Abstract
Electron cryotomography (ECT) is an emerging technology that allows thin samples such as small bacterial cells to be imaged in 3-D in a nearly native state to “molecular” (∼4 nm) resolution. As such, ECT is beginning to deliver long-awaited insight into the positions and structures of cytoskeletal filaments, cell wall elements, motility machines, chemoreceptor arrays, internal compartments, and other ultrastructures. Here we briefly explain ECT, review its recent contributions to microbiology, and conclude with a discussion of future prospects.
Introduction
After over a century of intense microbiological research, basic metabolism is now largely understood and the complete sequences of nearly 1000 bacterial species are available. Light microscopy has shown us the marvelous diversity of microbial cell shapes and, more recently through fluorescent tags, the approximate locations of numerous proteins and gene loci within those shapes. Higher resolution techniques like X-ray crystallography and NMR spectroscopy have produced atomic models of thousands of important macromolecules. Given this remarkable progress, our persistent ignorance about many of even the most fundamental microbial “cell biological” processes is surprising. We still don't know, for instance, how bacteria generate and maintain their characteristic shapes, establish polarity, organize their genomes, segregate their chromosomes, divide, and in some cases move. The problem in large part has been the lack of a technology that allows us to “see” these processes and their key underlying structures: the resolution gap between light microscopy and atomic models is the realm of electron microscopy (EM), but the “traditional” EM specimen preparation methods of chemical fixation, plastic embedment, sectioning, and staining that have been used in the past failed to preserve the needed details. In just the last few years, the emergence of ECT has opened a new window into microbial ultrastructure [1,2] that promises to revolutionize bacterial cell biology.
Electron cryotomography
Briefly, in ECT suspensions of intact cells are spread into thin films across EM grids, plunge-frozen in liquid ethane, transferred into an electron cryomicroscope, and imaged iteratively while being tilted incrementally. Plunge-freezing prevents ice crystallization, immobilizing the proteins and other cellular structures in their native states and locations. Recording a series of images at different angles allows a full, three-dimensional (3-D) reconstruction (“tomogram”) of the specimen to be calculated. Thus ECT produces 3-D images of intact cells in a nearly native state, without the artifacts of fixation, dehydration, plastic-embedment, sectioning, or staining. Resolutions of a few nanometers are typically achieved, though the resolution and interpretability are critically influenced by specimen thickness and crowdedness. Thus for slender bacteria, the positions and even orientations of individual ribosomes can be detected, but larger cells such as Bacillus subtilis are prohibitively thick. For thicker cells, biofilms, or tissues, the alternative specimen preparation technique “high-pressure freezing” rapidly freezes samples up to 0.5 mm in thickness under pressures of ∼2000 bar to suppress ice crystal formation. While still far from routine, techniques are being developed to “cryosection” such frozen-hydrated material, allowing it to also be imaged in a nearly-native, vitreous state [3].
One of the challenges in ECT is to unambiguously identify structures of interest in tomograms. Unfortunately there are as yet no “GFP-like,” genetically-encodable markers. Instead, some structures have been identified by imaging mutant cells where candidate proteins have been overexpressed, depleted, or deleted, and noting which structures are affected [4,5]. Correlative light and cryoelectron microscopy techniques have also been developed in which fluorescence optical images and cryo-EM images are recorded of the same cell [6-9]. Finally, structures have been identified by their structural “signatures” such as their shapes [10], subunit spacings [11], or bilayer structure [12,13].
Two years ago we reviewed the contributions and potential of ECT to microbiology [1] and noted that while the first paper reporting cryotomograms of prokaryotic cells was published just ten years ago [14], by 2006 there had been thirteen others from 4 different groups. As predicted, the activity in the field in the last two years has rapidly expanded: in just the last two years an additional 20 papers have been published reporting work by 10 different groups. Here we review those papers, including the increasingly promising applications of cryosectioning. The results cover diverse subcellular structures including cytoskeletal filaments, cell envelopes and cell walls, surface appendages, internal compartments, and the nucleoid as well as their functions in the processes of cell division, motility, chemotaxis, pathogenity, and cell-cell interaction.
