiting translation [11–13].
A series of two-cistron plasmids (bicistronic vectors) have been successfully used to overcome translational inhibition of mRNAs with stable secondary structures [14–18]. In such a system, the first/upstream cistron is generally an A/T-rich sequence, which can minimize local secondary structure(s) thereby allowing efficient translation initiation [15,16], while the second/downstream cistron containing the coding sequence of the target gene is translationally coupled to the first/upstream cistron. Therefore, protein production from a two-cistron system is theoretically dependent upon the efficiency of translation of the first/upstream cistron [19]. Two models have been proposed in terms of the mechanism of translational coupling [19–21]. One is that a ribosome translating the upstream cistron disrupts the inhibitory secondary structure thereby making the RBS of the downstream cistron accessible to other ribosomes which can initiate translation of the coupled downstream gene. The other is that the same translating ribosome can re-initiate and continue to translate the downstream cistron.
Mycobacterium tuberculosis, the causative agent of tuberculosis, is a high-G/C gram-positive bacterium with a genomic G/C content of approximately 65%. The high-G/C content results in a markedly different pattern of codon usage from that of E. coli. A number of rare codons for E. coli, widely used in the genome of M. tuberculosis, might be one reason for the poor expression of mycobacterial genes in E. coli, even in the presence of strong E. coli promoters [1,22,23]. Moreover, stable secondary structures of mRNA surrounding the TIR can be formed due to the high-G/C content and may be responsible for the poor expression. A number of expression systems have been developed for expressing mycobacterial proteins in bacteria that are phylogenetically closer to M. tuberculosis, particularly Mycobacterium smegmatis [24–28]. However, the success of these expression systems is still limited to a few genes [24–28]. Thus a versatile efficient expression system is still an urgent need.
The sequence analysis of the genome of M. tuberculosis [29] allowed us to identify three putative DNA glycosylase genes of the Fpg/Nei family, Rv2464c, Rv2924c and Rv3297. The Rv2924c gene encodes a 32.0 kDa formamidopyrimidine (Fpg) DNA glycosylase (MtuFpg1). The Rv2464c and Rv3297 genes encode a 29.7 kDa endonuclease VIII (MtuNei1) and a 28.5 kDa endonuclease VIII (MtuNei2) respectively. The DNA glycosylases of the Fpg/Nei family recognize and remove oxidized DNA bases as a first step in the base excision repair (BER) process responsible for removing endogenous oxidative damages from the DNA [30]. As shown in Salmonella typhimurium [31] and Helicobactor pylori [32], the BER pathway may be involved in pathogen proliferation or colonization conferring a virulence advantage for the microorganisms, and if this is true for M. tuberculosis, it could provide a target for future therapy.
The potential role of codon usage and mRNA secondary structure(s) in regulating gene expression was tested using the mycobacterial Rv3297 gene as an example. Although both are regarded as potential causes of poor expression of M. tuberculosis genes in E. coli, our results suggest that mRNA secondary structure(s), rather than codon usage, is the primary determinant in influencing translation efficiency. To our knowledge, this is the first report where a mycobacterial gene has been overexpressed in high-levels in E. coli, and the bicistronic vector designed here should be generally applicable to improve translation efficiency of mRNAs and to achieve high-level gene expression of heterologous genes in E. coli.
Materials and methods
Materials
The genomic DNA of M. tuberculosis H37Rv was kindly provided by Dr. Karin Eiglmeier (Unité de Génétique Moléculaire Bactérienne, Paris, France). The DNA sequences were retrieved from GenBank for M. tuberculosis H37Rv (Rv2924c: gi|15610061; Rv2464c: gi|15609601; Rv3297: gi|1877352). The primers were chemically synthesized by Operon Biotechnologies, Inc. (Germantown, MD) and Midland Certified Reagent Company, Inc. (Midland, TX). Restriction enzymes were purchased from New England Biolabs (NEB, Beverly, MA). Cloned Pfu DNA polymerase was purchased from Stratagene (Cedar Creek, TX). Expression vectors pET30a (Novagen, Madison, WI) and the bicistronic pET vector (pET30a/ORF6) constructed in this paper were used to overexpress the three mycobacterial genes in E. coli. Recombinant plasmids were amplified in E. coli TOP10 cells (Invitrogen, Carlsbad, CA) and plasmid DNA was purified using Wizard Plus Midiprep DNA Purification System (Promega, Madison, WI). BL21-Gold (DE3) (Stratagene) was used as the host strain for protein expression in E. coli.
Construction of the bicistronic vector
A 74-mer oligonucleotide (5′GGAATTCCATATGAAAATCGAAGCAGGTAAACTGGTACAGaaggagATTAACTGATATCGGATCCCTCGAGCGG) and its complementary strand were designed and chemically synthesized by Midland Certified Reagent Company, Inc that included NdeI and XhoI restriction sites (underlined) at the N and C-termini, an EcoRV restriction site (italics) for cloning the target gene and a RBS (lowercase/bold) for the target gene. An equal amount of the oligonucleotide and its complementary strand were annealed in NEBuffer 4 (50 mM potassium acetate, 20 mM Tris acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9) and then cleaved with NdeI and XhoI. The resulting fragment was purified from a 1% agarose gel with β-agarase (NEB) and then cloned between the NdeI and XhoI sites of pET30a vector to create the pET30a-ORF6 vector.
Cloning of the target Rv3297 gene into the pET30a vector
The nucleotide sequence of the Rv3297 gene was optimized with E. coli preferred codons and chemically synthesized by Gene-Script Corporation (Piscataway, NJ) resulting in the synthesized Rv3297 (sRv3297) gene. Using two primers: sRv3297-Fwd1 (5′GA TCCAATCATATGCCGGAAGGTGATACCGTGTGGC) and sRv3297-Rev1 (5′ TGGCTCGAGACGCTGGCACGCCGGGCACCAATAGC), the gene was amplified and cloned between NdeI and XhoI sites in pET30a vector as described below resulting in pET30a/sRv3297-His expression vector.
