Abstract
Anomalous choline metabolic patterns have been consistently observed in vivo using Magnetic Resonance Spectroscopy (MRS) analysis of patients with neurodegenerative diseases and tissues from cancer patient. It remains unclear; however, what signaling events may have triggered these choline metabolic aberrancies. This study investigates how changes in choline and phospholipid metabolism are regulated by distinct changes in the mitochondrial electron transport system (ETS). We used specific inhibitors to down regulate the function of individual protein complexes in the ETS of SH-SY5Y neuroblastoma cells. Interestingly, we found that dramatic elevation in the levels of phosphatidylcholine metabolites could be induced by the inhibition of individual ETS complexes, similar to in vivo observations. Such interferences produced divergent metabolic patterns, which were distinguishable via principal component analysis of the cellular metabolomes. Functional impairments in ETS components have been reported in several central nervous system (CNS) diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD); however, it remains largely unknown how the suppression of individual ETS complex function could lead to specific dysfunction in different cell types, resulting in distinct disease phenotypes. Our results suggest that the inhibition of each of the five ETS complexes might differentially regulate phospholipase activities within choline metabolic pathways in neuronal cells, which could contribute to the overall understanding of mitochondrial diseases.
Keywords: NMR, Pattern recognition, Metabolomics, SH-SY5Y, Mitochondrial disease
1 Introduction
Choline (Cho) and related lipid metabolites are vital for many aspects of cellular structure and function. In addition to being an important precursor for the synthesis of acetylcholine, a neurotransmitter, it is also required for the synthesis of phosphatidylcholine, sphingosylphosphorylcholine and other phospholipids, which are important for maintaining cell membrane integrity and facilitating signal transduction. Furthermore, Cho is an important methyl-group donor for the synthesis of S-adenosine methionine, an essential metabolite for the methylation of DNA and protein molecules. It is now clear that the deregulation of Cho metabolic pathways has a profound effect on cellular physiology. Abnormal Cho metabolism has been reported in several diseases where mitochondrial dysfunction is implicated (Farber et al. 2000; Michel et al. 2006). In many cases such alterations are characterized by an increase in the amount of total choline metabolite (tCho), which is increasingly utilized as one of the biomarkers for mitochondrial dysfunction (Jenkins et al. 2005; Meyerhoff et al. 1994). For instance, brain cells in AD show membrane defects associated with increased phospholipid turnover, which is characterized by a decrease in Cho and phosphorylcholine (PC) andan increase in glycerophosphorylcholine (GPC) and cytidine diphosphate choline (CDP-Cho) (Farber et al. 2000).
The exact relationship between Cho metabolism and mitochondrial dysfunction has not been well understood, but there is increasing evidence that they are functionally connected. Clinical studies using MRS have reported elevations in both tCho and PC in numerous human solid tumors (Negendank 1992; Podo 1999) and in animal models including breast, ovarian (Aboagye and Bhujwalla 1999) and brain tumors (Barker et al. 1993). Mitochondrial functional adaptation to hypoxia has been well documented in various cancer models (Glunde et al. 2006). Recently, hypoxia inducible factor-1α (HIF-1α) was shown to induce choline kinase, a key enzyme responsible for the synthesis of PC from Cho in metastatic cancer cells (Glunde et al. 2008). In addition to cancer, MRS studies have also reported changes in Cho metabolites in several CNS diseases associated with mitochondrial abnormalities. Higher tCho/creatine ratios were reported in the posterior mesial gray matter in AD versus control patients (MacKay et al. 1996). Similarly, a significant increase in tCho metabolites was also observed in basal ganglia (Clarke and Lowry 2000). Furthermore, tCho metabolites were found to be increased in a rodent model of Huntington’s disease (HD) (Jenkins et al. 1993). In order to understand the functional significance of Cho metabolism in mitochondrial dysfunction, we embarked on a metabolomics study of SH-SY5Y neuroblastoma cells treated with specific mitochondrial ETS complex inhibitors. This work stems from the fact that complex-specific ETS inhibitors have been shown to induce symptoms in animal models resembling those observed in different human CNS diseases (Brouillet et al. 1999; Kanthasamy et al. 1994; Tetrud and Langston 1989). For example, Complex I inhibition within dopaminergic neurons has been associated with the development of PD (Schmidt and Ferger 2001; Sherer et al. 2002); while a decrease in Complexes II/III activities have been seen in select populations of HD patients (Beal 1998). In addition, Complex IV defects have been observed in animal models of AD (Ohta and Ohsawa 2006).
We report here that inhibition of mitochondrial ETS complexes alters Cho catabolic and anabolic pathways, producing NMR spectral patterns similar to those observed by MRS in vivo. In addition, distinct metabolomic patterns following inhibition of individual ETS complexes can be differentiated using 1H-NMR analysis of cellular metabolites. In particular, we found that the relative levels of three Cho metabolites, Cho, PC and GPC, were among the most dramatically affected cellular metabolites by the inhibition of individual ETS complex. Differential induction of a choline-metabolizing lipase, choline kinase, has been detected following the inhibition of specific ETS complexes. Finally, we discuss a possible relationship between Cho turnover and mitochondrial dysfunction via phospholipase modulation.
2 Materials and methods
2.1 Materials
A human neuroblastoma SH-SY5Y cell line was obtained from American Type Culture Collection (MD, USA). Dulbecco’s modified Eagle’s medium (DMEM), trypsin-MEM (0.05%) and penicillin/streptomycin were purchased from Mediatech Inc. (VA, USA). Glucose-free DMEM and phosphate-buffered saline (PBS) were obtained from Gibco (Grand Island, NY). Fetal bovine serum (FBS) was obtained from TC Biologicals (Tulare, CA) Deuterium oxide (D2O, 99.996%), 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP), deuterium chloride (DCl, 99% D), sodium deutoroxide (NaOD, 99.5% D), 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), 3-nitropropionic acid (3-NP), oligomycin, antimycin-A, sodium azide, ferulic acid, choline chloride, acetylcholine and phosphorylcholine chloride were purchased from Sigma-Aldrich (St. Louis, MS). Perchloric acid (70–72%) was purchased from JT Baker (Phillipsburg, NJ).