Cytoskeleton
The discovery of the bacterial cytoskeleton is a fascinating tale, considering that not long ago cytoskeletal proteins were thought to exist exclusively in eukaryotic cells. It is now known that there are homologs of all three main classes of eukaryotic cytoskeletal proteins (tubulin, actin, and intermediate filaments) in bacteria (FtsZ, MreB, and CreS, respectively). Indeed it is now apparent that the bacterial cytoskeleton is not only complex, but probably ubiquitous, and is involved in numerous cellular functions [15]. Each filament's subcellular localization has been investigated mainly by immuno-EM, immunofluorescence and GFP-based light microscopy. For example, FtsZ was first localized to the mid-cell division site by immuno-EM [16] and then visualized as a ring structure by fluorescence light microscopy [17]. Traditional EM (involving chemical fixation, dehydration, embedding and staining) failed to reveal actual filaments, however, probably because the polymers were dynamic and depolymerized somehow during specimen preparation. Applying whole-cell ECT on dividing C. crescentus cells, we visualized short, arc-like filaments close to the cytoplasmic membrane at the constriction site (Fig. 1A and Supplementary Movie) [5]. The filaments' position, orientation, time of appearance, and correlation in numbers and length to expression levels and stabilities in several mutants all showed they were in fact composed of FtsZ. The 3-D images of FtsZ filaments in wild-type cells strongly supported a force-generating role of FtsZ filaments during cell division first proposed by Erickson [18].
Figure 1. Cytoskeletal filaments.
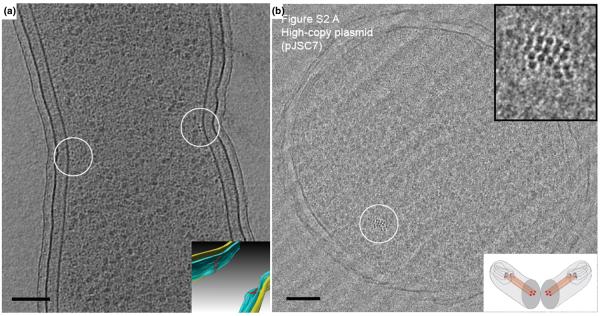
(A) 8-nm tomographic slice of a dividing Caulobacter crescentus cell, showing the filaments in cross-section (small dark dots near the center of the circles). Since the reconstruction can be visualized in 3-D, these filaments can be segmented (Inset). Filaments (red), outer membrane (yellow), and inner membrane (blue). Scale bar 100 nm. Adapted from [5] with permission from Nature Publishing Group. (B) Vitreous cryo-section of an E. coli cell carrying a high-copy ParMRC-bearing plasmid. The nominal thickness of the section is 50 nm. A bundle of filaments are shown in the circle. Top inset: enlarged view. Bottom inset: illustration showing the cellular plane (grey) seen in the image. Scale bar 100 nm. Adapted from [11] with permission from the American Association for the Advancement of Science.
While numerous other filament bundles have now been visualized in cryotomograms of bacteria (those reviewed in [1], [19-21]), so far only a few are identified. Recently Salje et al. cryosectioned E. coli cells over-expressing ParM, the actin-like protein that segregates R1 plasmids, and saw filament bundles (Fig. 1B) [11]. ECT of vitreous sections containing the filament bundles revealed a diffraction pattern similar to that derived from in vitro purified and assembled ParM protofilaments, thus identifying the filament bundles as ParM. In wild-type (low-copy number) strains, the authors observed bundles of three to five intracellular ParM filaments localized within the periphery of the nucleoid, strongly supporting the model that only one filament is needed to separate each plasmid pair.
Cell Envelope and Peptidoglycan
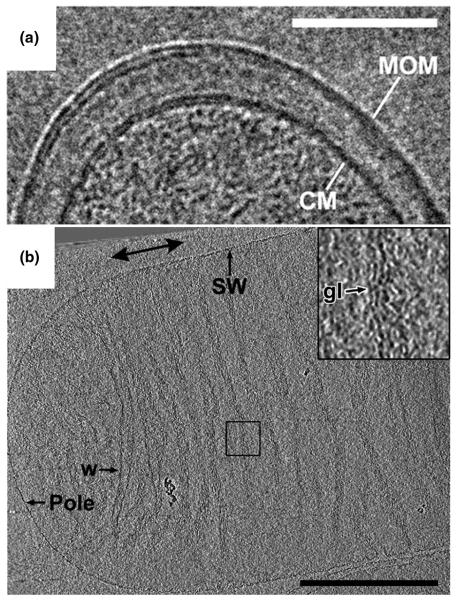
Bacterial cell envelopes shield the cytoplasm and genetic material from the environment. The mycobacterial cell envelope is of paramount medical interest because it constitutes a permeability barrier for antibiotics and is essential for virulence. Hoffmann et al. applied both cryosectioning and whole-cell ECT to investigate the cell envelope structure of Mycobacterium smegmatis, Mycobacterium bovis BCG, and Corynebacterium glutamicum [12]. With close-to-focus images and tomograms showing paired individual leaflets, they proved that the mycobacterial outer layer is indeed a bilayer structure, which they labeled the “mycobacterial outer membrane” (Fig. 2A). By further imaging a mycolic acid-deficient C. glutamicum mutant, the authors observed the absence of the outer membrane and concluded that mycolic acids are constituents of the outer membrane. In a similar study, Zuber et al. cryosectioned the same species and mutants and obtained similar results, although the authors proposed a different model of the mycobacterial outer membrane [13]. These studies highlighted how important it can be to preserve specimens in their native (in this case frozen-hydrated) state.