The Rv3297 gene was also cloned from the genomic DNA using two primers: Rv3297-F (5′AGATATACATATGCCGGAGGGCGACACCGTCTGGCAC) and pETRv3297Rev-2 (5′CCGCTCGAGGCGCTGGCAGGCCGGGCACCAATACC) to yield the cloned Rv3297 (cRv3297) gene. The PCR reaction was carried out with cloned Pfu DNA polymerase in an Air Thermo-Cycler (Idaho Technology, Salt Lake City, UT) for an initial denaturation for 5 min at 94 °C, 40 cycles with 94 °C for 15 s, 60 °C for 45 s and 72 °C for 2.5 min and an additional extension for 5 min at 72 °C. The PCR product was cleaved with NdeI and XhoI restriction enzymes, purified from a 1% agarose gel with β-agarase and then inserted between the NdeI and XhoI sites of pET30a vector to yield pET30a/cRv3297-His vector.
Additionally, in order to express MtuNei2 fused to a 37 amino acid N-terminal His/thrombin/S-tag, the cRv3297 gene was sub-cloned into pET30a vector between the KpnI and XhoI sites using a forward primer Fwd-5 (5′GAAGGAGATGGTACCATGCCGGAGGGCGACACCG) and pETRv3297Rev-2 to yield pET30a/His-cRv3297.
Cloning of the genes into the bicistronic pET30a-ORF6 vector
To improve expression of the Rv3297 in E. coli, the target gene was amplified and cloned into the biscistronic pET30a-ORF6 vector as follows. Forward primers, Rv3297-F (5′GCCGGAGGGCGACACCGTCTGGCAC) and sRv3297-Fwd2 (5′GCCGGAAGGTGATACCGTGTGGC) missing the ‘AT’ residues in the initiation codon were used with pETRv3297Rev-2, sRv3297-Rev1, respectively, to amplify the target gene as described above. The purified and XhoI cleaved PCR products were subsequently cloned into EcoRV–XhoI digested pET30a-ORF6 resulting in pET30a-ORF6/cRv3297-His and pET30a-ORF6/sRv3297-His expression vectors.
Similarly, the cloned Rv2924c (cRv2924) gene was amplified and sub-cloned into the pET30a-ORF6 vector using two primers: Rv2924F (5′pGCCGGAGCTGCCTGAAGTCGAGG) and pETRv2924c R-2 (5′CCGCTCGAGTTTACGTGGACGTGGCTGGCAACGC) to yield pET30a-ORF6/cRv2924. Also the synthesized Rv2464c (sRv2464) gene with E. coli preferred codons was also sub-cloned into the pET30a-ORF6 vector using two primers: Rv2464c Fwd-4 (5′GCC GGAGGGTCATACGCTGCATCGG) and sRv2464-Rev1 (5′CCGCTCGAGGGTCTGGCACACCGGGCACCAAAACACG) resulting in pET30a-ORF6/sRv2464.
The nucleotide sequences of all the vectors constructed above were verified by DNA sequencing (Vermont Cancer Center DNA core facility, Burlington, VT) and confirmed using Sequencher 4.2.2 (Gene Codes Corporation, Ann Arbor, MI).
mRNA secondary structures
All the mRNA secondary structures were predicted by using Mfold 3.2 [33]. The structures with the lowest energy were chosen for further comparison and analysis.
Protein expression
The pRARE2 plasmid, which carries seven rare codon tRNA genes for overcoming codon usage bias, was isolated from Rosetta 2(DE3) competent cells (Novagen) and co-transformed with each of the expression plasmids when necessary. The parental vector of the pRARE2 plasmid pACYC184, which does not carry any tRNA genes, was used as a control.
Expression vectors carrying the target gene and tRNA plasmids were co-transformed into BL21-Gold (DE3) and selected on LB agar plates containing kanamycin (50 μg/mL). Transformed colonies were inoculated into 60 mL of Luria Broth medium (LB) containing kanamycin (50 μg/mL) and chloramphenicol (34 μg/mL) and grown at 37 °C. Since Fpg/Nei family DNA glycosylases generally contain a zinc-finger motif, in order to facilitate the proper folding of the target protein, cultures were supplemented with 10 μM of ZnSO4 after reaching an OD600 of 0.2. To further increase the probability of correct folding, the cultures were then incubated at 22 °C with shaking until an OD600 of 0.5 [34]. After reaching the desired cell density, 30 mL of the cell culture was aliquoted and induced in a 250 mL flask with 1 mM IPTG at 22 °C for 18–20 h.
To test for protein induction, cells were harvested by centrifugation and re-suspended in Buffer A (50 mM sodium phosphate buffer (pH 8.0), 100 mM NaCl, 10% (v/v) glycerol, 5 mM β-mercaptoethanol) supplemented with 1 mM PMSF, 10 mM Benzamidine and 1% deoxyribonuclease I (Invitrogen). After sonication, the whole cell lysates were collected, boiled with 1× SDS-loading dye (50 mM Tris buffer (pH 8.0), 2% (w/v) SDS, 0.1% (w/v) Bromophenol Blue, 10% (v/v) glycerol, 100 mM β-mercaptoethanol) for 5 min and analyzed on a 12% SDS–PAGE. The protein bands of interest were then visualized by staining the gel with GelCode Blue Stain Reagent (Pierce, Rockford, IL) and quantitated with Quantity One (Bio-Rad, Hercules, CA). N-terminal protein sequencing (Biomolecular Resource Facility, The University of Texas Medical Branch) was used to confirm identity of the expressed proteins.
