Abstract
We used a proteomic approach to identify novel proteins that may regulate metabotropic glutamate receptor 5 (mGluR5) responses by direct or indirect protein interactions. This approach does not rely on the heterologous expression of proteins and offers the advantage of identifying protein interactions in a native environment. The mGluR5 protein was immunoprecipitated from rat brain lysates; co-immunoprecipitating proteins were analyzed by mass spectrometry and identified peptides were matched to protein databases to determine the correlating parent proteins. This proteomic approach revealed the interaction of mGluR5 with known regulatory proteins, as well as novel proteins that reflect previously unidentified molecular constituents of the mGluR5-signaling complex. Immunoblot analysis confirmed the interaction of high confidence proteins, such as phosphofurin acidic cluster sorting protein 1, microtubule-associated protein 2a and dynamin 1, as mGluR5-interacting proteins. These studies show that a proteomic approach can be used to identify candidate interacting proteins. This approach may be particularly useful for neurobiology applications where distinct protein interactions within a signaling complex can dramatically alter the outcome of the response to neurotransmitter release, or the disruption of normal protein interactions can lead to severe neurological and psychiatric disorders.
Keywords: mass spectrometry, metabotropic glutamate receptor, protein interaction, proteomics
Glutamate is the principal excitatory amino acid neurotransmitter in the central nervous system (CNS) and activates two distinct families of glutamate receptors (GluRs). The ionotropic glutamate receptor (iGluR) family is composed of N-methyl-d-aspartate (NMDA), alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate receptors that mediate fast synaptic transmission, while the metabotropic glutamate receptors (mGluRs) modulate cell excitability and synaptic transmission. Group I mGluRs (1 and 5) couple to the Gq signaling pathway, whereas Group II mGluRs (2 and 3) and Group III mGluRs (4, 6, 7 and 8) couple to Gi/Go signaling pathways in heterologous expression systems (Conn and Pin 1997). mGluR signaling is regulated by several mechanisms, including direct and indirect protein interactions, and the goal of this study was to identify novel regulatory proteins that are molecular constituents of the mGluR5-signaling complex. mGluR5 is expressed in postsynaptic (Romano et al. 1995; Shigemoto et al. 1997) and presynaptic membranes (Gereau and Conn 1995; Croucher et al. 2001), interneurons (van Hooft et al. 2000), glial cells (van den Pol et al. 1995) and, more recently, in nuclei where they may mediate intranuclear signaling pathways (O'Malley et al. 2003). This diverse distribution is consistent with the role of mGluR5 in the control of an array of key signaling events, including roles in the adaptive changes needed for long-term depression or potentiation of neuronal synaptic connectivity (for review see Hermans and Challiss 2001). In addition to playing critical physiological roles within the brain, Group I mGluRs are of considerable importance because of their potential as drug targets for the treatment of a variety of neurological and psychiatric disorders.
Several studies have defined a major site for regulation of mGluR signaling at the intracellular carboxy terminal domain (CTD) that, together with the second intracellular domain, comprise the G protein-binding site (Pin et al. 1994). For example, phosphorylation of the CTD by protein kinase C (PKC) acts in concert with NMDA-mediated calcineurin dephosphorylation of the CTD to regulate Group I mGluR signaling in neurons (Gereau and Heinemann 1998; Alagarsamy et al. 1999). Several proteins bind directly to the CTD of Group I mGluRs to regulate signaling, including Homer (Brakeman et al. 1997; Tu et al. 1998), the G proteins Gq alpha (Abe et al. 1992) and Go alpha (McCool et al. 1998), calmodulin (Minakami et al. 1997), beta-tubulin (Ciruela et al. 1999), Siah1A (Ishikawa et al. 1999) and protein phosphatase 1C (Croci et al. 2003). In addition, previous data support an indirect interaction between mGluR5 and the inositol trisphosphate receptor (IP3R) (Tu et al. 1998; Kammermeier et al. 2000), phospholipase C (PLC) beta (Kim et al. 1997), Shank (Ehlers 1999), arrestin (Mundell et al. 2001) and Src-family protein tyrosine kinases (Heuss et al. 1999). The functional roles of mGluRs within a cell may reflect their unique interactions with regulatory proteins within a signaling complex. For example, mGluR5 and mGluR1 are co-expressed in the same neuron yet mediate distinct physiological responses (Valenti et al. 2002; Pellegrini-Giampietro 2003). Classical approaches to study specific protein interactions include yeast two-hybrid screens or the use of glutathione-S-transferase (GST)-fusion proteins to identify interacting proteins. However, these approaches may not detect ternary or weak protein interactions, or protein interactions that are mediated by post-translational modifications such as lipid modification (Ashman et al. 2001) or the phosphorylation/dephosphorylation state of the protein (Slepnev et al. 1998). Thus, to identify novel mGluR5-interacting proteins, we utilized a proteomic approach that does not rely on the heterologous expression of proteins and offers the advantage of identifying protein interactions in a native environment.
The term ‘proteome’ was coined in 1995 (Wasinger et al. 1995) and defines the large-scale characterization of the entire protein complement of a cell line, tissue or organism. Proteomic studies have expanded greatly in the last decade, largely due to two significant scientific advancements: (i) increased sensitivity of analysis and accuracy of results for mass spectrometry protein identification such that proteins in the femtomolar range can now be detected; and (ii) advances in large-scale nucleotide sequencing of expressed sequence tags and genomic DNA that provides necessary information for protein identification. The first use of a proteomic approach to identify neuronal protein–protein interactions focused on the NMDA receptor complex (Husi et al. 2000). This study included an analysis by mass spectrometry of proteins that were co-immunoprecipitated with the NMDA receptor; the experiments revealed the identity of (at least) 77 proteins ranging from receptors to adaptors, signaling molecules and cytoskeletal proteins, as well as several novel proteins that comprise the NMDA receptor complex. Similar proteomic studies have been used to identify major proteins in the postsynaptic density (PSD) (Walikonis et al. 2000), novel proteins that interact with the purinergic P2X receptor (Kim et al. 2001) and membrane proteins that interact with the CTD of the serotonin 5-HT(2C) receptor (Bécamel et al. 2002). To identify proteins that interact with mGluR5, we immunoprecipitated the receptor from rat brain lysates, subjected the samples to liquid chromatography and mass spectrometry, and identified the protein by comparing the resulting data with protein databases. This approach revealed the interaction of mGluR5 with known regulatory proteins, as well as novel interacting proteins that may reflect previously unidentified molecular constituents of the mGluR5-signaling complex.
Materials and methods
Antibodies
The following primary antibodies were used in these experiments: rabbit anti-mGluR1a and rabbit anti-mGluR5 (Upstate, Lake Placid, NY, USA), mouse anti-PACS1a (phosphofurin acidic cluster sorting protein 1; BD Transduction Laboratories, Lexington, KY, USA), mouse anti-MAP2 (microtubule-associated protein 2; Sigma, St Louis, MO, USA) and mouse anti-dynamin (Upstate). All secondary antibodies were obtained from Bio-Rad (Hercules, CA, USA) or Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-Cascade Blue (CB; Molecular Probes, Eugene, OR, USA) was used as the negative control antibody in all of these experiments as the mGluR5 and the CB antibody are both rabbit IgG antibodies. CB hydrazide is an analog of the bright blue fluorescent tracer methoxypyrenetrisulfonic acid (Molecular Probes), and the rabbit anti-CB antibody was developed to characterize the morphology of neurons that have been filled with CB. As mammalian cells do not make CB, the proteins that are isolated by this antibody represent non-specific protein interactions with components used in the experimental protocol.