2.2 Cell culture and ETS inhibition
SH-SY5Y cells were seeded in poly-d-lysine coated 150 cm2 polystyrene culture dishes and grown in DMEM containing 10% FBS, penicillin (100 units/ml), streptomycin (100 µg/ ml) and non-essential amino acids (NAA) and incubated at 37°C in a humidified atmosphere of 5% CO2 and 95% air. For each metabolic reaction analysis, 8 × 107 cells were used. It was expected that the interferences of the ETS Complexes I to V by increasing concentrations of complex specific inhibitors would decrease cellular metabolism, promote reactive oxygen species production and eventually lead to cell death, therefore, at the inhibitor concentrations chosen for this study, we aimed to achieve approximately 80% cell viability after each treatment. This biological response-based normalization undertook considerations of differential inhibitor transport, metabolism and inhibition kinetics, allowing the distinction of cellular metabolomes and the comparison of complex-specific metabolomic patterns. Once 80–90% confluence was reached, cells were incubated with fresh DMEM containing 10% FBS, penicillin (100 units/ml), streptomycin (100 µg/ml) and NAA plus one of the following ETS inhibitors: 1 mM MPTP, 5 mM 3-NP, 4 µg/ml antimycin-A, 2 mM sodium azide or 1 µg/ml oligomycin. All ETS inhibition reactions were carried out for 24 h, except 3-NP, which was for only 12 h. Cells were counted after each ETS inhibition and overall cell viability was maintained at around 80%. A control cell culture was included along with each ETS inhibition reaction. All experiments were carried out in triplicate for NMR and with five repetitions for mass spectrometry (MS).
2.3 1H-NMR spectroscopy
After the incubation, the culture medium was removed and the cells were washed twice with ice-cold PBS and frozen immediately. Cells were scraped from the culture dishes and the metabolites were extracted with ice-cold perchloric acid (12%) and neutralized with 5 M KOH. The suspension was centrifuged at 15,000 × g to remove the insoluble KClO4 formed and the remaining cell debris and supernatants were stored at −80°C following lyophilization. Prior to NMR analysis, the samples were dissolved in 300 µl of D2O and pD was adjusted to 7.0–7.3 with DCl and NaOD. 1H-NMR spectra were recorded on an 800 MHz Bruker Avance spectrometer (Nuclear Magnetic Resonance Core Facility, RPI, Troy, NY) with a 5-mm ultra sensitive HCN inverse triple resonance cryoprobe. Each spectrum was acquired over 256 scans, and water suppression was achieved by a presaturation pulse sequence, based on the start of the NOESY pulse sequence at 25°C. Each spectrum was recorded with 10 s repetition time, 8,621 Hz spectral width and 32 K data size. Chemical shifts were referenced to the methyl protons of lactate at 1.32 ppm.
2.4 Quantification of metabolites by NMR
The relative amounts of cellular metabolites were calculated from the fully relaxed 1H-NMR spectra of the cell extracts using (trimethylsilyl)propionic-2,2,3,3,d4-acid (TSP) as the external standard. Chemical shift assignments were confirmed both by performing additional 2D NMR experiments and by comparison with the reported chemical shifts and coupling constants of the metabolites (Govindaraju et al. 2000). 1H-NMR spectra were phased and baseline corrected, then integrated between 0.5 and 4.5 ppm and binned into 0.04 ppm integral regions with MestrecC (MESTRELAB RESEARCH SL, Santiago de Compostela, Spain). Individual integral regions were normalized to the total spectra excluding the water region, so each bin was normalized to the total metabolite concentration in each sample. The data were then transferred to the SIMCA package (Umetrics, Umea, Sweden) and pre-processed using pareto-scaling. This processing procedure weighs each integral region by (1/Sk)1/2, where Sk represents the standard deviation of the variables and increases the representation of lower concentration metabolites while minimizing the noise (Pears et al. 2005). Statistical analysis of individual metabolites among the ETS inhibition group means was calculated using an unpaired two-tailed student’s t-test using Microsoft Excel, with P ≤ 0.05 considered significant. The relative changes are presented as mean ± SD.
2.5 Principal component analysis (PCA)
SIMCA package was used to simplify NMR spectral data comparison and to identify the differences among sets of NMR data (n = 3 for each treatment, total 6 groups). The unsupervised method was used to analyze the data. Two types of plots are usually generated with this method. The score plot shows the similarities and differences among the data sets. The data sets exhibiting similarities are clustered together and those that are different are placed further apart; therefore, one can quickly judge the degree of similarity among the data sets. The second plot is called a loadings plot and shows the variables/buckets responsible for the variation within the dataset. This plot helps to pinpoint the exact metabolites or NMR spectral features that are the differentiating factors for the separation of clusters. The PCA score values were subjected to ANOVA in order to evaluate the statistical differences among sample groups.
2.6 Mass spectrometry and quantification of Cho and PC
Subsequent to treating the SH-SY5Y neuroblastoma cells with each of the five ETS inhibitors, the metabolites were extracted with 250 µl of ice-cold perchloric acid (12%). The supernatants were neutralized with 250 µl of 1.5 MKOH and centrifuged at 14,000 rpm to remove insoluble materials. Metabolite analysis was performed on an Applied Biosystems Inc. 4800 Proteomics Analyzer matrix-assisted laser desorption/ionization mass spectrometer (MALDI-TOF MS), where MS spectra (m/z 50–300) were acquired in positive ion and reflectron mode with external mass calibration. Each spectrum was averaged over 2200 laser shots. The matrix solution composition was 10 mg/ml ferulic acid in 30% acetonitrile plus 0.1% trifluoroacetic acid. One ml of matrix was spiked with 200 pmol of acetylcholine as the internal standard for quantification. Two micro liters from each sample was mixed with an equal volume of the matrix solution and four aliquots of 0.5 µl each were spotted onto an Opti-TOF MALDI target plate. In order to compensate for analytical and biological variation, Cho (m/z 104.1) and PC (m/z 184.1) ion counts were first divided by the acetylcholine ion signals. The metabolites were then normalized against the total metabolite ion counts. Each treatment group had five repetitions. The relative changes are presented as mean ± SD relative to the controls. Microsoft Excel was used to calculate the P values using unpaired two-tailed student’s t-tests.