Figure 2. Cell envelope.
(A) Vitreous cryo-section of a Mycobacterium bovis BCG cell. The section has a nominal thickness of 35 nm. Scale bar 100 nm. Adapted from [12] with permission. © 2008 by The National Academy of Sciences of the USA. (B) 10-nm tomographic slice through an E. coli sacculus. The individual glycan strand densities lie in the plane of the sacculus, roughly perpendicular to the saccular long axis. Scale bar 500 nm. Adapted from [22] with permission. © 2008 by The National Academy of Sciences of the USA.
The mechanical strength of bacterial cell walls arises from a molecular “bag-like” exoskeleton of peptide-crosslinked glycan strands called the sacculus. While the chemical composition and subunit structure of peptidoglycan has been known for decades, the overall architecture of the sacculus was unclear. Two fundamentally different models of 3-D peptidoglycan organization had been suggested, namely the “Layered” and “Scaffold” models. Gan et al. applied ECT to intact sacculi purified from two Gram-negative bacteria, E. coli and C. crescentus [22]. In cryotomograms of both preparations, sacculi were seen to contain a single peptidoglycan layer in which the individual glycan strands were oriented in the plane of the sacculus perpendicular to the long axis of the cell and spaced approximately 5-8 nm apart (Fig. 2B). This observation ruled out the “Scaffold” model and instead established a “Disordered, Circumferential, Layered” model.
Motility and Surface Appendages
Bacterial cells exhibit wonderfully diverse modes of motility including swimming, swarming, twitching, and gliding. The most studied mode of motility, swimming, is driven by one or more flagella which are rotated by a membrane-embedded motor. In spirochetes, the flagella are located in the periplasmic space between the outer and cytoplasmic membranes. Three spirochetes have now been imaged with ECT. In a recently cultured termite gut spirochete, Triponema primitia, Murphy et al. revealed novel structures including bowls, arches, fibrils, and two layers of peptidoglycan sandwiching the flagella between them (Fig. 3A) [23]. These results, combined with the earlier ECT structure of the flagellar motor from the same species [24], are consistent with the “rolling cylinder” model of spirochete motility originally proposed by Berg [25]. ECT of the pathogenic spirochete Treponema denticola revealed periplasmic flagella, cytoplasmic filaments and plate-like structures, and a patella-like periplasmic structure at one polar tip [20]. The cytoplasmic filaments, earlier suggested as CfpA (a unique protein in spirochetes) [26], were visible in both wild-type and aflagellate strains. Finally, Charon et al. reported the structure of the Lyme disease spirochete Borrelia burgdorferi [27]. In contrast to the stacked bundle observed previously by traditional EM [28], by ECT it was seen that the periplasmic flagella adopt a flat-ribbon configuration.
Figure 3. Surface appendages.
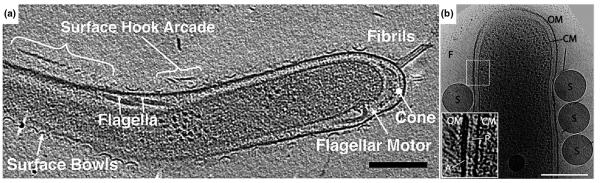
(A) 10-nm tomographic slice through a Treponema primitia cell, showing novel surface appendage structures. Scale bar 200 nm. Adapted from [23] with permission from Blackwell Publishing Ltd. (B) 3-nm tomographic slice through the middle of a Flavobacterium johnsoniae cell, showing the outer membrane (OM), cytoplasmic membrane (CM), cell surface filaments (F) and added latex spheres (S). Inset: enlarged view of the boxed area showing the peptidoglycan layer (P) and a patch (A) underneath the outer membrane. Scale bar 300 nm. Adapted from [29] with permission from the American Society of Microbiology.
The gliding bacterium Flavobacterium johnsoniae was shown to possess tufts of ∼5-nm-wide cell surface filaments emanating from the inner surface of the outer membrane (Fig. 3B) [29]. These filaments were absent in a non-motile gldF mutant cell but were restored in the same mutant complemented with plasmid-encoded GldF, a component of a putative ATP-binding cassette transporter. The cell surface filaments are unlikely to be composed of any Gld proteins, however, since the Gld proteins are known to localize inside the outer membrane.