Brain lysate preparation
Frozen normal brains from adult male Sprague-Dawley rats (250–300 g) were purchased from Harlan Bioproducts (Indianapolis, IN, USA). For one experiment, adult male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA, USA) were subjected to middle cerebral artery occlusion (MCAO) by the suture method for 30 min and reperfused for 24 h (Shimizu et al. 2001) to stimulate glutamate release in vivo. Each brain was homogenized in 10 mL cold lysis buffer [25 mm HEPES, 140 mm NaCl, 15 mm EGTA, 0.5% Triton X-100 and complete protease inhibitor (Roche, Basel, Switzerland) that includes chymotrypsin (1.5 μg/mL), thermolysin (0.8 μg/mL), papain (1 μg/mL), pronase (1.5 μg/mL), pancreatic extract (1.5 μg/mL) and trypsin (0.002 μg/mL)]. The homogenate was incubated on a rotary shaker for 30 min then centrifuged at 24 000 g for 15 min. The lysates were pre-cleared with 70 μL of a 1 : 1 (v/v) slurry of protein A-sepharose (PAS)/mL lysate and 3 μg of the CB antibody/mg lysate protein for 2 h, and then centrifuged again at 24 000 g for 15 min. The protein concentrations of the cleared lysates were determined using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Co-immunoprecipitation
Individual tubes containing 60 μg primary antibody (experimental anti-mGluR5 or control anti-CB antibody for the immunoprecipitations; anti-Dyamin1, anti-MAP2 and anti-PACS1 for the reverse co-immunoprecipitations)/60 mg of lysate protein were incubated overnight at 4°C, and 100 μL of a 1 : 1 (v/v) PAS slurry were added to each tube and incubated for an additional 3 h with shaking. The tubes were spun at 800 g for 2 min, and the pellet was resuspended in wash buffer (25 mm HEPES, 140 mm NaCl, 15 mm EDTA, 0.1% Triton X-100) and centrifuged at 800 g for 2 min for a total of three washes; the residual supernatant fluid was removed from the PAS beads using a loading gel pipette tip.
Sample preparation
Two protocols were used to prepare the proteins that co-immunoprecipitated with mGluR5 and proteins that bound to the CB antibody, the negative control, for mass spectrometry. The proteins to be analyzed were either (i) removed en masse from the beads and digested with trypsin, or (ii) removed from the beads, separated by one-dimensional (1D) gel electrophoresis and digested with trypsin in gel. The first protocol consisted of en masse removal of the co-immunoprecipitated proteins from the PAS beads as indicated in the text and Tables 1–3 (#). Thus, the PAS bead–protein complex was resuspended twice in 30 μL citric acid (pH 2.0) at 37°C for 15 min to elute the immunoprecipitated proteins. The eluates were transferred to an Eppendorf tube and the pH was adjusted to 7.5 using 5 N NaOH. The samples were dialyzed into 0.1 m NH4HCO3 (pH 8), followed by the addition of urea to 2 m final concentration. The samples were incubated at 90°C for 20 min, returned to 37°C and brought to 10 mm dithiothreitol, and further incubated at 37°C for 1 h. The samples were then brought to 40 mm iodoacetamide, incubated in the dark at 37°C for 1 h and brought to 1 mm CaCl2. At this point, the proteins were trypsinized by the addition of 2 μg trypsin (approximately 0.1 μg/μL final concentration) and incubated at 37°C for 24 h. Complete trypsin digestion was assessed by silver stained 1D sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and if necessary, the trypsin digestion was repeated. Finally, the pH of the samples was lowered with trifluoroacetic acid (3% final concentration). The samples were then reduced to 100 μL in a speed vacuum and stored at − 20°C.
Table 1.
Known high and low confidence molecular constituents of mGluR5-protein complexes
| Name | Rattus Gi number | Protocol | PPID (Separation) |
|---|---|---|---|
| Homer 3** | 16758964 | B## | A0104 (1) |
| GTP-binding protein alpha q (Gq)** | 9296968 | B## | A0041 (1) |
| GTP-binding protein alpha o (Go)** | 8394152 | B## | A0041 (1) |
| Calmodulin** | 1334203 | A,B## | A0008 (3) |
| Inositol 1,4,5-triphosphate receptor 1 (IP3R1)** | 111838 | A,B#,## | A0154 (2) |
| Inositol 1,4,5-triphosphate receptor 2 (IP3R2) * | 111839 | A# | No PPID |
| Phospholipase C-beta-1 (PLC-beta-1)* | 130223 | B# | A0330 (2) |
| Shank 1a* | 13929054 | A# | A0075 (2) |
Identified by at least one high confidence peptide.
Identified by multiple peptides that did not meet the filtering criteria.
En masse off beads,
1D gel.
Table 3.