2.7 Western blot
After the treatments, cells were washed with ice-cold PBS and lysed in lysis buffer (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton × 100) plus protease inhibitors (Sigma, St. Louis, USA). Twenty micrograms of proteins from each sample was resolved over a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane (Millipore, USA). The membrane was first blocked in the blocking buffer (5% nonfat dry milk, 0.1% Tween 20; in PBS) for 1 h at room temperature and then incubated with an antibody against choline kinase (ab38290, Abcam, Cambridge, USA) in blocking buffer for overnight at 4°C. After washing, the membrane was incubated with an HRP-conjugated goat anti-rabbit secondary antibody (Bio-Rad, Hercules, USA) for 1 h at room temperature and detected by chemiluminescence. Equal protein loading was confirmed by stripping the blot and re-probing with a β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, USA).
3 Results
3.1 1H-NMR analysis of SH-SY5Y cell metabolome
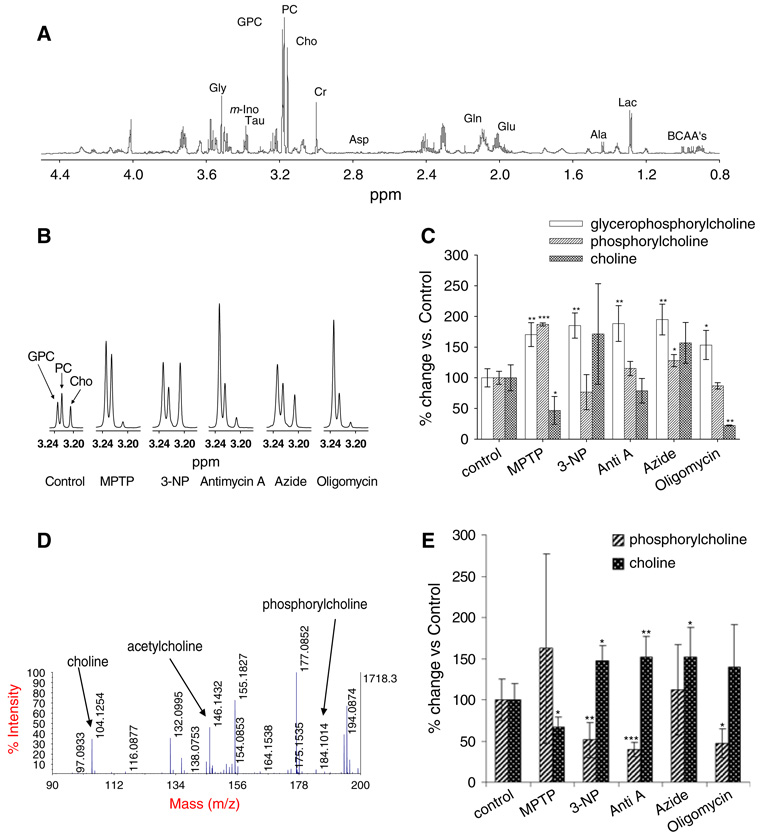
Metabolites derived from SH-SY5Y cells treated with each inhibitor were extracted with perchloric acid and analyzed by 1H NMR. An 1H-NMR spectrum of a control sample is shown in Fig. 1a. The high-resolution 800 MHz 1H-NMR spectra enabled us to assign the spectral signals to various metabolites with high confidence (Govindaraju et al. 2000). Fifteen known metabolites were quantified after manual phasing and baseline corrections for deviations (Table 1).
Fig. 1.
Changes in choline metabolite levels following ETS complex inhibition. (a) An 1H-NMR spectrum of SH-SY5Y neuroblastoma cell metabolites in the absence of ETS inhibition. Metabolites were extracted with 12% perchloric acid. BCAA’s: branched chain amino acids; Lac: lactate; Ala: Alanine; Glu: Glutamate; Gln: Glutamine; Asp: Aspartate; Cr: Creatine; Cho: Choline; PC: Phosphorylcholine; GPC: Glycerophosphorylcholine; Tau: Taurine; m-Ino: Myo-inositol; Gly: Glycine. The chemical shifts were assigned according to Govindaraju et al. (2000). (b) Select regions within representative 1H NMR spectra where Cho, PC and GPC were detected. Values are given as % over control, mean ± SD (n = 3, *P < 0.05, **P <0.01, ***P < 0.001). All ETS complex inhibitors resulted in a combined increase in total choline metabolites (tCho). (c) Quantitative analysis of NMR spectral changes in specific choline metabolites following each ETS complex inhibitor treatment. (d) A MS spectrum (m/z 50–300) of a control sample. The masses for Cho and PC are monoisotopic. (e) Quantitative analysis of Cho and PC by MALDI-TOF MS. Values are given as % over control, mean ± SD (n = 5, *P < 0.05, **P < 0.01, ***P < 0.001)
Table 1.