Chemotaxis
Bacteria sense nutrient gradients through an array of proteins that form a complex at the cell pole. As for the structures described above, despite rich information about individual components and their functions, the overall architecture of the complex was unclear. Two contradicting models regarding its organization had been suggested based on crystallographic and other data [30]. Following work with negatively stained samples, through ECT of wild-type E. coli and mutant strains over-expressing the serine chemoreceptor Tsr, Zhang et al. visualized striations at the poles and identified them as chemoreceptor arrays [31]. Briegel et al. confirmed that similar structures in C. crescentus cells were in fact chemoreceptor arrays by correlating fluorescent light microscopic images (where the receptors had been labeled with a fluorescent tag) with electron cryotomograms of the same, lightly fixed cells (Fig. 4A) [9]. Analysis of the wild-type Caulobacter array structures revealed that the receptors were arranged in a 12-nm hexagonal lattice with “trimers of receptor dimers” at each vertex. A concurrent study by Khursigara et al. confirmed the lattice spacing and the trimers-of-dimers model in the same species, but emphasized that the arrays were not perfectly hexagonal [21]. In an exciting combination of tomographic, crystallographic, and “single-particle-like” techniques, Khursigara et al. went on to show that the HAMP domains in E. coli mutants over-expressing Tsr undergo conformational changes due to ligand binding and methylation (Fig. 4B and C) [32].
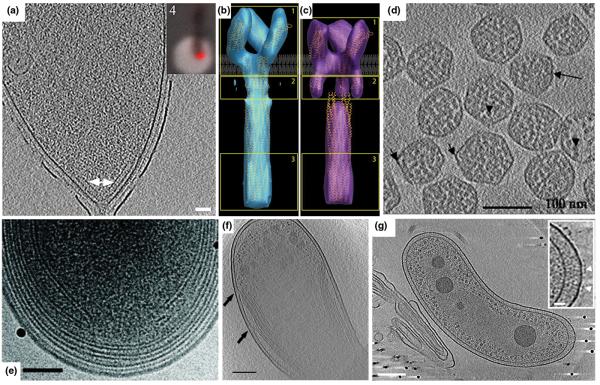
Figure 4. Intracellular structures.
(A) 15-nm tomographic slice of a C. crescentus cell showing a polar chemoreceptor array (arrow). Inset: fluorescence light microscopy image of the same cell showing the co-localization of McpA-mCherry with the polar array seen by ECT. Scale bar 50 nm. Adapted from [9] with permission from Blackwell Publishing Ltd. (B, C) ‘Compact’ (B) and ‘Expanded’ (C) conformations of the E. coli chemoreceptor Tsr visualized by ECT and sub-volume averaging. Adapted from [32] with permission. © 2008 by The National Academy of Sciences of the USA. (D) 4-nm tomographic slice of isolated Halothiobacillus neapolitanus carboxysomes. Scale bar 100 nm. Adapted from [34] with permission from Elsevier Ltd. (E) Tomographic slice of a Prochlorococcus MIT9313 cell showing the intracytoplasmic membrane. Scale bar 100 nm. Adapted from [36] with permission from the American Society of Microbiology. (F) 20-nm tomographic slice of a Rhodopseudomonas viridis cell exhibiting photosynthetic membranes. Scale bar 200 nm. Adapted from [37] with permission from Elsevier Ltd. (G) 8-nm slice of a Bdellovibrio bacteriovorus cell exhibiting a distinct nucleoid region. Scale bar 150 nm. Inset scale bar 25 nm. Adapted from [19] with permission from the American Society of Microbiology.
Carboxysomes
The carboxysome, a proteinaceous shell that encapsulates bacterial ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO), is the most well characterized member of a family of polyhedral bodies termed bacterial microcompartments that encapsulate key enzymes [33]. In cyanobacteria and many chemoautotrophic bacteria, the encapsulated RuBisCO catalyzes the first step of carbon fixation in the Calvin-Benson-Bassham cycle. The structure of carboxysomes has been proposed to increase the local concentrations of substrate and enzyme and sequester the reaction from useless side reactions. Schmid et al. first reported the structure of carboxysomes by ECT from Halothiobacillus neapolitanus (Fig. 4D), showing that they were regular icosahedra but with different sizes. These authors averaged sub-classes and suggested that the different sizes might be a result of different packing arrangements of the shell proteins [34]. Iancu et al. imaged carboxysomes from Synechococcus strain WH8102, showing that they too were regular icosahedra, but suggested instead that the different sizes arose from different T-numbers [35]. Iancu et al. went on to show through simulation that the concentric shells of RuBisCO seen in both studies could be explained by simple close packing.