Novel, high and low confidence molecular constituents of mGluR5-protein complexes, identified in single experiments
| Name | Rattus Gi number | Protocol | PPID (Separation) |
|---|---|---|---|
| 14-3-3 beta** | 9507243 | A## | A0467 (4) |
| 14-3-3 eta** | 6981710 | B## | A0633 (3) |
| Adducin, alpha** | 1083589 | B## | No PPID |
| Adducin, beta** | 10720378 | B## | A0587 (4) |
| Aggrecan 1* | 11990616 | A# | No PPID |
| Aldolase A** | 6978487 | B## | A1923 (ND) |
| Aldolase C** | 6978489 | B## | A0428 (3) |
| Bassoon* | 9506427 | A# | A0285 (ND) |
| Calretinin** | 16758892 | B## | A0437 (ND) |
| Casein kinase II, beta** | 13591930 | B## | A0753 (4) |
| Clathrin heavy chain** | 9506497 | B## | A0119 (4) |
| Citron* | 2745840 | A# | A0276 (4) |
| Destrin** | 7441446 | B## | No PPID |
| Dynamin 1** | 18093102 | B## | A0095 (3) |
| Dynamin 2** | 1083647 | B## | A1927 (ND) |
| Endophilin 1** | 10720268 | B## | A0256 (3) |
| Fibronectin** | 9506703 | A# | A1466 (3) |
| Glutamate receptor interacting protein (GRIP)* | 14091754 | A# | A0301 (5) |
| Histamine receptor H1** | 8393564 | B## | No PPID |
| Myristoylated alanine-rich C-kinase substrate (MARCKS)** | 266495 | B## | No PPID |
| Microtubule-associated protein 1b (MAP1b)** | 1083716 | B## | A0625 (3) |
| Mitogen-activated protein kinase 12* | 3023701 | A# | A0293 (4) |
| NBC (Na/HCO3)-like protein* | 9437326 | B## | No PPID |
| N-ethylmaleimide sensitive factor (NSF)* | 13489067 | B## | No PPID |
| Na+,K ± ATPase alpha 1** | 6978543 | B## | A0340 (4) |
| Na+,K ± ATPase alpha 2** | 6978545 | B## | A0341 (ND) |
| Na+,K ± ATPase alpha 3** | 19855078 | B## | A0342 (ND) |
| Na+,K ± ATPase beta 1** | 6978549 | B## | A0344 (ND) |
| Na+,K ± ATPase beta 2** | 6978551 | B## | A0345 (ND) |
| N-methyl d-aspartate (NMDA) receptor 2a* | 31377498 | A# | A0002 (3) |
| Neural cell adhesion molecule L1 (N-CAM L1)** | 13928706 | B## | A0290 (4) |
| Nuclear factor regulated by interleukin 3** | 16758542 | A# | No PPID |
| Osteopontin** | 6981580 | A# | A1526 (5) |
| Periaxin* | 18677712 | A# | No PPID |
| Phosphatidylethanolamine binding protein** | 406294 | B## | A1328 (5) |
| Phospholipase C-delta | 130228 | A# | No PPID(PLC-delta)* |
| Phospholipase C-epsilon | 16758594 | A,B# | No PPID(PLC-epsilon)* |
| Plasma membrane calcium-transporting ATPase (PMCA1)** | 14286099 | B## | No PPID |
| Profilin II** | 13540707 | B## | A0524 (3) |
| Ras-related small GTP binding protein 3a (RAB3a)** | 6981452 | B## | A0037 (4) |
| Ras-related small GTP binding protein 10 (RAB10)** | 420269 | B## | A0621 (6) |
| Ras-related small GTP binding protein 27b (RAB27b)** | 16758202 | B## | A0360 (ND) |
| RAB GDP dissociation inhibitor** | 1707888 | B## | A0567 (5) |
| RABphilin 3a* | 19424162 | B## | A0212 (4) |
| Reelin* | 31543579 | A# | A1119 (5) |
| SNAP-25-interacting protein (SNIP)* | 9507127 | A# | No PPID |
| Solute carrier family 1, member 3** | 9507115 | B## | No PPID |
| Solute carrier family 12, member 5** | 19705463 | B## | No PPID |
| Synaptogyrin 1** | 9507167 | B## | A0288 (ND) |
| Synaptojanin 1** | 8134729 | B## | A0120 (3) |
| Synaptopodin** | 11067429 | B## | No PPID |
| Utrophin* | 6981696 | A,B# | A0752 (3) |
| Vacuolar ATP Synthase F* | 1718093 | A# | A0405 (6) |
| Vesicle associated membrane protein (VAMP) 2b** | 4894188 | B## | No PPID |
| Voltage-dependent calcium channel, alpha 1a** | 6978579 | B## | No PPID |
| Voltage-dependent calcium channel, L-type beta* | 16758716 | B# | No PPID |
| Voltage-activated calcium channel, T-type alpha 1* | 21687094 | A## | No PPID |
Identified by at least one high confident peptide.
Identified by multiple peptides that did not meet the filtering criteria.
En masse off beads,
1D gel.
The en masse sample preparation protocol was modified because at times, the complexity of the sample was problematic for the mass spectrometry analysis. Thus, the proteins were removed from the beads and separated by 1D gel electrophoresis, as indicated in the text and Tables 1–3 (##). In the modified procedure, the PAS bead–protein complex was resuspended twice in 30 μL citric acid (pH 2.0) at 37°C for 15 min to elute the immunoprecipitated proteins. The eluates were transferred to an Eppendorf tube and the pH was adjusted to 7.5 using 5 N NaOH. At this point, SDS loading buffer was added to the samples [final concentration 1×: 0.0625 m Tris-HCl (pH 6.8), 2% (w/v) SDS, 5% (v/v) β-mercaptoethanol, 10% (v/v) glycerol, 0.002% bromophenol blue] and separated by 1D gel electrophoresis on a 4–15% criterion gel (Bio-Rad). Protein visualization was obtained using either the PlusOne silver staining kit (Amersham Biosciences, Piscataway, NJ, USA) without glutardialdehyde, or the SilverQuest silver staining kit (Invitrogen Corporation, Carlsbad, CA, USA). The mGluR5 and the CB control lanes were each cut into 10 fragments, with the fragment regions from the mGluR5 matching the identical region in the CB control, and each individual fragment was subjected to mass spectrometry.
Immunoblot analysis
The proteins to be analyzed by immunoblot were removed from the beads and separated by 1D gel electrophoresis as described above (Sample preparation). The proteins in the gel were transferred to polyvinylidenedifluoride (PVDF) membrane (Immobilon-P; Millipore, Billerica, MA, USA). The resultant protein blot was incubated with 5% (w/v) milk in Tris-buffered saline (TBS) containing 0.1% Tween (TBST) for 1.5 h at room temperature, followed by incubation with the primary antibody diluted into 5% (w/v) milk in TBST for 1.5 h at room temperature. The blot was rinsed four times for 5 min each in TBST without milk and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad) for 1 h at room temperature. The blot was rinsed once in TBST and four times in TBS, and the proteins were detected using an enhanced chemiluminescent (ECL) western blotting kit (Amersham Biosciences) and Kodak BioMax film (Eastman Kodak Co., Rochester, NY, USA).
Liquid chromatography (LC) and mass spectrometry (MS)
Two protocols at two proteomic facilities were used to identify the proteins that co-immunoprecipitated with mGluR5, and proteins that bound to the CB antibody, the negative control.
Protocol (A) Mass Spectrometry Laboratory, Oregon State University
All samples were mixed 1 : 1 (v/v) with solvent A [0.1% formic acid (FA), 0.005% trifluoroacetic acid (TFA) and 3% acetonitrile (ACN)], and 6 μL of this solution were injected onto a column. Solvent B contained 0.1% FA and 0.005% TFA in 80% ACN. A 5 mm × 0.32 mm C18 trap (LC Packing, San Francisco, CA, USA) was used to de-salt and concentrate each sample. A 10 cm long, 75 μm inner diameter PicoFrit column (New Objective, Woburn, MA, USA) was packed in-house with Jupiter C18 (Phenomenex, Inc., Torrance, CA, USA). The LC conditions started with 3% Solvent B for 5 min to wash the sample, followed by a gradient to 40% Solvent B over 40 min, 70% Solvent B at 50 min, 90% Solvent B at 52 min, and held at 90% Solvent B until 60 min. A Waters CapLC system (Waters Corporation, Milford, MA, USA) was used with a flow rate estimated to be 300 nL/min. The mass spectrometer used for electrospray ionization (ESI) tandem mass spectrometry (MS/MS) was a Quadrupole Time-of-Flight (Q-TOF) Global Ultima system (Micromass, Ltd, Manchester, UK) operated with a spray voltage of 3.5 kV. Data-dependent MS/MS was used with a 0.5 s survey scan and 2.5 s MS/MS scans on the three most abundant peaks in the MS survey scan.