Relative changes of each metabolite following ETS complex inhibitions
| ETS complex Inhibitor |
I MPTP |
II 3-NP |
III Antimycin-A |
IV Azide |
V Oligomycin |
|---|---|---|---|---|---|
| Val | 74.1 ± 6.1** | 77.0 ± 7.0** | 80.7 ± 6.5** | 74.9 ± 0.5*** | 78.1 ± 7.3** |
| Leu | 58.2 ± 6.4*** | 65.8 ± 4.8*** | 66.2 ± 3.9*** | 61.1 ± 2.5*** | 60.4 ± 7.9** |
| Ile | 65.7 ± 11.3* | 80.6 ± 7.8 | 74.9 ± 7.6* | 70.5 ± 2.4** | 67.4 ± 10.6* |
| Lac | 91.6 ± 10.2 | 68.4 ± 4.4** | 107.9 ± 4.8 | 81.0 ± 6.5* | 90.1 ± 4.7 |
| Ala | 40.4 ± 5.3* | 42.6 ± 2.3* | 27.4 ± 2.9* | 30.2 ± 6.3* | 31.2 ± 5.7* |
| Glu | 95.9 ± 10.2 | 88.3 ± 14.8 | 100.0 ± 7.9 | 77.6 ± 5.3 | 88.4 ± 11.8 |
| Gln | 81.7 ± 8.0* | 84.0 ± 1.9** | 101.7 ± 4.3 | 64.4 ± 8.3** | 82.8 ± 7.2* |
| Cr, PCr | 65.8 ± 1.1** | 60.1 ± 4.2** | 87.7 ± 8.7 | 42.4 ± 5.9*** | 69.6 ± 11.8* |
| Cho | 46.8 ± 22.7* | 171.5 ± 81.9 | 78.9 ± 19.9 | 156.9 ± 33.3 | 22.2 ± 0.8** |
| PC | 187.0 ± 2.5*** | 76.6 ± 28.6 | 115.2 ± 11.6 | 128.0 ± 9.9* | 86.8 ± 5.1 |
| GPC | 170.4 ± 19.4** | 185.2 ± 20.5** | 188.4 ± 29.0** | 195.0 ± 25.1** | 153.5 ± 23.8* |
| Tau | 70.3 ± 4.7*** | 54.6 ± 12.8** | 118.2 ± 15.4 | 58.3 ± 5.2*** | 92.5 ± 5.6 |
| m-Ins | 113.0 ± 23.5 | 136.2 ± 27.5 | 138.1 ± 1.2** | 95.7 ± 7.1 | 126.2 ± 3.6* |
| Gly | 90.8 ± 1.4* | 129.4 ± 12.3* | 137.3 ± 14.0* | 104.7 ± 8.7 | 85.5 ± 4.9* |
| Asp | 245.5 ± 31.9** | 163.8 ± 113.0 | 237.9 ± 14.8*** | 228.6 ± 36.7** | 235.7 ± 46.0* |
The percentage ratios were calculated by dividing the integration of the respective peak areas in 1H-NMR spectra against the values obtained from the controls. The relative changes are given as means ± SD (n = 3)
P < 0.05
P < 0.01
P < 0.001
3.2 Distinct changes in choline metabolites
Inhibition of specific ETS complexes resulted in the distinct changes in several Cho metabolites (Fig. 1). Because there were only three replicates for the NMR analysis as a result of long data acquisition time required on the NMR, changes in Cho and PC could not be confidently determined for several treatments. Therefore, we also analyzed Cho and PC by MS as a complimentary analysis, to improve statistical significance of the measurements (Fig. 1d, e; Supplemental Table 1). Although all five of the ETS complex inhibitors showed significant increases in GPC levels (Fig. 1c), only 3-NP, antimycin-A and azide treatments produced elevated free Cho (Fig. 1e). In contrast, MPTP treatment resulted in decrease of free Cho, (Table 1, Fig. 1c, e). PC levels were elevated by MPTP and azide treatments, respectively (Table 1, Fig. 1c). On the other hand, 3-NP, antimycin-A and oligomycin treatments caused a decrease in PC (Fig. 1e).
3.3 Distinct metabolomic patterns following ETS inhibition
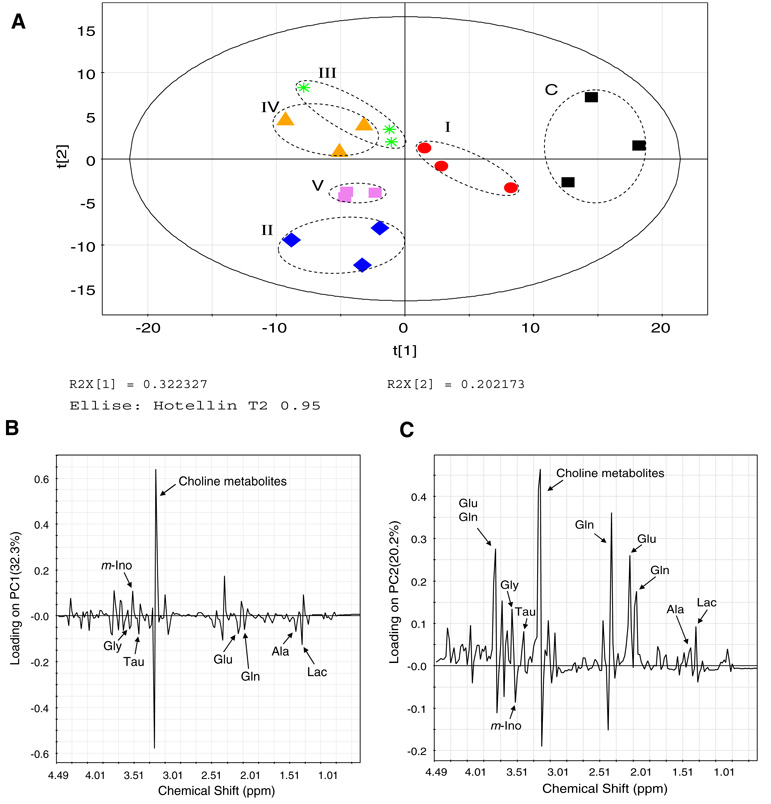
Treatments of SH-SY5Y neuroblastoma cells with MPTP, 3-NP, antimycin-A, azide or oligomycin resulted in the inhibition of mitochondrial ETS Complexes I through V, respectively. This inhibition created perturbations in a number of metabolites. 1H-NMR spectra was divided into 0.04 ppm buckets and PCA analysis was applied for all the treatments. Patterns from PCA analysis of the chemical shift intensity information within all NMR spectra revealed that different inhibitors showed clear separations among one another and with the control cells (Fig. 2), meaning that the 1H-NMR spectral signatures of each ETS complex inhibition group were distinguishable. The clustering of each sample group is depicted with the PCA score plot (Fig. 2a), where each treated cell group produced significantly distinct NMR patterns, resulting in spatial separations among the treatment groups in the plot. On the other hand, some metabolic similarities and PCA overlapping were seen for antimycin-A and azide-treated cells (Complexes III and IV inhibition). Studying the principal component 1 and 2 loading plots and the respective spectra showed that the primary differentiating buckets belonged to Cho metabolites, including free Cho (3.20 ppm), PC (3.22 ppm) and GPC (3.24 ppm) (Fig. 2b, c). In addition, some alterations in glutamate, glutamine and branched chain amino acids were also observed (Fig. 2b, c).
Fig. 2.