Photosynthetic membranes
Two ECT studies have characterized bacterial photosynthetic membranes [36,37]. Ting et al. applied ECT to both frozen-hydrated cells and sections of two closely related, ecologically important cyanobacteria (strains of Prochlorococcus) [36]. Compared to the MIT9313 strain, the MED4 strain (which has one of the smallest genomes (1.66 Mbp) of any known photosynthetic organisms) had a smaller cell volume, a smaller carboxysome, less intracytoplasmic lamellae, and a minimal cell wall architecture (Fig. 4E).
Comparative genomic analyses found differences in genes between the two strains including those involved in peptidoglycan synthesis. Konotry et al. applied whole-cell ECT to the anaerobic purple photosynthetic bacterium Rhodopseudomonas viridis (Fig. 4F), documenting the tunnel-like structures connecting the photosynthetic membranes to the inner membrane and the highly packed arrangement of the flattened sacs [37].
Nucleoid and cell-cell interaction
In traditional EM images, the nucleoid, which is supposed to contain the compacted genome, was often seen as a lighter area within the cytoplasm. In higher detail and reliability, cryotomograms of thin bacterial cells often show a central ribosome-free area that has an obviously finer texture than the rest of the cytoplasm. A particularly clear example is seen in the highly bent Bdellovibrio bacteriovorus (Fig. 4G) [19]. ECT of frozen-hydrated sections of Gemmata obscuriglobus (a member of the phylum Planctomycetes) revealed a network of double-membrane compartments that probably enclose packed chromatin, an organization speculated to increase the radiation tolerance of this species [38]. ECT also revealed the structure of the contact site between two symbiotic bacteria, Ignicoccus hospitalis and Nanoarchaeum equitans [39], and the 3-D ultrastructure of an uncultivated, ultra-small archaeon in acid mine drainage (AMD) biofilm samples [40].
Future prospects
In addition to these dominantly cryotomographic studies, others have begun to include ECT reconstruction as just one of a set of approaches to characterize cells and processes [41-44]. Thus in combination with other new imaging and experimental methods, ECT is precipitating a new era in bacterial cell biology. Improvements in both the quality and number of cryotomograms being produced should be expected. Beginning with the sample, further improvements in methods for cryosectioning will make cryotomography of serial sections possible for larger cells and even biofilms. The development of genetically-encodable, electron-dense tags to unambiguously identify specific macromolecules in cryotomograms is sorely needed [45-47], as are further improvements in the technologies for correlating light and electron cryomicroscopy [6-9].
Instrumentally, the development of direct electron detectors, phase plates, and aberration correctors could dramatically improve image quality [48,49]. Automation of image acquisition, tilt-series alignment, and reconstruction [50,51], coupled with the development of databases for the management and recovery of the large datasets produced by ECT [52](J. Ding et al., in preparation) will facilitate the imaging of very large numbers of cells, as is sometimes necessary. Computational advances such as contrast-transfer-function correction and improved methods for denoising, template-matching, sub-volume averaging, and segmentation will all enhance the interpretability of the tomograms [53] [54]. Finally, as more and more institutions acquire the expensive microscopes needed and more and more practitioners are trained, the numbers of studies applying this technology will also obviously increase.
Supplementary Material
The movie is a three-dimensional tour through the ECT reconstructions of four cells: two NA1000 cells and two cells over-expressing FtsZG109S. The NA1000 cells are introduced with a fLM image of YFP-labeled FtsZ in NA1000 cells (labeled “Caulobacter crescentus PvanA::ftsZ-eyfp (MT196)” in the movie), showing that FtsZ normally localizes as a ring at midcell. Next a cryo-EM image of a dividing NA1000 cell is shown, zooming in on the division site, which will be reconstructed. The original tilt-series of images is shown (labeled “Tilt Series”), followed by the full 3-D reconstruction (labeled “Tomogram”) slice-by-slice. The segmented inner membrane (blue), outer membrane (gold), and FtsZ filaments (red) are then shown from a range of views while the raw reconstruction is flashed on and off to give the viewer a sense for the interpretability of the data. Next a second NA1000 cell is shown (labeled “Another cell more deeply constricted”) in a similar but abbreviated fashion without the tilt series or full reconstruction. The two cells over-expressing FtsZG109S are introduced with another fLM image using FtsZ-YFP (labeled “FtsZ G109S”), showing that FtsZ accumulates in the extended division sites. An additional fly-around of one particular filament and its connection to the cell wall is included (labeled “Connections to peptidoglycan”). Finally the second cell over-expressing FtsZG109S is shown (labeled “Another mutant cell”), wherein an additional cytoplasmic cytoskeletal filament bundle (orange) is seen. Three particularly well-resolved, long filaments that could almost be traced over the top and bottom of the cell were organized as interlaced spirals. Adapted from [5] with permission from Nature Publishing Group.