Protocol (B) Proteomics Facility, Fred Hutchinson Cancer Research Center
Prior to proteolytic in-gel digestion, the stain from silver-stained gel slices was removed by mixing equal volumes of 30 mm potassium ferricyanide [K3Fe(CN)6] with 100 mm sodium thiosulfate (Na2S2O3) and adding a volume of this mixture to cover the gel slices. The de-staining solution was removed after approximately 5 min and the gel slices were washed repeatedly with 100 mm ammonium bicarbonate (NH4HCO3) until they were clear. Gel slice digestions were then performed as described in Shevchenko et al. (1996). Following digestion, samples were de-salted using a microC18 ZipTip (Millipore, Bedford, MA), dried, and resuspended in 7 μL of 0.1% TFA. The mass spectrometer used for LC/ESI MS/MS was a LCQ DECA XP mass spectrometer (ThermoElectron, San Jose, CA, USA) as described in Gatlin et al. (1998). Data were collected in a data-dependent mode in which an MS scan was followed by MS/MS scans of the three most abundant ions from the preceding MS scan.
Identification of proteins using database search
Mass spectrometry data were searched against a rat protein database (a subset of proteins from the NCBI non-redundant protein database) using the software search algorithms mascot (Matrix Science Ltd, London, UK), sequest (ThermoElectron) or comet (Institute for Systems Biology, Seattle, WA, USA). These searches resulted in the identification of both known and novel mGluR5-interacting proteins.
Files appropriate for mascot (pkl files) were created using Masslynx software (Waters, Milford, MA, USA) with a function that smoothes, calculates centroids and assesses the quality of data. mascot calculates one statistical parameter to validate protein identification, the Mowse Score. This score is based on the probability (p) that a peptide identified from the experimental fragment matches a peptide in a protein database, and is calculated as: Mowse score = − 10 × log (p). Thus, a random match will have a high probability value and low Mowse score, while a valid match will have a low probability value and a high Mowse score. mascot then ranks the quality of the peptide matches and sums the scores of acceptable peptides to calculate a total protein score. The default total protein score is > 25. Thus, all proteins with a mascot total protein score > 25 were considered as real, high confidence (**) proteins that were identified in the mGluR5-signaling complex.
Peptide identification results from the sequest (Eng et al. 1994) algorithm were filtered using three result parameters: cross-correlation score (Xcorr), delta correlation value (ΔCn) and ion percentage (%ions). Peptides that were identified with a Xcorr > 2, ΔCn > 0.1 and %ions > 30 were considered to be high confidence peptides. Regardless of whether a single high confidence peptide or multiple high confidence peptides identified a protein, that protein was considered to be a real, high confidence (**) protein, identified in the mGluR5-signaling complex.
A recently developed computer algorithm called comet was also used for protein identification. The filtering criteria utilized the score, Z-score, dN and ion% parameters from comet. The score is the dot product between an experimental spectrum and a theoretical spectrum, with the resulting score scaled to 1000. The Z-score is the number of standard deviations away from the mean for the top scoring peptide compared with the top 500 clustered peptide scores. The difference between the normalized scores is represented by the dN parameter, and ion% is the percentage of matched fragment ions over the total number of expected fragment ions for the best matching peptide. Peptides in which + 1 ions had scores greater than 200, + 2 ions had scores greater than 300 and + 3 ions had scores greater than 300, and each of these ions had Z-scores greater than 4.0, dN greater than 0.1 and ion% greater than 30, were considered as real, high confidence peptides. Proteins that were identified with high confidence peptides were considered to be high confidence proteins in the mGluR5-signaling complex.
In this paper, we present 10 proteins that were identified in multiple experiments with multiple high confidence peptides as novel, high confidence (**) molecular constituents of mGluR5-protein complexes (Table 2). In addition, we present proteins that were identified in only one experiment and by at least one high confidence peptide (Table 3). We also include several proteins that were identified by multiple peptides that did not meet the filtering criteria and are identified as low confidence (*) proteins in Tables 1 and 3. The proteins that are low confidence may represent low abundance proteins that result in tandem mass spectra of poorer quality. However, they are included here because of their potential biological relevance as mGluR5-signaling partners based on literature support for either a direct or indirect interaction.
Table 2.
Novel, high confidence molecular constituents of mGluR5-protein complexes, identified in multiple experiments
| Name | Rattus Gi number | Protocol | PPID (Separation) |
|---|---|---|---|
| 14-3-3 gamma** | 9507245 | A,B## | A0362 (4) |
| 14-3-3 theta** | 6981712 | B## | A0907 (5) |
| 14-3-3 zeta** | 13487931 | A, B## | A0361 (3) |
| Adaptor-related protein complex (AP2), beta subunit** | 18034787 | A,B#,## | A0253 (4) |
| Cell division cycle 10 (CDC10), rat homolog** | 12018296 | A,B#,## | No PPID |
| CDCrel-1a** | 8953677 | B#,## | No PPID |
| Microtubule-associated protein 1a (MAP1a)** | 13591886 | B#,## | A0085 (3) |
| Microtubule-associated protein 2 (MAP2)** | 547890 | A,B ## | A0134 (3) |
| Phosphofurin acidic cluster sorting protein 1a (PACS1)** | 19705535 | A,B## | No PPID |
| Phosphofurin acidic cluster sorting protein 1b (PACS1b)** | 3347955 | B## | No PPID |
Identified by at least one high confidence peptide.
En masse off beads,
1D gel.
Protein–Protein Interaction Database analysis of mGluR5-interacting proteins
The Protein–Protein Interaction Database (PPID; http://www.anc.ed.ac.uk/mscs/PPID) is an evolving database that arose from the need to interpret proteomic datasets such as those generated by analyzing the NMDA-receptor complex (Husi et al. 2000). The PPID includes protein sequence and protein-binding information, together with literature references, that can be queried by online browsing, batch-submission, or by using software like mascot. For these studies, the PPID was queried online to determine the degree of separation of the mGluR5 (PPID #A0327) and putative interacting proteins identified in this study. Where a PPID number exists for any given protein, the database provides useful information concerning the likelihood of protein interactions and their linkages based on previously reported data.
Results and Discussion
Immunoprecipitation of mGluR5 from rat brain lysate
The mass spectrometry data depend on the successful immunoprecipitation of mGluR5 from the rat brain lysate under these experimental conditions. Thus, mGluR5 and the closely related mGluR1a were immunoprecipitated from rat brain lysate using subtype-selective antibodies. Parallel experiments were also performed using the negative control anti-CB antibody. The immunoprecipitated proteins from each sample were separated by their molecular mass using 1D gel electrophoresis and transferred to PVDF membrane; the resultant immunoblots were then probed with an mGluR5-selective polyclonal antibody (anti-mGluR5). Figure 1(a) shows that mGluR5 is detected in the total rat brain lysate and the mGluR5 immunoprecipitate, but is not detected in the CB immunoprecipitate. In addition, Fig. 1(b) shows that mGluR5 is detected in the total rat brain lysate and the mGluR5 immunoprecipitate, but is not detected in the mGluR1a immunoprecipitate. These data show that mGluR5 is specifically immunoprecipitated from rat brain lysates under these experimental conditions.
Fig. 1.