Protein purification
His-tagged proteins from 1 l of induced cultures were purified on a 5 mL chelating HP column (GE Healthcare, Piscataway, NJ) using ÄKTA purifier (GE Healthcare) as previously described [35]. Briefly, cell lysates were prepared as described above and the soluble fraction harvested after centrifugation was loaded onto the column using Buffer A. The target protein was eluted with a linear gradient of 0–100% Buffer B (50 mM sodium phosphate buffer (pH 8.0), 150 mM NaCl, 500 mM Imidazole (pH 8.0), 10% (v/v) glycerol, 5 mM β-mercaptoethanol) in 20 column volumes. Fractions containing target protein were identified on an SDS–PAGE, pooled and dialyzed into the storage buffer containing 25% glycerol (20 mM HEPES–NaOH (pH 7.6), 150 mM NaCl, 5 mM DTT, 25% (v/v) glycerol). After subsequent dialysis into the storage buffer containing 50% glycerol, the protein preparations were quantitated using the Bradford Assay (Bio-Rad), aliquoted and stored at −20 °C until use.
Results
Overcoming codon bias does not enhance the expression of MtuNei2 in E. coli
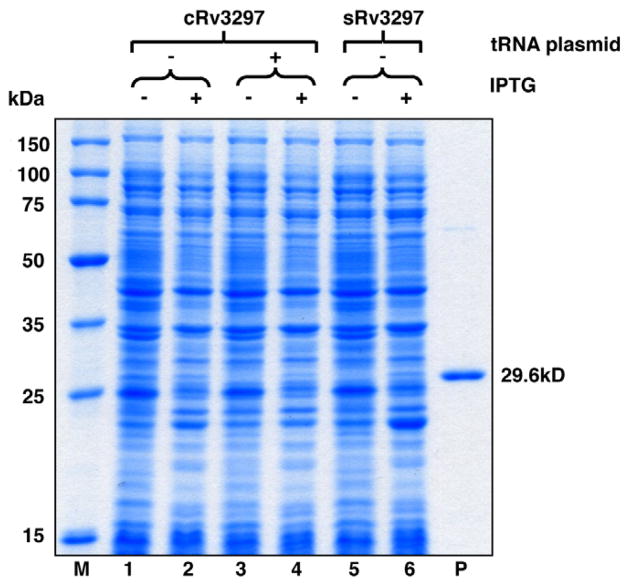
A number of E. coli rare codons, such as GGA (Gly), CUA (Leu), CCC (Pro), AGG (Arg), CGG (Arg), and CGA (Arg) are widely used in the genome of M. tuberculosis. The total usage of rare codons is 12.9%, and the CGG codon accounts for 6.6% (Table 1) in the cRv3297 gene. Tandem repeats of CGG codons which can inhibit translation are also present in the coding sequence of the cRv3297 gene [36]. We initially attempted to express cRv3297 fused to a C-terminal hexa-his tag using pET30a vector in E. coli. No detectable expression was observed upon induction with IPTG for 18 h (Fig. 1, compare lanes 1 and 2). Similar observations were also made when expressing the cloned Rv2924c and the cloned Rv2464c genes in the pET30a vector (data not shown).
Table 1.
Comparison of rare codon usage in the cloned and synthesized Rv3297 (MtuNei2) genes.
| Amino acid | Rare codon | pRARE2 | Percentage of total codons in the genes |
|
|---|---|---|---|---|
| Cloned | Synthesized | |||
| Gly | GGA | + | 0.78 | 0.00 |
| Leu | CUA | + | 0.78 | 0.00 |
| Ile | AUA | + | 0.00 | 0.00 |
| Pro | CCC | + | 0.78 | 0.00 |
| Arg | AGG | + | 0.39 | 0.00 |
| AGA | + | 0.00 | 0.00 | |
| CGG | + | 6.64 | 0.00 | |
| CGA | − | 1.95 | 0.00 | |
| Total | 12.89 | 0.00 | ||
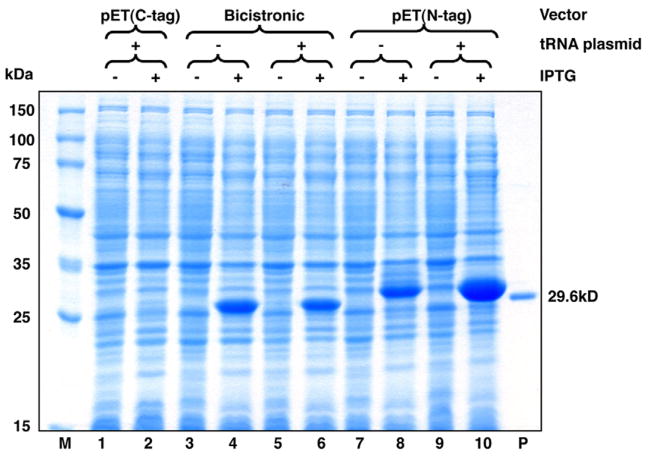
Fig. 1.
Expression of Rv3297 (MtuNei2) in the pET30a vector in the presence of pRARE2 tRNA plasmid. Uninduced (lanes 1, 3 and 5) and induced (lanes 2, 4 and 6) whole cell lysates of transformed BL21-Gold (DE3) cells were analyzed on a 12% SDS–PAGE gel. Lane M, Novagen Perfect Protein™ Marker (15–150 kDa). Lane P, purified MtuNei2-His (29.6 kDa).