Acknowledgements
We wish to thank Alasdair McDowall, Lu Gan, Morgan Beeby and Ariane Briegel for their help in improving the draft. Ultrastructural research in the Jensen lab is supported by the Howard Hughes Medical Research Institute, NIH grant R01 AI067548 to GJJ, the Beckman Institute at Caltech, and gifts to Caltech from the Gordon and Betty Moore Foundation and Agouron Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1•.Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. Two years ago this review discussed ECT as an emerging technology that had just begun to reveal prokaryotic ultrastructure. Covers all relevant papers before 2007. [DOI] [PubMed] [Google Scholar]
- 2.Lucić V, Förster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 3.Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlén LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 5••.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. Arc-like filaments were seen for the first time at the division plane of C. crescentus cells and identified as the cell division protein FtsZ. Images of the filaments in curved and straight conformations lead to a model wherein FtsZ itself generates force. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Lucić V, Kossel AH, Yang T, Bonhoeffer T, Baumeister W, Sartori A. Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography. Journal of Structural Biology. 2007;160:146–156. doi: 10.1016/j.jsb.2007.08.014. Reports the development of a technique to correlate light and electron microscopy images. [DOI] [PubMed] [Google Scholar]
- 7•.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. Journal of Structural Biology. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. Also reports the development a technique to correlate light and electron microscopy images. See also [8]. [DOI] [PubMed] [Google Scholar]
- 8•.Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. Journal of microscopy. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. See comment to reference 7 above. [DOI] [PubMed] [Google Scholar]
- 9•.Briegel A, Ding H, Li Z, Werner J, Gitai Z, Dias D, Jensen R, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. By immobilizing fixed C. crescentus cells onto EM support grids and then imaging them by both fluorescence light microscopy and ECT, the authors identify chemoreceptor arrays and show they are hexagonally packed with a lattice spacing of 12 nm. This leads to a model for the architecture of chemoreceptor arrays. See also [21]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. ECT and a template-matching were used to reveal the arrangement of 70S ribosomes in translating polysomes from E. coli-derived translation lysates. The arrangement was suggested to increase the efficiency of nascent polypeptide folding. [DOI] [PubMed] [Google Scholar]
- 11••.Salje J, Zuber B, Lowe J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509–512. doi: 10.1126/science.1164346. Vitreous sectioning and ECT were used to show that the ParM filaments responsible for segregating R1-derived plasmids are arranged in small bundles in the periphery of the nucleoid. [DOI] [PubMed] [Google Scholar]
- 12••.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. ECT and vitreous sectioning revealed that the outer layer of the mycobacterial cell envelope was in fact a lipid bilayer membrane. See also [13]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffé M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. Journal of Bacteriology. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. Vitreous sectioning of mycobacteria and corynebacteria revealed their outer membrane structures. See also [12]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm R, Singh H, Rachel R, Typke D, Zillig W, Baumeister W. Electron tomography of ice-embedded prokaryotic cells. Biophys J. 1998;74:1031–1042. doi: 10.1016/S0006-3495(98)74028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pogliano J. The bacterial cytoskeleton. Current Opinion in Cell Biology. 2008;20:19–27. doi: 10.1016/j.ceb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 17.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 18.Erickson HP. FtsZ, a tubulin homologue in prokaryote cell division. Trends in Cell Biology. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 19•.Borgnia MJ, Subramaniam S, Milne JL. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. Journal of Bacteriology. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. ECT revealed the 3-D architecture and cell shape deformation of predator Bdellovibrio bacteriovorus cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. Journal of Structural Biology. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. ECT revealed the native cellular architecture of the pathogenic spirochete Treponema denticola. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Khursigara C, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: trimers of receptor dimers in a partially ordered hexagonally packed array. Journal of Bacteriology. 