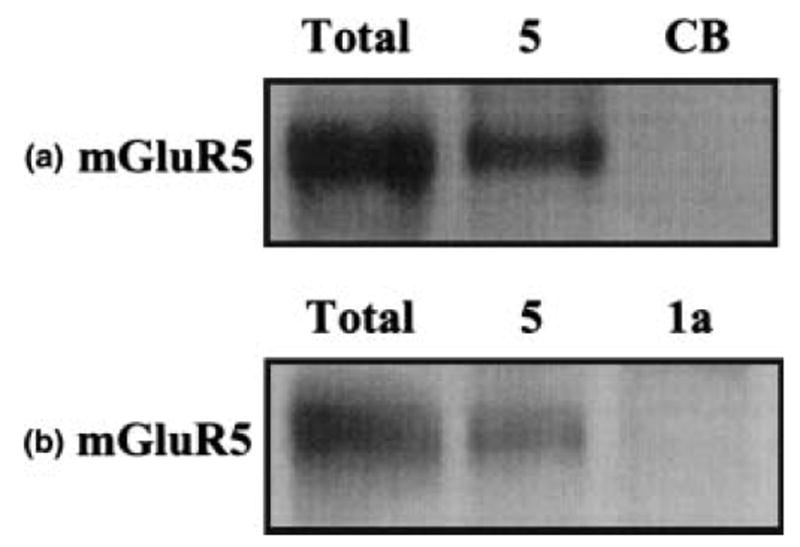
Immunoprecipitation of mGluR5 from rat brain lysate. (a) Total rat brain lysate (Total), mGluR5 (5) and Cascade Blue (CB) immunoprecipitates probed with anti-mGluR5. (b) Total rat brain lysate (Total), mGluR5 (5) and mGluR1a (1a) immunoprecipitates probed with anti-mGluR5.
Mass spectrometry identification of mGluR5 protein
The mass spectrometry results were analyzed for the presence of mGluR5 to prove that the protein is detected by the mass spectrometry, and Fig. 2 shows the LC/MS/MS spectrum that identified mGluR5. Figure 2(a) shows the total ion chromatogram for the proteins that co-immunoprecipitated with mGluR5, Fig. 2(b) shows the mass spectrum of the sample at 25.56 min retention time, and Fig. 2(c) shows the fragmentation mass spectrum of the peak at 782.1 from Fig. 2(b). The software search algorithm SEQUEST identified a total of four peptides at high confidence levels that correspond to peptide sequences in the amino terminal domain of mGluR5, as depicted in Fig. 2(d) (1: DSLISSEEEEGLVR, 2: GLAGEFLLLGSDGWADR, 3: EAVGGITIK and 4: GEVSCCWTCTPCKENEYVFDEYTCK). These data show that mGluR5 is immunoprecipitated from the rat brain lysates under our experimental conditions, and that mGluR5 peptides are identified by LC/MS/MS with high confidence.
Fig. 2.
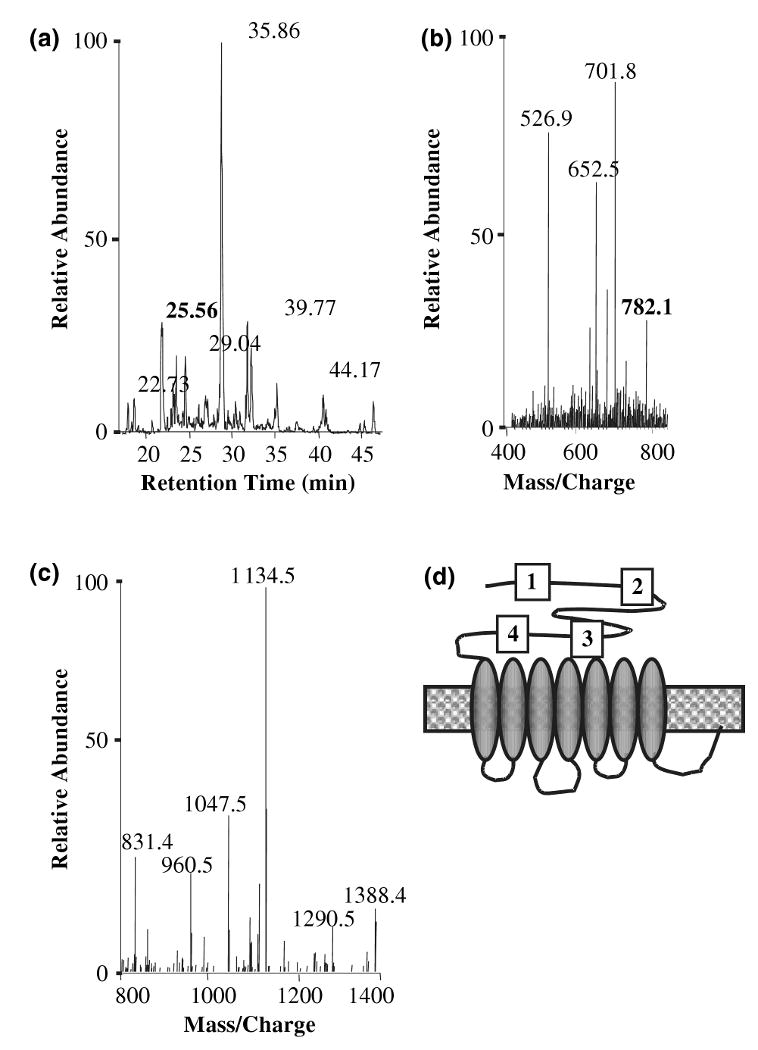
Identification of mGluR5 peptides by mass spectrometry. (a) Total ion chromatogram for the mGluR5 sample. (b) Mass spectrum at 25.56 min retention time. (c) Tandem mass spectrum of m/z 782.1 from Fig. 2(b). (d) Diagramatic representation of peptides identified in the second mass spectrometry.
Silver stain analysis of proteins that co-immunoprecipitate with mGluR5 and Cascade Blue separated by one-dimensional gel electrophoresis
The proteins that co-immunoprecipitated with mGluR5 were visually compared with those isolated by the negative control, anti-CB antibody. Figure 3 shows that the most abundant protein bands detected in both the mGluR5 and CB immunoprecipitates are the (A) heavy (50 kDa) and the (B) light (25 kDa) immunoglobulin chains (arrows). However, several protein bands that are unique to the mGluR5 immunoprecipitate (Fig. 3, mGluR5) are not detected by the naked eye in the CB immunoprecipitate (Fig. 3, CB). These data show that the mGluR5 sample contains several protein bands that are not detected in the CB negative control, and suggest that these proteins are specifically isolated as molecular constituents of the mGluR5-signaling complex.
Fig. 3.