If translation of the cRv3297 gene was limited by the rare codons, supplementing the tRNA pool with rare codon tRNAs during expression in E. coli or optimizing codon usage should enhance MtuNei2 expression. To test if rare codon usage is the reason for the poor translation of the cRv3297 gene in E. coli, we then co-expressed pET30a/cRv3297-His in the presence of pRARE2 tRNA plasmid, which carries seven rare codon tRNA genes (Table 1). No difference in expression was observed in the presence of the pRARE2 plasmid (Fig. 1, compare lanes 3 and 4 to lanes 1 and 2). Alternatively, Rv3297 gene optimized with E. coli preferred codons (sRv3297) was synthesized, cloned into pET30a and expressed in E. coli. As a control, since no rare codons are present in the sRv3297 gene, the pACYC184 vector missing the tRNA genes was co-transformed with pET30a/sRv3297-His into E. coli. Analysis of the whole cell lysates on a SDS-PAGE gel showed little or no expression of MtuNei2 even after optimizing codon usage in the synthetic MtuNei2 gene (Fig. 1, compare lanes 5 and 6). These results indicate that rare codon usage is not the principal factor contributing to the poor expression of MtuNei2 in E. coli.
The bicistronic vector enhances the expression of MtuNei2
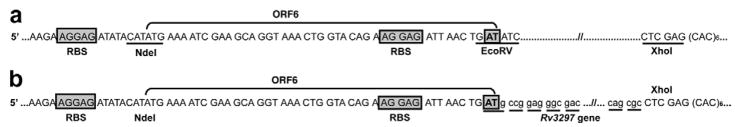
Another reason for the poor expression of heterologous genes in E. coli is that mRNA secondary structure(s) surrounding the TIR sequesters the accessibility of the RBS and/or the translation initiation codon thereby inhibiting efficient translation. Several two-cistron expression systems have been successfully applied to enhance translation efficiency thus achieving high-level expression of mammalian and archaeal genes [14–16,18–20,37–39]. To test if secondary structure at the TIR was responsible for the poor expression of MtuNei2 in E. coli, we designed a bicistronic vector in which a leader sequence or an open reading frame (ORF) preceded the target gene and included the RBS for the downstream gene (Fig. 2a). Also, the last nucleotide in the stop codon for the ORF overlaps with the initiation codon of the downstream gene thereby coupling the translation of the target gene with the upstream ORF sequence (Fig. 2b). Further, inclusion of the RBS for the downstream gene in the upstream ORF sequence should prevent any secondary structure(s) thereby facilitating efficient translation initiation of the target gene. The sequence for the ORF used here was derived from the first 15 amino acids of the maltose binding protein (MBP) gene and was optimized to prevent translational frame-shifts especially near the RBS in the ORF sequence (data not shown). Finally, two restriction sites (EcoRV and XhoI) were used to aid in cloning the target genes readily into the biscistronic vector. Restriction digestion with EcoRV leaves a blunt end and provides the ‘AT’ nucleotides in the initiation codon (Fig. 2a). Amplification of the target gene with an N-terminal primer missing the start codon and with a ‘G’ overhang restores the initiation codon for the target gene after cloning into the bicistronic vector (Fig. 2b). The cloned and synthesized Rv3297 genes were amplified and sub-cloned into pET30a and the bicistronic vector as described above resulting in plasmids, pET30a-ORF6/cRv3297-His and pET30a-ORF6/sRv3297-His, respectively.
Fig. 2.
Schematic of the bicistronic vectors used in this study. (a) Parental pET30a-ORF6 vector, (b) Rv3297 cloned into the pET30a-ORF6. RBS for the upstream leader sequence and the downstream target gene are boxed. The cloning sites for the Rv3297 (MtuNei2) gene are underlined in the parental vector. Nucleotides ‘AT’ of the EcoRV recognition sequence overlap with the stop codon for the upstream ORF6 and the initiation codon for the downstream target gene.
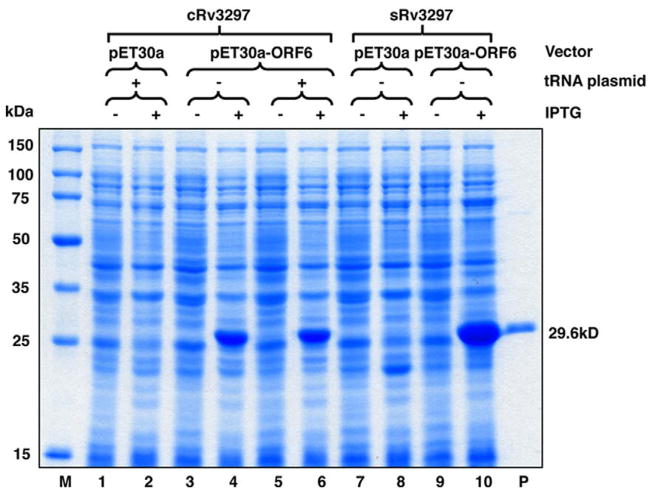
To compare their translational efficiency, E. coli cells were transformed with the bicistronic expression vectors carrying the cloned or synthesized Rv3297 genes in the presence of pRARE2 tRNA plasmid or pACYC184 plasmid missing the tRNA genes, respectively. Analysis of induced cultures by SDS–AGE showed greater than thirty-fold increased expression from the bicistronic vectors only carrying the cloned (Fig. 3, lanes 4 and 6) or the synthesized Rv3297 gene (Fig. 3, lane 10). Interestingly, the bicistronic vector carrying the cRv3297 gene showed expression upon IPTG induction both in the absence (Fig. 3, lane 4) and in the presence (Fig. 3, lane 6) of the tRNA plasmid. These data support the hypothesis that disrupting the mRNA secondary structure at the TIR improves the translation efficiency of the target gene as evidenced by expression of the cRv3297 gene even in the absence of the tRNA plasmid (Fig. 3, lane 4) and the sRv3297 with no rare codons using bicistronic vector only (Fig. 3, compare lanes 8 and 10). Also, these data suggest that the tRNA for the rare arginine CGG codon is not the rate-limiting step for the poor expression of cRv3297 in E. coli (Fig. 3, compare lanes 4 and 6 with and without the tRNA plasmid, respectively). Finally, the enhanced expression observed with the bicistronic vector carrying the sRv3297 gene (Fig. 3, lane 10) also suggests that overcoming rare codon usage significantly improves protein expression even though the mRNA secondary structure determines the translation efficiency at the TIR. The solubility of the MtuNei2 protein was further improved by using pET30a-ORF6/sRv3297-His in Arctic Express (DE3) cells (Stratagene), which express cold-adapted chaperonins (data not shown). Apparently, the enhancement of the overall protein expression is a pre-requisite for the further improvement of soluble expression.