2008:22. doi: 10.1128/JB.00640-08. This paper confirmed the organization of C. crescentus chemoreceptors reported in [9], but emphasized that the arrays were not perfectly hexagonal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. ECT of Gram-negative sacculi revealed that the glycan strands lie in a single layer parallel to the cell membranes, roughly perpendicular to the long axis of the cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol. 2008;67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. ECT of a recently cultured termide gut spirochete Treponema primitia revealed rich subcellular structures. These results, together with the in situ structure of the flagellar motor from the same species reported in [24], support the “rolling cylinder” model of spirochete motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 25.Berg HC. How spirochetes may swim. J Theor Biol. 1976;56:269–273. doi: 10.1016/s0022-5193(76)80074-4. [DOI] [PubMed] [Google Scholar]
- 26.Izard J, Samsonoff WA, Limberger RJ. Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J Bacteriol. 2001;183:1078–1084. doi: 10.1128/JB.183.3.1078-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N. The flat ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. Journal of Bacteriology. 2008 doi: 10.1128/JB.01288-08. ECT of the Lyme disease spirochete Borrelia burgdorferi revealed that the periplasmic flagella form a right-handed ribbon rather than a bundle, as was previously suggested by traditional EM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Liu J, McBride MJ, Subramaniam S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. Journal of Bacteriology. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. ECT and genetic analysis revealed tufts of ∼5-nm-wide cell surface filaments that appeared to be anchored to the inner surface of the outer membrane in wild-type cells. These filaments are absent in nonmotile gldF mutant cells, suggesting that these filaments are involved in gliding motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weis RM. Inch by inch, row by row. Nat Struct Mol Biol. 2006;13:382–384. doi: 10.1038/nsmb0506-382. [DOI] [PubMed] [Google Scholar]
- 31••.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. This paper used ECT to directly visualize chemotaxis receptor arrays in wild-type E. coli and mutant strains over-expressing the serine chemoreceptor Tsr. Combined with biochemical data, the authors demonstrated that the signaling molecules CheA and CheW are required for functional chemotaxis arrays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci USA. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. ECT and 3-D image averaging revealed two distinct conformations of the HAMP domain of Tsr chemoreceptors, suggesting a possible molecular mechanism in chemotaxis signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 34•.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. Journal of Molecular Biology. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. ECT revealed the 3-D structure of carboxysomes from Halothiobacillus neapolitanus, showing that they are regular icosahedra but with different sizes. See also [35]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. Journal of Molecular Biology. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. ECT revealed the 3-D structure of carboxysomes from Synechococcus Strain WH8102, showing that they too are regular icosahedra but suggesting that the different sizes arise from different T-numbers. See also [34]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. Journal of Bacteriology. 2007;189:4485–4493. doi: 10.1128/JB.01948-06. This paper applied whole-cell ECT, ECT of vitreous sections, and comparative genomic analysis to characterize two closely related strains of cyanobacterium Prochlorococcus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Konorty M, Kahana N, Linaroudis A, Minsky A, Medalia O. Structural analysis of photosynthetic membranes by cryo-electron tomography of intact Rhodopseudomonas viridis cells. Journal of Structural Biology. 2008;161:393–400. doi: 10.1016/j.jsb.2007.09.014. ECT of the purple photosynthetic bacterium Rhodopseudomonas viridis revealed closely packed, flattened photosynthetic membranes connected to the inner membrane by tunnel-like invaginations. [DOI] [PubMed] [Google Scholar]
- 38•.Lieber A, Leis A, Kushmaro A, Minsky A, Medalia O. Chromatin organization and radio-resistance in the bacterium Gemmata obscuriglobus. J Bacteriol. 2008 doi: 10.1128/JB.01513-08. ECT of frozen-hydrated sections of Gemmata obscuriglobus (a member of the phylum Planctomycetes) revealed a network of double-membrane compartments that probably enclose packed chromatin, an organization speculated to increase the radiation tolerance of this species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Junglas B, Briegel A, Burghardt T, Walther P, Wirth R, Huber H, Rachel R. Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch Microbiol. 2008;190:395–408. doi: 10.1007/s00203-008-0402-6. This paper describes the 3-D ultrastructure of Ignicoccus hospitalis and Nanoarchaeum equitans and their cell-cell interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 2009;3:159–167. doi: 10.1038/ismej.2008.99. This paper describes the 3-D ultrastructure of an uncultivated, ultra-small archaeon in acid mine drainage (AMD) biofilm samples. [DOI] [PubMed] [Google Scholar]