Principal component analysis of the metabolomes following all six treatments. (a) A PCA score plot distinguishes the metabolic profiles of SH-SY5Y neuroblastoma cells treated with: I: 1 mM MPTP; II: 5 mM 3-NP; III: 4 µg/ml antimycin-A; IV: 2 mM sodium azide and V: 1 µg/ml oligomycin. C: Control cells treated with saline only. The solid ellipse denotes the 95% significance limit (Hotelling T2) and the dotted ellipse indicates the differentially inhibited sample groups (the dotted lines do not have statistical significance). (b) Loading plot for principal component 1 (PC1). (c) Loading plot for principal component 2 (PC2). The PC1 calculated by this method represents the largest variation among the data sets and the subsequent PCs represent lesser variations by comparison. Usually, the first three PCs should be able to explain 50–80% of the total variation. The resulting PCA scores were subjected to one-way analysis of variance (ANOVA) in order to test the level of statistical difference among the sample groups. Differences were considered significant when P ≤ 0.05
3.4 Induction of choline kinase by ETS inhibition
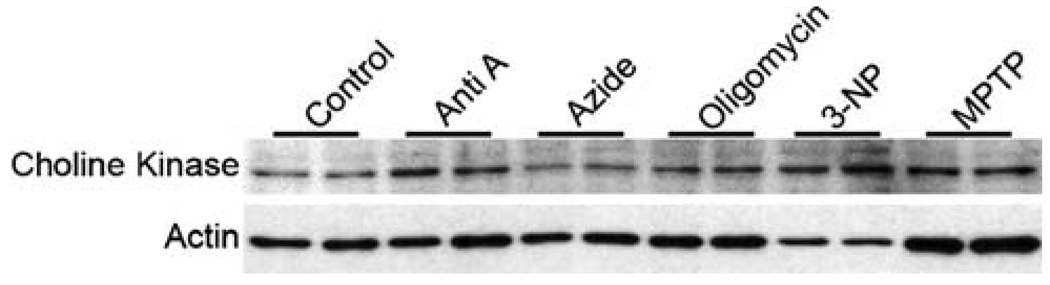
In order to establish the causal relationship between ETS complex inhibition and the observed alterations of choline metabolites, we performed a western blot analysis of choline kinase following ETS complex inhibition. Other than azide inhibition of Complex IV, choline kinase was substantially induced by all of the ETS inhibitors, albeit to different degrees (Fig. 3).
Fig. 3.
Western blot analysis of choline kinase following ETS Complex inhibition. Twenty micrograms of proteins derived from SH-SY5Y cells treated with each of five ETS complex inhibitors were loaded on a 12% SDS-PAGE gel and probed with a anti-choline kinase antibody (ab38290, Abcam). The results suggest that other than azide, choline kinase was induced by all of the other inhibitors of ETS complexes, albeit to a varying degrees compared to untreated SH-SY5Y cells
4 Discussion
Cho is an essential precursor for the synthesis of phosphatidylcholine (PtdCho), an integral component of cell membranes. It is derived either from diet or from the catabolism of PtdCho and acetylcholine (Li and Vance 2008). Within normal cells, over 95% of Cho is present as PtdCho, with 5% as Cho, PC, GPC and other metabolites (Zeisel and Blusztajn 1994). Although Cho metabolism is known to occur both at the membrane and the cytosol, changes in Cho metabolite levels have been suggested to indicate differential turnover of cellular membranes (Farooqui et al. 2000), including mitochondrial membranes. In this study, we used high resolution 800 MHz 1H-NMR to ascertain whether inhibition of individual ETS complexes in SH-SY5Y neuroblastoma cells could induce the abnormal Cho metabolic patterns that are reported in clinical studies of CNS diseases. Indeed, careful examination of the NMR spectra revealed a dramatic overall increase in tCho, (Fig. 1b, all three peaks combined) following inhibition of each of the five ETS complexes compared with control. There were also clear Cho metabolic differences, however, among the five different inhibitors (Fig. 1c, e). To the best of our knowledge, this is the first time that differential choline metabolic patterns have been directly linked to ETS complex inhibition. PtdCho and Cho metabolism involves a network of metabolic enzymes (Fig. 4). The mechanisms governing how the inhibition of each ETS complex might regulate different Cho metabolic enzymes, however, are presently unknown. Earlier studies have provided some clues that either the activation of mitochondrial uncoupler (Farber et al. 2000) or hypoxia (Glunde et al. 2008) is able to selectively modulate Cho metabolic enzymes and produce Cho metabolic profiles similar to those observed in AD patients or in cancer cells. Although it is not well documented that ETS inhibition directly regulate the metabolism of phospholipids in general or of PtdCho in particular, it is plausible that inhibition of specific ETS complexes could differentially activate selective phospholipid metabolic pathways. Consequently, the resulting metabolites could serve as indicators for dysfunction of individual ETS complexes. More importantly, these bio-active compounds might be responsible for instigating the signaling events that lead to the manifestation of ETS complex-specific mitochondrial dysfunction.
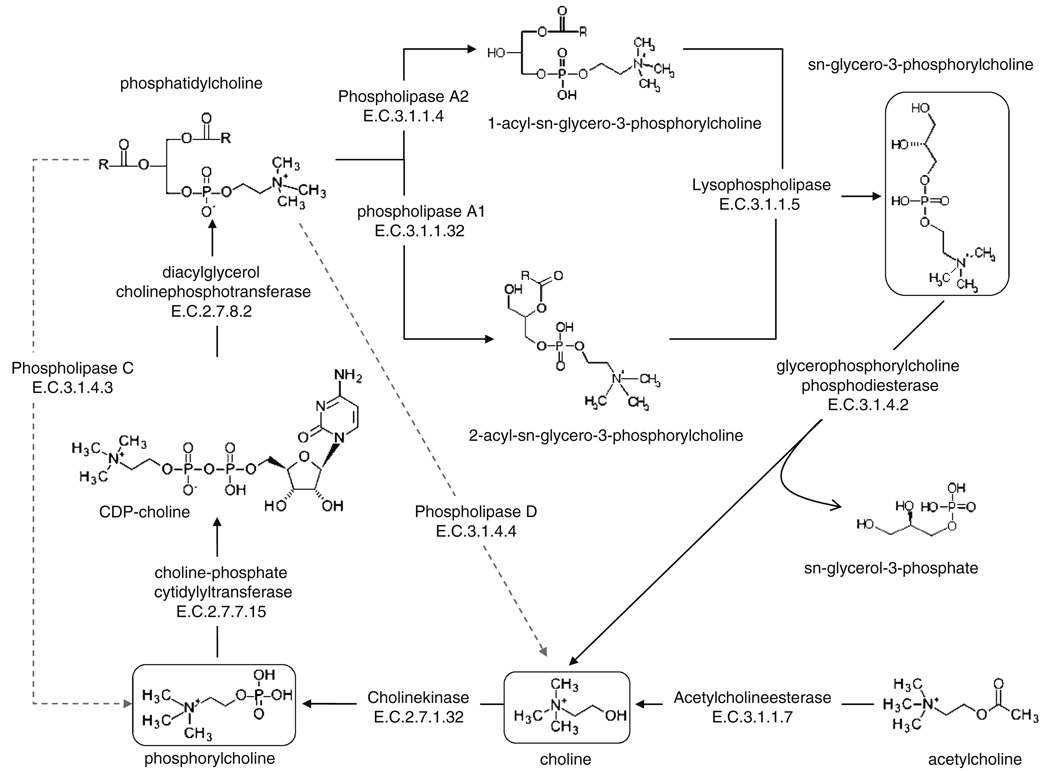
Fig. 4.