Fig. 3.
Induction of Rv3297 (MtuNei2) expression in pET30a and the pET30a-ORF6 bicistronic vector in the presence of pRARE tRNA plasmid or pACYC184 missing the tRNA genes. Uninduced (lanes 1, 3, 5, 7 and 9) and induced (lanes 2, 4, 6, 8 and 10) whole cell lysates of transformed BL21-Gold (DE3) cells were analyzed on a 12% SDS–PAGE. Lane M, Novagen Perfect Protein™ Marker (15–150 kDa). Lane P, purified MtuNei2-His (29.6 kDa).
Translational coupling using the bicistronic vector inhibits mRNA secondary structure(s) surrounding the TIR of the target gene
In principle, optimization of codon usage also alters the G/C content of a gene which could influence accessibility and mRNA secondary structures at or near the translation initiation site (TIR). To investigate this possibility, we analyzed the G/C content of the nucleotide sequences of both the cloned and the synthesized Rv3297 genes. Whereas the overall G/C content was similar for both, the G/C content of the N-terminal first 50 nucleotides in the synthesized gene was 63% compared to 74% for the cloned Rv3297 gene. While this difference in the N-terminal G/C content might explain the enhanced expression of the synthesized Rv3297 gene in the bicistronic vector (Fig. 3, lane 10), this does not account for lack of expression of the same gene when present in the pET30a vector (Fig. 3, lane 8).
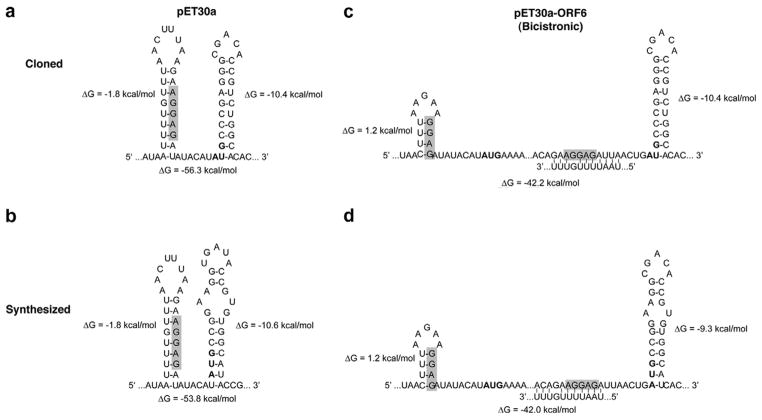
To further address this question, we analyzed the potential mRNA conformations of the first 165 nucleotides transcribed for the cloned and synthesized Rv3297 in pET30a and bicistronic vectors using Mfold 3.2 [33]. For the bicistronic vector constructs, the 165 nucleotide sequence included not only the RBS and initiation codon for the ORF6 sequence but also the first 50 nucleotides of the target Rv3297 where the G/C content of the cloned and synthesized Rv3297 genes differed by more than 10%. Fig. 4 illustrates the most energetically favored potential stem-loop structures involving the RBS or the AUG codons within the first 165 nucleotides. The average free energy values (ΔG) of the secondary structures in the two pET30a constructs are −56.3 kcal/mol and −53.8 kcal/mol, respectively (Fig. 4a and b), whereas for the bicistronic vector constructs the values are −42.23 kcal/mol and −42.03 kcal/mol (Fig. 4c and d). This decrease in the ΔG values by more than −10 kcal/mol is consistent with the observed expression of both the cloned and synthesized Rv3297 genes in bicistronic vector constructs only. Accessibility of the RBS (shaded) in the original pET30a vector (Fig. 4a and b) is potentially weak since the RBS in both constructs are involved in a long-range stem-loop structure with ΔG values of −1.8 kcal/mol. The existence of stable secondary structures can potentially block the efficient binding of ribosomes and further melting of the TIR. In contrast, the mRNA secondary structures at the RBS for the ORF6 sequence in pET30a-ORF6 constructs (Fig. 4c and d) are involved in short stem-loop structures and are thermodynamically less favored with positive ΔG values of 1.2 kcal/mol. Although the free energy values of the mRNA secondary structures in all four constructs surrounding the RBS and initiation codon of the target Rv3297 gene are similar (Fig. 4, ΔG = −9.3 to 10.6 kcal/mol), our data suggest that expression of the target gene in the bicistronic constructs is improved by the effective translation initiation of the leader ORF6 sequence and by the translational coupling mechanism [19–21].
Fig. 4.
Partial mRNA conformations of the first 165 nucleotides transcribed in pET30a and the pET30a-ORF6 constructs. (a) pET30a/cRv3297-His, (b) pET30a/sRv3297-His, (c) pET30a-ORF6/cRv3297-His, (d) pET30a-ORF6/sRv3297-His. Only secondary structures near the RBS (shaded) and AUG start codons (bold) are shown. The free energy (ΔG) values for the individual stem-loop structures (aside) and for the folded 165 nt mRNA (below) are shown.