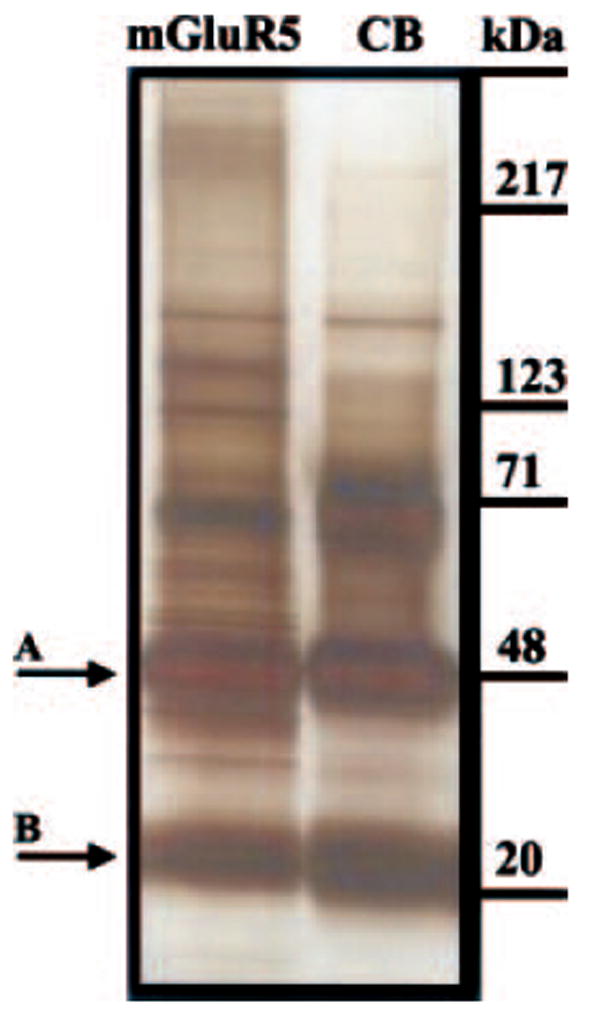
Silver stain analysis of proteins that co-immunoprecipitate with mGluR5 and Cascade Blue separated by one-dimensional gel electrophoresis. Arrows indicate (A) heavy and (B) light chain immunoglobulin subunits of the primary antibodies.
Identification of known, high confidence and low confidence molecular constituents of mGluR5-protein complexes
The proteomic approach identified several proteins in the mGluR5 immunoprecipitated samples that are known to directly interact with mGluR5, or that are mGluR5-signaling intermediates, and were not identified in the CB negative control samples (Table 1). Many of these proteins were identified as high confidence (**) proteins based on mass spectrometry filtering criteria. However, several were identified as low confidence (*) proteins (see Table 1). Previous studies support an interaction between each of these proteins and mGluR5. For example, there is a direct interaction between Group I mGluRs and Gq alpha (Abe et al. 1992), Go alpha (McCool et al. 1998), calmodulin (Minakami et al. 1997) and Homer proteins (Brakeman et al. 1997; Tu et al. 1998). Homer proteins link Group I mGluRs to the inositol trisphosphate receptor (IP3R) (Tu et al. 1998; Kammermeier et al. 2000), regulate trafficking of mGluR5 (Roche et al. 1999) and provide a link between mGluR5 and Shank (Ehlers 1999; Tu et al. 1999). Interestingly, the interaction between mGluR5 and Homer, and mGluR5 and Gq/Go, was only identified with confidence in the tolerant rat brain lysate, suggesting that receptor activation leads to altered protein interactions. Finally, previous data support an indirect interaction between mGluR5 and phospholipase C (PLC) beta (Kim et al. 1997). Thus, the proteins identified by a proteomic approach are consistent with previous protein interaction studies and PPID queries.
Identification of novel, high confidence molecular constituents of mGluR5-protein complexes in multiple experiments
In addition to known mGluR5-interacting proteins, this proteomic approach also identified 10 mGluR5-interacting proteins in several experiments, at one or both proteomic facilities (Table 2), in both normal and tolerant rat brain lysate. These proteins are considered to be valid, high confidence (**) molecular constituents of the mGluR5-signaling complex, whether direct or indirect, because the identifying peptides met the mass spectrometry filtering criteria for high confidence and were not found in the CB negative control sample. Thus, based on the mass spectrometry results that suggest these novel proteins are valid mGluR5-interactings proteins, we investigated two proteins further to confirm their interaction with mGluR5.
Confirmation of novel, high confidence mGluR5–protein interactions by immunoblot
The identification of mGluR5-interacting proteins is the result of mass spectrometry-identified peptide fragments searched against a rat protein database using the software search algorithms to correlate experimental peptide fragments with theoretical peptide fragments. To validate that the peptide fragments correlated with the identified high confidence interacting proteins, we used immunoblots to specifically test for the presence of these proteins. Figure 4 shows individual immunoblots that contain total rat brain lysate (50 μg protein), mGluR5 immunoprecipitated samples, or CB immunoprecipitated samples probed with antibodies to (a) PACS1 or (b) MAP2. The results show that these high confidence mGluR5-interacting proteins are detected in the total rat brain lysate and the mGluR5 immunoprecipitate, but are not detected in the CB sample, consistent with the mass spectrometry data. Literature searches support potential interactions between mGluR5 and each of these proteins. For example, Group I mGluRs are down-regulated after agonist treatment, possibly due to endocytosis by clathrin-coated pits (Luis Albasanz et al. 2002), and in the substantia nigra, more than 80% of immunoreactivity for mGluR5 is intracellular while the majority of mGluR1a is located on the plasma membrane (Hubert et al. 2001). This distinct subcellular localization of each Group I mGluR suggests that trafficking and internalization may contribute to the functional role of these receptors in neurons (Lujan et al. 1997). A major role for PACS1 is to facilitate the trafficking of proteins between the plasma membrane and the cytosol, and PACS1 is required to maintain phosphorylated furin molecules in a cycling loop between early endosomes and the plasma membrane (Molloy et al. 1998). In addition, PACS1 facilitates the movement of furin and the mannose-6-phosphate receptor from the endosome to the Golgi by joining cytoplasmic domain acidic amino acid clusters of the proteins to the adaptor-protein complex-1 (AP-1) in endosomal clathrin-coated membrane pits (Wan et al. 1998; Crump et al. 2001; Scott et al. 2003). As the intracellular carboxy terminal domain of mGluR5 contains several acidic amino acid clusters (969EAEE972 and 1000DDD1002) and phosphorylation sites (see Hermans and Challiss 2001), PACS1 may play a role in the trafficking of mGluR5 between the plasma membrane and the cytosol.
Fig. 4.
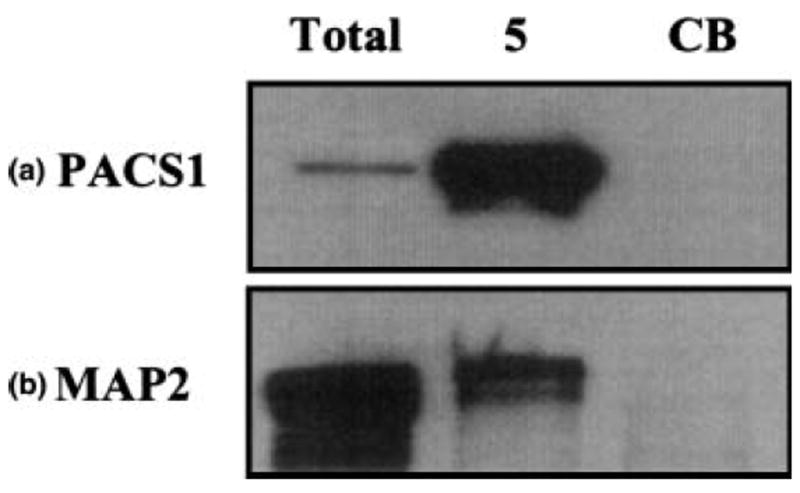
Confirmation of novel, high confidence mGluR5–protein interactions by immunoblot. Total rat brain lysate (Total), mGluR5 (5) and CB (CB) immunoprecipitates probed with (a) anti-PACS1 and (b) anti-MAP2.