Choline metabolic pathways. The scheme shows the structures of individual choline metabolic intermediates and the enzymes that take part in the choline metabolic flux. Metabolites observed in this study are highlighted
Phospholipids are essential components of the plasma membrane and the membranes of cellular organelles. In addition to their structural roles, metabolites derived from phospholipids can also be important signaling molecules, regulating a wide range of cellular functions from apoptosis to stress response. Among the many signaling molecules that can be derived from PtdCho, one of the most abundant phospholipids in eukaryotic membranes, are lyso-PtdCho, lysophosphatidic acid, phosphatidic acid, diacylglycerol and arachidonic acid (AA), a precursor to eicosanoids and endocannabinoids (van Meer et al. 2008). The production of these signaling molecules is regulated in a localized fashion by different classes of phospholipases, including A1, A2, C and D, based on the specific bonds hydrolyzed within the phospholipids. Among the most widely studied is phospholipase A2 (PLA2), a group of enzymes that catalyzes the hydrolysis the sn-2 acyl bond of phospholipids and can yield AA and lysophospholipids. The released AA can then be converted by cyclooxygenases into eicosanoids, which are crucial proinflammatory molecules. There are over 20 different isoforms of PLA2, including cytosolic, Ca+2-dependent, Ca+2-independent, secretory and mitochondrial isoforms. Each is uniquely regulated and can participate in many different signaling pathways. Membrane localization and functional activation of PLA2, for example, is modulated by elevated cellular calcium concentration and by mitogen-activated protein kinase (MAPK) phosphorylation. Regulation of mitochondrial function by PLA2 variants has been well documented. Recently, Zhu et al. demonstrated that PLA2 was capable of mediating amyloid-β peptide-induced mitochondrial swelling in astrocytes (Zhu et al. 2006). Interestingly, Sharpley et al. reported that the activity of NADH:ubiquinone oxireductase of ETS Complex I was enhanced by direct interaction with PtdCho (Sharpley et al. 2006), suggesting that PtdCho catabolism could be associated with Complex I inhibition. On the other hand, how ETS inhibition might modulate PLA2 is not well understood. Inhibition of ETS complexes has been shown to cause elevations of reactive oxygen species (ROS), which may in turn activate PLA2 via MAPK pathways. We observed a significant increase in GPC from 50% to 100% with all five ETS inhibitors, suggesting that accelerated membrane catabolism could have occurred as a results of ETS complex inhibition (Podo 1999). Indeed, elevated GPC has been reported as an indication for increased phospholipid turnover and membrane defects observed in AD patients (Farber et al. 2000). Since, we observed overall elevation of GPC, the end product of combined activities of PLA1, PLA2 and lysophospholipase, with all five ETS complex inhibitors (Fig. 4), these lipases may be activated by reduced mitochondrial ATP synthesis or elevated ROS. ROS elevation associated with ETS dysfunction might also compromise membrane integrity; consequently, Ca2+ level could increase, which in turn would activate PLA2 and other lipases. Activation of PLA2 in mitochondria has been shown to play a role in phospholipid repair and the reduction of lipid peroxidation and mitochondrial swelling (Kinsey et al. 2007).
In addition to GPC, Cho and PC are the two metabolites to be most varied among ETS complex inhibitors (Fig. 1c, e). This suggests that additional Cho metabolic pathway enzymes, including phospholipases C and D, choline kinase and GPC phosphodiesterase (GPC-PDE) might also be differently affected by the five ETS inhibitors (Fig. 3, Fig. 4), resulting in metabolic profiles specific to the ETS complex inhibited. Although phospholipase C (PLC) is usually associated with the release of phosphoinositols from phosphatidylinositol, a choline-specific PLC has been reported (Wolf and Gross 1985). From PtdCho, PLC can produce PC directly, while phospholipase D (PLD) generates Cho (Fig. 4). In addition, Cho can be phosphorylated by choline kinase to synthesize PC. We found that the elevation of PC following MPTP treatment could be the result of choline kinase (Fig. 3) and/or choline-specific PLC activation. On the other hand, the specific decrease of Cho as seen following MPTP treatment could be due to the inactivation of PLD and/or GPC-PDE, along with the activation of choline kinase. PLD is broadly expressed in many eukaryotic cell types and generates Cho and phosphatidic acid from PtdCho. A wide range of signals, including ROS and calcium has been shown to cause the activation of PLD (Steinhour et al. 2008). In addition, PLD isoforms PLD1 and PLD2 have been shown to be regulated by small GTPases (Oude Weernink 2007). Recently a novel PLD distantly related to PLD1 and PLD2 at amino acid sequence level was found to be localized in the mitochondrial outer membrane and to promote organelle fusion (Choi et al. 2006), by producing fusogenic lipids.
The functional relationships between phospholipases and mitochondria are complex, but they may be crucial for understanding specific mitochondrial dysfunction. Overall, it appears that the inhibition of ETS complexes promotes rapid turnover of membrane phospholipids, possibly regulating mitochondrial membrane dynamics that include fusion and fission activities. Such changes might reflect the ability of cells to compensate for functional deficits following the inhibition of ETS complexes. Furthermore, the lipid metabolites produced by activation of phospholipases following ETS inhibition might also modulate a host of signal transduction activities, which could contribute to mitochondrial remodeling and adaptation to ETS inhibition.