Furthermore, the difference in the expression levels of the cloned and synthesized Rv3297 genes in the bicistronic vector constructs (Fig. 3, compare lanes 6 and 10) is somewhat surprising since the constructs did not show distinct differences in the overall ΔG values of the secondary structures in the first 165 nt of the target genes (Fig. 4c and d). However, the ΔG values for the stem-loop structures near the initiation codon of the downstream target genes in the bicistronic vector constructs are −10.4 kcal/mol and −9.3 kcal/mol, respectively (Fig. 4c and d). This difference in the free energy values near the initiation codon of the target gene along with efficient translational initiation of the upstream ORF6 sequence may explain enhanced expression of the synthesized gene in the bicistronic vector (Fig. 4d). Also, these data suggest that ribo-some binding may be very sensitive to small changes in the stability of secondary structures resulting from decreased G/C content of the N-terminus of the target gene. Additionally, it is likely that codon usage is also a contributing factor to enhanced expression of the synthesized Rv3297 expression in the bicistronic vector.
The bicistronic vector is convenient for expressing proteins in their native forms
To test the hypothesis that secondary structure(s) at or near TIR is influencing expression, the cRv3297 gene was sub-cloned between the KpnI and XhoI sites of the pET30a vector resulting in pET30a/His-cRv3297 for expression of MtuNei2 fused to a long 37 amino acid N-terminal His/thrombin/S tag (His-MtuNei2, 32.7 kDa). If mRNA conformation surrounding the TIR is important in controlling translational efficiency, fusion of the target gene to the long N-terminal tag should lead to efficient expression of Mtu-Nei2. As shown in Fig. 5, the N-terminal tagged cRv3297 was expressed at high-levels in the presence of the tRNA plasmid (Fig. 5, lane 10). Expression of the cRv3297 was also observed in the absence of tRNA (Fig. 5, lane 8) consistent with our earlier observation (Fig. 3, lane 4) that disrupting mRNA secondary structure at the TIR is a pre-requisite for translational efficiency. However, MtuNei2 is non-functional with an N-terminal tag since the N-terminal proline of Fpg/Nei family DNA glycosylases is the catalytic residue that initiates excision of oxidized bases from DNA [40]. On the other hand, the cloned and synthesized Rv3297 genes were efficiently expressed (Fig. 3, lanes 6 and 10) by using the pET30a-ORF6 bicistronic vector and the soluble proteins catalyze excision of 5-hydroxyuracil from single-stranded DNA containing this lesion (data not shown).
Fig. 5.
Expression of cloned Rv3297 (MtuNei2) as N- or C-terminal hexa-his tagged fusion proteins. Uninduced (lanes 1, 3, 5, 7 and 9) and induced (lanes 2, 4, 6, 8 and 10) whole cell lysates of transformed BL21-Gold (DE3) cells were analyzed on a 12% SDS–PAGE. Lane M, Novagen Perfect Protein™ Marker (15–150 kDa). Lane P, purified MtuNei2-His (29.6 kDa).
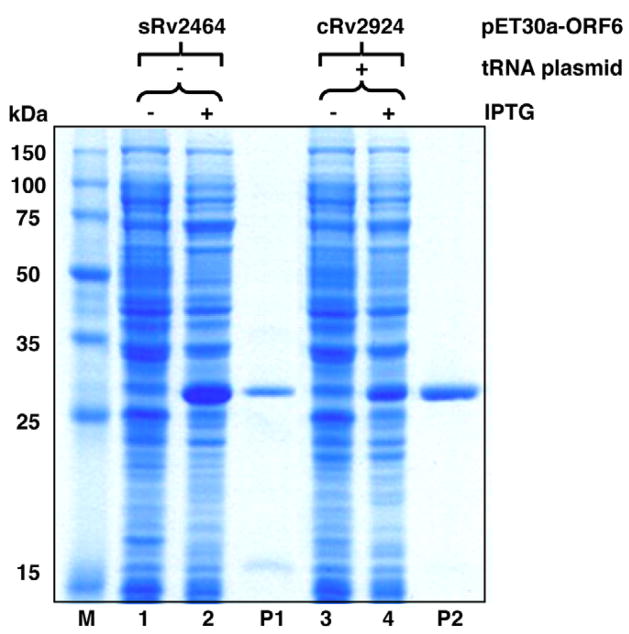
The bicistronic vector was also successfully used to overexpress the other two mycobacterial DNA glycosylases in this family, MtuNei1 (encoded by sRv2464) and MtuFpg1 (encoded by cRv2924) (Fig. 6). Both proteins catalyze excision of oxidized bases from DNA (data not shown). This further demonstrates that the bicistronic vector described here should be generally applicable to achieve high-level expression of heterologous genes in E. coli.
Fig. 6.
Induction of sRv2464 (MtuNei1) and cRv2924 (MtuFpg1) expression in the pET30a-ORF6 bicistronic vector. Uninduced (lanes 1 and 3) and induced (lanes 2 and 4) whole cell lysates of transformed BL21-Gold (DE3) cells were analyzed on a 12% SDS–PAGE. Lane M, Novagen Perfect Protein™ Marker (15–150 kDa). Lane P1, purified MtuNei1-His (30.8 kDa). Lane P2, purified MtuFpg1-His (33.0 kDa).
Discussion
Codon usage vs. efficiency of translational initiation
Theoretically, gene expression can be improved by overcoming codon bias. However, to our knowledge, there is only one group who showed improved expression of M. tuberculosis antigen 85B by replacing rare codons with E. coli preferred codons through silent mutations [41]. A greater than 50-fold improvement of 85B expression was achieved after two pairs of substitutions, one of which was located at nucleotides 7–15 within the TIR. Numerous examples of increased protein expression through altered codon usage have been reported. In many of these examples, codon usage at the N-terminal region was found to be of particular importance [9,42]. Taken together, these observations suggest that the codon optimization associated alteration in mRNA conformations should not be ignored.