There is also support for an interaction between mGluR5 and MAP2, a postsynaptic, neuron-specific, microtubule-associated protein that can bind to both microtubules and F-actin, and plays a key role in specific cytoskeletal rearrangements that control the transition from an undifferentiated state to neurite-bearing morphology (for review see Dehmelt and Halpain 2004). MAP2 is found mainly in dendritic shafts (Kaech et al. 1997) where it may facilitate transport of membrane-bound receptors, such as NMDA receptors or mGluR5, to the postsynaptic density. Interestingly, glutamate can produce a biphasic change in MAP2 whereby a rapid, transient increase in phosphorylation is mediated by Group I mGluRs and a persistent dephosphorylation of MAP2 is mediated by NMDA receptors (Quinlan and Halpain 1996). The opposing actions of glutamate on the phosphorylation state of MAP2 regulate its interaction with microtubules and actin filaments, and suggests that glutamatergic regulation of MAP2 phosphorylation may transduce neural activity into modifications in dendritic structure.
Identification of novel, high confidence and low confidence molecular constituents of mGluR5-protein complexes in single experiments
This proteomic approach identified many novel high confidence (**) mGluR5-interacting proteins in a single experiment that were not detected in the CB negative control sample, and approximately 50% of these identifications came from the tolerant rat brain lysate. We also included several novel low confidence (*) mGluR5-interacting proteins identified in several experiments that were not detected in the CB negative control sample. Inclusion of the low confidence proteins is supported by data in Table 1, which lists several known mGluR5-interacting proteins that are designated as low confidence proteins based on the filtering criteria, but are known to be valid mGluR5-interacting proteins. In addition, prior studies support a potential role for many of the proteins in Table 3 as mGluR5-interacting proteins, and they may represent valid signaling intermediates or effectors in the mGluR5-signaling complex. For example, Citron and Group I mGluRs are both expressed in distinct hippocampal interneuron populations (Zhang et al. 1999; van Hooft et al. 2000) where Citron may contribute to the regulation of Group I mGluR-mediated release of GABA from these cells. In addition, Bassoon, a 420 kDa protein localized at the active zone of presynaptic nerve terminals that is involved in cytomatrix organization at the neurotransmitter release site (tom Dieck et al. 1998), and electron microscopy reveals mGluR5 immunoreactivity on presynaptic axon terminals (Romano et al. 1995). Physiology studies suggest that a presynaptic mGluR is responsible for the induction of long-term depression (LTD) in hippocampal CA1 (Faas et al. 2002) and interestingly, this mGluR-induced LTD can be completely blocked by (2-methyl-6-(phenylethynyl)-pyridine (MPEP), a specific mGluR5 antagonist. Thus, mGluR5 regulation of presynaptic neurotransmitter release from hippocampal synapses may involve mGluR5 protein interactions with Bassoon.
Confirmation of a novel, high confidence mGluR5–protein interaction, identified in a single experiment, by immunoblot
To validate that these high confidence peptide fragments identified in a single experiment correlated with the identified protein and interacted with mGluR5, we used immunoblot to specifically analyze the protein interaction. Figure 5 shows an immunoblot that contains total rat brain lysate (50 μg protein), mGluR5 immunoprecipitated sample, or CB immunoprecipitated sample probed with an antibody to Dynamin1. The results show that this mGluR5-interacting protein, identified in only one experiment, is detected in the total rat brain lysate and the mGluR5 immunoprecipitate, but is not detected in the CB sample, consistent with the mass spectrometry data. There is support for the interaction of mGluR5 and Dynamins. Dynamins interact with the Src homology 3 (SH3) domain of PLCγ (Seedorf et al. 1994; Okamoto et al. 1997), and the pleckstrin homology (PH) domain of Dynamin binds phosphatidylinositol-4,5-bisphosphate (PIP2) (Zheng et al. 1996). Both PIP2 and G protein βγ subunits can regulate Dynamin I GTPase activity (Lin and Gilman 1996), although sequestering Gβγ subunits inhibits clathrin-coated endocytosis (Lin et al. 1998). Recently, Dynamin was shown to be necessary for endocytosis (for review see Sever 2002) and Dynamin 2 contributes to the clathrin-independent endocytosis of mGluR5 in COS-7 cells (Fourgeaud et al. 2003). In addition, Dynamin 3 is found in complex with Homer and mGluR5 (Gray et al. 2003), and there is a direct interaction between Dynamin 2 and Shank (Okamoto et al. 2001) and members of the Shank/ProSAP family of post-synaptic density scaffolding proteins. These data are consistent with a role for Dynamins in glutamate receptor down-regulation, and support the interaction between mGluR5 and Dynamin in our proteomic studies.
Fig. 5.
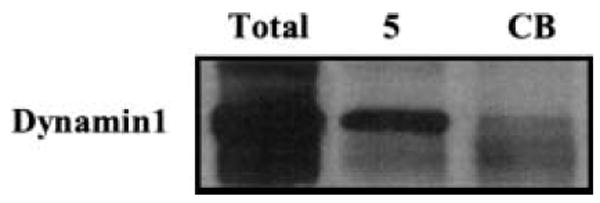
Confirmation of a novel, high confidence mGluR5–protein interaction, identified in a single experiment, by immunoblot. Total rat brain lysate (Total), mGluR5 (5) and CB (CB) immunoprecipitates probed with anti-Dynamin1.
Confirmation of mGluR5–protein interactions by reverse immunoblot
To strengthen the existing data for an interaction between mGluR5 and Dynamin, MAP2 and PACS1, we used reverse co-immunoprecipitation of each reported protein and probed for mGluR5 immunoreactivity. Figure 6 shows a reverse immunoblot that contains total rat brain lysate (50 μg protein) and Dynamin1, MAP2 or PACS1 immunoprecipitated samples, as well as the CB immunoprecipitated sample probed with an antibody to mGluR5. The results show that a weak band was detected in the Dynamin 1 immunoprecipitate, and a stronger band was detected in the MAP2 immunoprecipitate, both at molecular weights that correspond to the dimer form of mGluR5. In contrast, there is a very strong band in the PACS1 immunoprecipitate at the molecular weight that corresponds to the monomer form of mGluR5. These findings are consistent with the mass spectrometry data. Dynamin was detected in the proteomic analysis of mGluR5 co-immunoprecipitates from ischemic pre-conditioned rat brain, representing activated receptor. MAP2 was detected in several experiments, and PACS1 was abundantly detected in virtually every co-immunoprecipitation that we analyzed. Thus, we feel that these data support true interaction between these novel proteins and mGluR5 in rat brain.