We suggest here that the inhibition of each ETS complex has specific biochemical consequences that are reflected by unique cellular metabolic profiles. These profiles can be used to distinguish the specific functional deficits associated with each ETS complex. Our observation of ETS complex-specific alterations in Cho metabolic patterns could offer a new understanding of mitochondrial dysfunction. Both nuclear and mitochondrial gene mutations as well as environmental factors are known to impair the function of specific ETS complexes in animals and humans; e.g., reduction of Complex I activity has been observed in PD patients (Schapira et al. 1989). Further evidence of the involvement of Complex I in PD pathogenesis comes from the use of MPTP treatment in animal models, which recapitulated PD-like symptoms (Bove et al. 2005). Another CNS disease, HD, is known to be caused by an expansion of the CAG trinucleotide repeats in the Huntington (Htt) gene. This is manifested by a poly-glutamine track expansion in the Htt protein. Although Htt is not a mitochondrial protein, there has been significant mitochondrial dysfunction reported for HD patients. Both Complexes II and III activities were found to be decreased by more than half, in addition to a one-third reduction in Complex IV activity in HD patients (Browne et al. 1997). 3-NP, an inhibitor of Complex II, was able to induce HD-like pathology in animal brains (Beal et al. 1993). Elevated tCho has been found in many of the CNS diseases, similar to our observation in neuroblastoma cells treated with ETS inhibitors. MRS technology for in vivo imaging, however, does not yet have the resolution to differentiate Cho from PC and GPC. Once clinical MRS technology is able to distinguish various Cho metabolites, it may be interesting to see whether ETS complex-specific dysfunction can be mapped in different CNS diseases. Several key questions remain: it is unclear how inhibition of different ETS complexes could selectively affect phospholipases associated with PtdCho and Cho pathways. Studies in cancer have already produced clues. Recently cholinekinase was shown to be induced by HIF-1α in human prostate cancer cells (Glunde et. al. 2008). Our data has indicated that similar signal transduction events could be propagated from pathways triggered by ETS complex inhibition (Fig. 3). Alternatively, the observed results could be due to non-specific off-target inhibition of choline metabolic enzymes by the ETS complex inhibitors. Each possibility should be dissected in details for future studies.
5 Concluding remarks
Choline metabolic dysregulation might play an important role in the manifestation of mitochondrial dysfunction. Such aberrant Cho metabolic patterns could be useful to distinguish specific abnormalities within individual ETS complex and to understand their downstream phospholipase signal transduction pathways.
Supplementary Material
Acknowledgments
This work is supported in part by an NIH grant NS046593 and the Foundation of UMDNJ.
Abbreviations
- AA
Arachidonic acid
- AD
Alzheimer’s disease
- ANOVA
Analysis of variance
- CDP-Cho
Cytidine diphosphate choline
- Cho
Choline
- CNS
Central nervous system
- ETS
Electron transport system
- GPC
Glycerophosphorylcholine
- GPC-PDE
Glycerophosphorylcholine phosphodiesterase
- HD
Huntington’s disease
- MPTP
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- HIF-1α
Hypoxia-inducible factor-1α
- Htt
Huntington gene
- MALDI-TOF
Matrix-assisted laser desorption/ionization
- MS
Mass spectrometry
- NAA
Non-essential amino acids
- NMR
Nuclear magnetic resonance spectroscopy
- 3-NP
3-Nitropropionic acid
- PCA
Principal component analysis
- PC
Phosphorylcholine
- PD
Parkinson’s disease
- PLA2
Phospholipase A2
- PLC
Phospholipase C
- PLD
Phospholipase D
- PtdCho
Phosphatidylcholine
- ROS
Reactive oxygen species
- Tau
Taurine
- tCho
Total choline
- TSP
3-(trimethylsilyl)propionic-2,2,3,3-d4 acid
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11306-008-0125-3) contains supplementary material, which is available to authorized users.
References
- Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Research. 1999;59:80–84. [PubMed] [Google Scholar]
- Barker PB, Glickson JD, Bryan RN. In vivo magnetic resonance spectroscopy of human brain tumors. Topics in Magnetic Resonance Imaging. 1993;5:32–45. doi: 10.1097/00002142-199300520-00006. [PubMed] [Google Scholar]
- Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. Biochimica et Biophysica Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. doi: 10.1016/S0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- Beal MF, Brouillet E, Jenkins BG, Ferrante RJ, Kowall NW, Miller JM, et al. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. The Journal of Neuroscience. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J, Prou D, Perier C, Przedborski S. Toxin-induced models of Parkinson’s disease. Neurotherapeutics: The Journal of the American Society for Experimental NeuroTherapeutics. 2005;2:484–494. doi: 10.1602/neurorx.2.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillet E, Conde F, Beal MF, Hantraye P. Replicating Huntington’s disease phenotype in experimental animals. Progress in Neurobiology. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. doi: 10.1016/S0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: Selective vulnerability of the basal ganglia. Annals of Neurology. 1997;41:646–653. doi: 10.1002/ana.410410514. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nature Cell Biology. 2006;8:1255–1262. doi: 10.1038/ncb1487. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Clarke CE, Lowry M. Basal ganglia metabolite concentrations in idiopathic Parkinson’s disease and multiple system atrophy measured by proton magnetic resonance spectroscopy. European Journal of Neurology. 2000;7:661–665. doi: 10.1046/j.1468-1331.2000.00111.x. doi: 10.1046/j.1468-1331.2000.00111.x. [DOI] [PubMed] [Google Scholar]
- Farber SA, Slack BE, Blusztajn JK. Acceleration of phosphatidylcholine synthesis and breakdown by inhibitors of mitochondrial function in neuronal cells: A model of the membrane defect of Alzheimer’s disease. The FASEB Journal. 2000;14:2198–2206. doi: 10.1096/fj.99-0853. doi: 10.1096/fj.99-0853. [DOI] [PubMed] [Google Scholar]