As was also reported by other groups [8], we found that either supplementing the rare tRNAs or using the codon-optimized gene failed to enhance the expression of the target protein in the pET30a vector (Fig. 1), although expression was enhanced in the bicistronic vector (Figs. 3 and 5). These results strongly suggest that factors, other than codon usage, play a dominant role in controlling translation efficiency.
mRNA secondary structure vs. efficiency of translational initiation
A prevalent mechanism for translational control of gene expression is through modulation of mRNA secondary structure(s) at the TIR [10]. The stable mRNA secondary structures involving the RBS or the AUG codons were not only observed in the pET30a construct containing MtuNei2 gene (cRv3297) but also in the pET30a constructs containing the MtuFpg1 (cRv2924) and MtuNei1 (sRv2464) genes (data not shown), which we believe to be responsible for the poor expression of these heterologous genes in E. coli as well. The stem-loop structure surrounding the translational start site as well as other potential stable mRNA secondary structures due to the high-G/C content should also be present in M. tuberculosis and may function as translational regulators. However, to our knowledge, there are no reports in the literature describing how M. tuberculosis may utilize these structures.
Many reports suggest that the possibility of inhibitory base-pairing in mRNAs surrounding the TIR could be decreased by designing a proper leader sequence as the first cistron in a bicistronic vector [14,16,17,37,38,43,44]. However, whether or not mRNA secondary structure(s) is necessarily involved in the translational coupling of bicistronic vectors remains unclear. Our results comparing the predicted mRNA secondary structures between the bicistronic vector and pET30a vector suggest that the decrease in the stability of the secondary structure surrounding the TIR using the bicistronic constructs is consistent with the enhancement of expression levels. Therefore, our results strongly support the hypothesis that mRNA secondary structure(s) surrounding the TIR, rather than codon usage, is the critical first step in effecting translation efficiency.
Based on our observations, we conclude that there are two rate-limiting steps during translation: the initiation step (ribosome binding) and the elongation step (protein synthesis). Translation efficiency first depends on the rate of translation initiation, where the limiting factor is the stability of mRNA secondary structure(s) surrounding the TIR. After efficient translational initiation, codon usage controls the elongation kinetics due to different tRNA availabilities.
Summary
Our results demonstrate that rare codon usage is not the primary barrier to efficient translation of the Rv3297 gene. Instead, mRNA secondary structures appear to play a significant role in controlling translational efficiency. The bicistronic vector, pET30a-ORF6, greatly enhanced the expression of MtuNei2 most likely by destabilizing the mRNA secondary structures surrounding the TIR thereby increasing translation initiation.
Although the versatility of bicistronic systems has been questioned [19] a number of bicistronic vectors have been successfully used to express foreign genes whose expression was originally poor in E. coli [14,18,19,37,38]. We show here that the bicistronic system is particularly useful to achieve expression of soluble and active proteins, which carry the proline residue at the P1′ position and require a free N-terminus. To our knowledge, this is the first report to show high-level expression of mycobacterial genes using a bicistronic vector in E. coli.
Acknowledgments
We thank Dr. Stewart Cole and Dr. Karin Eiglmeier of Unité de Génétique Moléculaire Bactérienne, Paris, France, for generously providing the genomic DNA of M. tuberculosis H37Rv and Melissa Batonick for technical assistance. We are grateful to Dr. Jeffery Bond for the helpful comments on the manuscript. The study was supported by National Institutes of Health Grant PO1 CA098993 awarded by the National Cancer Institute. Yin Guo was supported by a research fellowship from DoE EPSCoR DE-F602-00ER45828.
Footnotes
Abbreviations used: TIR, translation initiation region; RBS, ribosome binding site; BER, base excision repair; LB, Luria Broth medium; ORF, open reading frame; MBP, maltose binding protein.
References
- 1.Andersson GE, Sharp PM. Codon usage in the Mycobacterium tuberculosis complex. Microbiology (Reading, England) 1996;142(4):915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 2.Makoff AJ, Oxer MD, Romanos MA, Fairweather NF, Ballantine S. Expression of tetanus toxin fragment C in E. coli: high level expression by removing rare codons. Nucleic Acids Research. 1989;17:10191–10202. doi: 10.1093/nar/17.24.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannig G, Makrides SC. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends in Biotechnology. 1998;16:54–60. doi: 10.1016/s0167-7799(97)01155-4. [DOI] [PubMed] [Google Scholar]
- 4.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Current Opinion in Biotechnology. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 5.Kurland C, Gallant J. Errors of heterologous protein expression. Current Opinion in Biotechnology. 1996;7:489–493. doi: 10.1016/s0958-1669(96)80050-4. [DOI] [PubMed] [Google Scholar]
- 6.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. Journal of Molecular Biology. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 7.Zahn K. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. Journal of Bacteriology. 1996;178:2926–2933. doi: 10.1128/jb.178.10.2926-2933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Z, Schnake P, Xiao L, Lal AA. Enhanced expression of a recombinant malaria candidate vaccine in Escherichia coli by codon optimization. Protein Expression and Purification. 2004;34:87–94. doi: 10.1016/j.pep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Griswold KE, Mahmood NA, Iverson BL, Georgiou G. Effects of codon usage versus putative 5′-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expression and Purification. 2003;27:134–142. doi: 10.1016/s1046-5928(02)00578-8. [DOI] [PubMed] [Google Scholar]