- 41.Burns DG, Janssen PH, Itoh T, Kamekura M, Li Z, Jensen G, Rodríguez-Valera F, Bolhuis H, Dyall-Smith ML. Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain. Int J Syst Evol Microbiol. 2007;57:387–392. doi: 10.1099/ijs.0.64690-0. [DOI] [PubMed] [Google Scholar]
- 42.Osman S, Moissl C, Hosoya N, Briegel A, Mayilraj S, Satomi M, Venkateswaran K. Tetrasphaera remsis sp. nov., isolated from the Regenerative Enclosed Life Support Module Simulator (REMS) air system. Int J Syst Evol Microbiol. 2007;57:2749–2753. doi: 10.1099/ijs.0.65137-0. [DOI] [PubMed] [Google Scholar]
- 43.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Nishino Y, Yasunaga T, Miyazawa A. A genetically encoded metallothionein tag enabling efficient protein detection by electron microscopy. J Electron Microsc (Tokyo) 2007;56:93–101. doi: 10.1093/jmicro/dfm008. These authors fused three repeats of metallothionein (3MT) to the 14-mer protein GroEL, expressed the protein in E. coli grown in Cd2+-containing medium, and then visualized the purified protein by cryoelectron microscopy, showing that MT can be an effective tag for EM. See also [46, 47]. [DOI] [PubMed] [Google Scholar]
- 46•.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. This paper showed that fusions of gold-labeled metallothionein to the maltose binding protein (MBP) were visible by electron microscopy, suggesting the potential of metallothionein as a clonable electron microscopy tag. See also [45, 47]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2008 doi: 10.1016/j.jsb.2008.11.009. Localized three different proteins in E. coli by fusing them to gold-labeled metallothionein. See also [45, 46]. [DOI] [PubMed] [Google Scholar]
- 48•.Jin L, Milazzo AC, Kleinfelder S, Li S, Leblanc P, Duttweiler F, Bouwer JC, Peltier ST, Ellisman MH, Xuong NH. Applications of direct detection device in transmission electron microscopy. J Struct Biol. 2008;161:352–358. doi: 10.1016/j.jsb.2007.10.007. This paper describes the first large area mosaic image and tomography dataset recorded on a direct electron detector. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Xuong NH, Jin L, Kleinfelder S, Li S, Leblanc P, Duttweiler F, Bouwer JC, Peltier ST, Milazzo AC, Ellisman M. Future directions for camera systems in electron microscopy. Methods Cell Biol. 2007;79:721–739. doi: 10.1016/S0091-679X(06)79028-8. Describes a prototype direct electron detector that improves both spatial resolution and signal-to-noise ratio. [DOI] [PubMed] [Google Scholar]
- 50.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 51•.Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M. Markov random field based automatic image alignment for electron tomography. Journal of Structural Biology. 2008;161:260–275. doi: 10.1016/j.jsb.2007.07.007. Describes a new algorithm for automatic tilt-series alignment. [DOI] [PubMed] [Google Scholar]
- 52.Martone ME, Zhang S, Gupta A, Qian X, He H, Price DL, Wong M, Santini S, Ellisman MH. The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics. 2003;1:379–395. doi: 10.1385/NI:1:4:379. [DOI] [PubMed] [Google Scholar]
- 53.Frangakis AS, Forster F. Computational exploration of structural information from cryo-electron tomograms. Curr Opin Struct Biol. 2004;14:325–331. doi: 10.1016/j.sbi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie is a three-dimensional tour through the ECT reconstructions of four cells: two NA1000 cells and two cells over-expressing FtsZG109S. The NA1000 cells are introduced with a fLM image of YFP-labeled FtsZ in NA1000 cells (labeled “Caulobacter crescentus PvanA::ftsZ-eyfp (MT196)” in the movie), showing that FtsZ normally localizes as a ring at midcell. Next a cryo-EM image of a dividing NA1000 cell is shown, zooming in on the division site, which will be reconstructed. The original tilt-series of images is shown (labeled “Tilt Series”), followed by the full 3-D reconstruction (labeled “Tomogram”) slice-by-slice. The segmented inner membrane (blue), outer membrane (gold), and FtsZ filaments (red) are then shown from a range of views while the raw reconstruction is flashed on and off to give the viewer a sense for the interpretability of the data. Next a second NA1000 cell is shown (labeled “Another cell more deeply constricted”) in a similar but abbreviated fashion without the tilt series or full reconstruction. The two cells over-expressing FtsZG109S are introduced with another fLM image using FtsZ-YFP (labeled “FtsZ G109S”), showing that FtsZ accumulates in the extended division sites. An additional fly-around of one particular filament and its connection to the cell wall is included (labeled “Connections to peptidoglycan”). Finally the second cell over-expressing FtsZG109S is shown (labeled “Another mutant cell”), wherein an additional cytoplasmic cytoskeletal filament bundle (orange) is seen. Three particularly well-resolved, long filaments that could almost be traced over the top and bottom of the cell were organized as interlaced spirals. Adapted from [5] with permission from Nature Publishing Group.