- Farooqui AA, Horrocks LA, Farooqui T. Glycerophospholipids in brain: Their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chemistry and Physics of Lipids. 2000;106:1–29. doi: 10.1016/s0009-3084(00)00128-6. doi: 10.1016/S0009-3084(00)00128-6. [DOI] [PubMed] [Google Scholar]
- Glunde K, Jacobs MA, Bhujwalla ZM. Choline metabolism in cancer: Implications for diagnosis and therapy. Expert Review of Molecular Diagnostics. 2006;6:821–829. doi: 10.1586/14737159.6.6.821. doi: 10.1586/14737159.6.6.821. [DOI] [PubMed] [Google Scholar]
- Glunde K, Shah T, Winnard PT, Jr, Raman V, Takagi T, Vesuna F, et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 alpha signaling in a human prostate cancer model. Cancer Research. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR in Biomedicine. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. doi :10.1002/1099-1492(200005)13:3<129::AID-NBM619>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Jenkins BG, Choi J-K, Beal MF. Magnetic resonance spectroscopy of neurodegenerative illness. In: Beal MF, Lang AE, Ludolph A, editors. Neurodegenerative Diseases. London: Cambridge University Press; 2005. [Google Scholar]
- Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- Kanthasamy AG, Borowitz JL, Pavlakovic G, Isom GE. Dopaminergic neurotoxicity of cyanide: Neurochemical, histological, and behavioral characterization. Toxicology and Applied Pharmacology. 1994;126:156–163. doi: 10.1006/taap.1994.1102. doi: 10.1006/taap.1994.1102. [DOI] [PubMed] [Google Scholar]
- Kinsey GR, McHowat J, Beckett CS, Schnellmann RG. Identification of calcium-independent phospholipase A2gamma in mitochondria and its role in mitochondrial oxidative stress. American Journal of Physiology. Renal Physiology. 2007;292:F853–F860. doi: 10.1152/ajprenal.00318.2006. doi: 10.1152/ajprenal.00318.2006. [DOI] [PubMed] [Google Scholar]
- Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. Jornal of Lipid Research. 2008;48:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- MacKay S, Ezekiel F, Di Sclafani V, Meyerhoff DJ, Gerson J, Norman D, et al. Alzheimer disease and subcortical ischemic vascular dementia: Evaluation by combining MR imaging segmentation and H-1 MR spectroscopic imaging. Radiology. 1996;198:537–545. doi: 10.1148/radiology.198.2.8596863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhoff DJ, MacKay S, Constans JM, Norman D, Van Dyke C, Fein G, et al. Axonal injury and membrane alterations in Alzheimer’s disease suggested by in vivo proton magnetic resonance spectroscopic imaging. Annals of Neurology. 1994;36:40–47. doi: 10.1002/ana.410360110. doi: 10.1002/ana.410360110. [DOI] [PubMed] [Google Scholar]
- Michel V, Yuan Z, Ramsubir S, Bakovic M. Choline transport for phospholipid synthesis. Experimental Biology and Medicine (Maywood, NJ) 2006;231:490–504. doi: 10.1177/153537020623100503. [DOI] [PubMed] [Google Scholar]
- Negendank W. Studies of human tumors by MRS: A review. NMR in Biomedicine. 1992;5:303–324. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- Ohta S, Ohsawa I. Dysfunction of mitochondria and oxidative stress in the pathogenesis of Alzheimer’s disease: On defects in the cytochrome c oxidase complex and aldehyde detoxification. Journal of Alzheimer’s Disease. 2006;9:155–166. doi: 10.3233/jad-2006-9208. [DOI] [PubMed] [Google Scholar]
- Oude Weernink PA, López de Jesús M, Schmidt M. Phospholipase D signaling: Orchestration by PIP2 and small GTPases. Naunyn-Schmiedeberg’s Archives of Pharmacology. 2007;374:399–411. doi: 10.1007/s00210-007-0131-4. doi: 10.1007/s00210-007-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pears MR, Cooper JD, Mitchison HM, Mortishire-Smith RJ, Pearce DA, Griffin JL. High resolution 1H NMR-based metabolomics indicates a neurotransmitter cycling deficit in cerebral tissue from a mouse model of Batten disease. The Journal of Biological Chemistry. 2005;280:42508–42514. doi: 10.1074/jbc.M507380200. doi: 10.1074/jbc.M507380200. [DOI] [PubMed] [Google Scholar]
- Podo F. Tumour phospholipid metabolism. NMR in Biomedicine. 1999;12:413–439. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. doi :10.1002/(SICI)1099-1492(199911)12:7<413::AID-NBM587>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. doi: 10.1016/S0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schmidt N, Ferger B. Neurochemical findings in the MPTP model of Parkinson’s disease. Journal of Neural Transmission. 2001;108:1263–1282. doi: 10.1007/s007020100004. doi: 10.1007/s007020100004. [DOI] [PubMed] [Google Scholar]
- Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH:ubiquinone oxi-doreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, Panov AV, et al. An in vitro model of Parkinson’s disease: Linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. The Journal of Neuroscience. 2002;22:7006–7015. doi: 10.1523/JNEUROSCI.22-16-07006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhour E, Sherwani SI, Mazerik JN, Ciapala V, O’Connor Butler E, Cruff JP, et al. Redox-active antioxidant modulation of lipid signaling in vascular endothelial cells: Vitamin C induces activation of phospholipase D through phospholipase A(2), lipoxygenase, and cyclooxygenase. Molecular and Cellular Biochemistry. 2008;315:97–112. doi: 10.1007/s11010-008-9793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetrud JW, Langston JW. MPTP-induced parkinsonism as a model for Parkinson’s disease. Acta Neurologica Scandinavica. Supplementum. 1989;126:35–40. doi: 10.1111/j.1600-0404.1989.tb01780.x. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nature Reviews. Molecular Cell Biology. 2008;9:112–124. doi: 10.1038/nrm2330. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf RA, Gross RW. Identification of neutral active phospholipase C which hydrolyzes choline glycerophospholipids and plasmalogen selective phospholipase A2 in canine myocardium. The Journal of Biological Chemistry. 1985;260:7295–7303. [PubMed] [Google Scholar]
- Zeisel SH, Blusztajn JK. Choline and human nutrition. Annual Review of Nutrition. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- Zhu D, Lai Y, Shelat PB, Hu C, Sun GY, Lee JC. Phospholipases A2 mediate amyloid-beta peptide-induced mitochondrial dysfunction. The Journal of Neuroscience. 2006;26:11111–11119. doi: 10.1523/JNEUROSCI.3505-06.2006. doi: 10.1523/JNEUROSCI.3505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.