- 10.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Molecular Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Hall MN, Gabay J, Debarbouille M, Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982;295:616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- 13.Iserentant D, Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980;9:1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- 14.Ishida M, Oshima T. Overexpression of genes of an extreme thermophile Thermus thermophilus in Escherichia coli cells. Journal of Bacteriology. 1994;176:2767–2770. doi: 10.1128/jb.176.9.2767-2770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoner BE, Belagaje RM, Schoner RG. Translation of a synthetic two-cistron mRNA in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8506–8510. doi: 10.1073/pnas.83.22.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoner BE, Belagaje RM, Schoner RG. Enhanced translational efficiency with two-cistron expression system. Methods in Enzymology. 1990;185:94–103. doi: 10.1016/0076-6879(90)85010-l. [DOI] [PubMed] [Google Scholar]
- 17.Schoner BE, Hsiung HM, Belagaje RM, Mayne NG, Schoner RG. Role of mRNA translational efficiency in bovine growth hormone expression in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5403–5407. doi: 10.1073/pnas.81.17.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spanjaard RA, van Dijk MC, Turion AJ, van Duin J. Expression of the rat interferon-alpha 1 gene in Escherichia coli controlled by the secondary structure of the translation-initiation region. Gene. 1989;80:345–351. doi: 10.1016/0378-1119(89)90298-9. [DOI] [PubMed] [Google Scholar]
- 19.Makoff AJ, Smallwood AE. The use of two-cistron constructions in improving the expression of a heterologous gene in E. coli. Nucleic Acids Research. 1990;18:1711–1718. doi: 10.1093/nar/18.7.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumperli D, McKenney K, Sobieski DA, Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982;30:865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- 21.Baughman G, Nomura M. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell. 1983;34:979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- 22.Alldread RM, Nicholls DJ, Sundaram TK, Scawen MD, Atkinson T. Overexpression of the Thermus aquaticus B malate dehydrogenase-encoding gene in Escherichia coli. Gene. 1992;114:139–143. doi: 10.1016/0378-1119(92)90720-a. [DOI] [PubMed] [Google Scholar]
- 23.de Miranda AB, Alvarez-Valin F, Jabbari K, Degrave WM, Bernardi G. Gene expression amino acid conservation, and hydrophobicity are the main factors shaping codon preferences in Mycobacterium tuberculosis and Mycobacterium leprae. Journal of Molecular Evolution. 2000;50:45–55. doi: 10.1007/s002399910006. [DOI] [PubMed] [Google Scholar]
- 24.Daugelat S, Kowall J, Mattow J, Bumann D, Winter R, Hurwitz R, Kaufmann SH. The RD1 proteins of Mycobacterium tuberculosis: expression in Mycobacterium smegmatis and biochemical characterization. Microbes and Infection/Institut Pasteur. 2003;5:1082–1095. doi: 10.1016/s1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 25.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Research. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harth G, Lee BY, Horwitz MA. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major Mycobacterium tuberculosis extracellular proteins considered to be leading vaccine candidates and drug targets. Infection and Immunity. 1997;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. Journal of General Microbiology. 1993;139:575–583. doi: 10.1099/00221287-139-3-575. [DOI] [PubMed] [Google Scholar]
- 28.Triccas JA, Parish T, Britton WJ, Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiology Letters. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 29.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 30.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chemical Reviews. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 31.Suvarnapunya AE, Lagasse HA, Stein MA. The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Molecular Microbiology. 2003;48:549–559. doi: 10.1046/j.1365-2958.2003.03460.x. [DOI] [PubMed] [Google Scholar]
- 32.O’Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2789–2794. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchko GW, Hess NJ, Bandaru V, Wallace SS, Kennedy MA. Spectroscopic studies of zinc(II)- and cobalt(II)-associated Escherichia coli formamidopyrimidine-DNA glycosylase: extended X-ray absorption fine structure evidence for a metal-binding domain. Biochemistry. 2000;39:12441–12449. doi: 10.1021/bi001377k. [DOI] [PubMed] [Google Scholar]
- 35.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods in Enzymology. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 36.McNulty DE, Claffee BA, Huddleston MJ, Porter ML, Cavnar KM, Kane JF. Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli. Protein Expression and Purification. 2003;27:365–374. doi: 10.1016/s1046-5928(02)00610-1. [DOI] [PubMed] [Google Scholar]
- 37.Birikh KR, Lebedenko EN, Boni IV, Berlin YA. A high-level prokaryotic expression system: synthesis of human interleukin 1 alpha and its receptor antagonist. Gene. 1995;164:341–345. doi: 10.1016/0378-1119(95)00488-r. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Tanaka Y, Ishida M, Ishizuka M, Yamagishi A, Oshima T. Screening of a mutant plasmid with high expression efficiency of GC-rich leuB gene of an extreme thermophile, Thermus thermophilus, in Escherichia coli. Journal of Biochemistry. 1997;121:1031–1034. doi: 10.1093/oxfordjournals.jbchem.a021690. [DOI] [PubMed] [Google Scholar]
- 39.Saito Y, Ishii Y, Niwa M, Ueda I. Direct expression of a synthetic somatomedin C gene in Escherichia coli by use of a two-cistron system. Journal of Biochemistry. 1987;101:1281–1288. doi: 10.1093/oxfordjournals.jbchem.a121992. [DOI] [PubMed] [Google Scholar]
- 40.Zharkov DO, Rieger RA, Iden CR, Grollman AP. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. The Journal of Biological Chemistry. 1997;272:5335–5341. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]
- 41.Lakey DL, Voladri RK, Edwards KM, Hager C, Samten B, Wallis RS, Barnes PF, Kernodle DS. Enhanced production of recombinant Mycobacterium tuberculosis antigens in Escherichia coli by replacement of low-usage codons. Infection and Immunity. 2000;68:233–238. doi: 10.1128/iai.68.1.233-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldman E, Rosenberg AH, Zubay G, Studier FW. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. Journal of Molecular Biology. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 43.Ishida M, Oshima T. Effective structure of a leader open reading frame for enhancing the expression of GC-rich genes. Journal of Biochemistry. 2002;132:63–70. doi: 10.1093/oxfordjournals.jbchem.a003199. [DOI] [PubMed] [Google Scholar]
- 44.Spanjaard RA, van Duin J. Translational reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Research. 1989;17:5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]