Fig. 6.
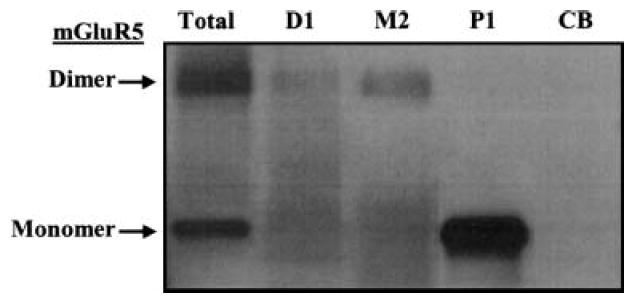
Confirmation of mGluR5–protein interactions by reverse immunoblot. Total rat brain lysate (Total), or Dynamin1 (D1), MAP2 (M2), PACS1 (P1) and CB (CB) immunoprecipitates probed with anti-mGluR5.
Identification of proteins detected in both mGluR5 and CB samples
This proteomic approach identified several proteins as mGluR5-interacting proteins at high confidence levels, but these proteins were also identified in the CB negative control samples. Most of these proteins contribute to the organization of subcellular structures and are involved in membrane skeleton organization. They include several isoforms of actin, actinin, ankyrin, cofilin, dynein, fibrillin, myosin, plecktin, spectrin, tubulin and vimentin, and their identification is consistent with previous studies that did not analyze a stringent negative control by mass spectrometry; their identification in the negative control may reflect their high abundance in the sample. For example, actin, actinin, spectrin and myosin were all reported as protein components of the NMDA receptor multiprotein complex (Husi et al. 2000). In addition, previous studies support interactions between cytoskeletal proteins and mGluRs. Actin binds to Cupidin (Homer 2a) that in turn binds to mGluR1a (Shiraishi et al. 1999), and actin polymerization/depolymerization regulates the movement of mGluR5 in the plasma membrane (Sérge et al. 2003). Actin also regulates the release of calcium from the endoplasmic reticulum in cultured hippocampal neurons (Wang et al. 2002), and actin-dependent changes in dendritic spines may play a role in neuronal plasticity (Matus and Shepherd 2000). Furthermore, studies show that mGluR5 binds to microtubules (Sérge et al. 2003), mGluR1a interacts with β-tubulin (Ciruela et al. 1999) and mGluR7a interacts with α-tubulin (Saugstad et al. 2002). Similarly, the muscarinic receptor agonist, carbachol, induces a rapid and transient translocation of tubulin to the plasma membrane, microtubule reorganization and a change in cell shape (Popova 2000). Thus, microtubules play a role in G protein-coupled receptor targeting and organization during synapse formation, and G protein-coupled receptors modulate cytoskeletal dynamics, intracellular trafficking and cellular architecture; the detection of cytoskeletal proteins in the CB negative control sample may simply reflect the high abundance of these proteins in the samples.
In conclusion, we utilized a proteomic approach to identify novel proteins that may regulate mGluR5 responses by direct or indirect protein interactions. This approach does not rely on the heterologous expression of proteins and offers the advantage of identifying protein interactions in a native environment. Thus, mGluR5 was immunoprecipitated from rat brain lysates, and the co-immunoprecipitating proteins were originally removed en masse from the protein A sepharose beads and subjected to liquid chromatography and mass spectrometry. However, this method sometimes resulted in a sample complexity that made the mass spectrometry analysis difficult and thus, the sample preparation procedure was modified to include separation of the protein samples by 1D gel electrophoresis; this is the preferred method for future studies. The peptide sequences identified by mass spectrometry were then matched to protein databases to identify correlating parent proteins. This proteomic approach revealed the interaction of mGluR5 with known regulatory proteins, as well as novel interacting proteins that may reflect previously unidentified molecular constituents of the mGluR5-signaling complex. As immunoblot analysis and reverse co-immunoprecipitation confirmed the interaction between high confidence mGluR5-interacting proteins, such as PACS1 and MAP2, and Dynamin, this approach has successfully led to the identification of novel proteins that may be molecular constituents in the mGluR5-signaling complex. Interestingly, the reverse immunoblots showed that Dynamin1 and MAP2 interact with the dimer form of mGluR5, while PACS1 interacts with the monomer form of mGluR5. These data are intriguing and may represent protein interactions at distinct subcellular regions. For example, immunogold studies revealed that in rat substantia nigra, mGluR1a immunoreactivity was primarily detected at the plasma membrane, while > 80% of mGluR5 immunoreactivity was intracellular (Hubert et al. 2001). As a major role for PACS1 is to facilitate the trafficking of proteins between the plasma membrane and the cytosol, the interaction of monomer mGluR5 with PACS may reflect sequestering of mGluR5 in the intracellular compartment. Alternatively, while little is known about the functional role for mGluR dimers (see Romano et al. 2001), it is possible that the interaction of Dynamin1 and MAP2 with the dimer form of mGluR5 may reflect protein interactions at the plasma membrane, based loosely on evidence that the extracellular ligand binding domain of mGluR1 can form homodimers (Kunishima et al. 2000).
In addition, PPID searches support a link between mGluR5 and several proteins identified in this study, and several of these proteins are now the focus of new studies in our laboratory to determine their role in mGluR5-signaling in the CNS. These data support the use of a proteomic approach to identify novel protein interactions in native tissue, which will be particularly useful for neurobiology applications where distinct protein interactions can dramatically alter the outcome of the response to neurotransmitter release, or the disruption of normal protein interactions can lead to severe neurological and psychiatric disorders. For example, there is evidence that a genetic defect in DISC1 (disrupted in schizophrenia 1) leads to the disruption of a trimolecular protein complex that may in part lead to the developmental onset of schizophrenia (Brandon et al. 2004). In addition, studies show that a physical interaction between NMDA receptors and dopamine receptors leads to increased plasma membrane insertion of dopamine receptors (Pei et al. 2004). Thus, disruption of NMDA receptor-mediated up-regulation of dopamine receptor function may serve an as underlying molecular mechanism that leads to the onset of schizophrenia. New opportunities now exist for the study of complex protein interactions due to significant advancements in mass spectrometry and protein identification provided by large-scale nucleotide sequencing, and these approaches can be used to study the molecular constituents of signaling complexes and the alterations in protein interaction that may underlie neuropsychiatric or neuropathological disorders.
Acknowledgments
The authors gratefully acknowledge financial assistance from the Rockefeller Brothers Fund Charles E. Culpeper Biomedical Pilot Initiative (02–175; CDF), the National Institute of Mental Health (R01-521635; JAS) and the National Institute of Environmental Health Sciences (P30 ES00210; DFB). The authors thank Sufang Yang for technical assistance, and Jimmy Eng (Institute for Systems Biology) for assistance with using comet and the analysis of protein identification results.
Abbreviations used
- ACN
acetonitrile
- AMPA
alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- CB
Cascade Blue
- CTD
carboxy terminal domain
- ESI
electrospray ionization
- FA
formic acid
- GluRs
glutamate receptors
- GST
glutathione-S-transferase
- HRP
horseradish peroxidase
- IP3R
inositol triphosphate receptor
- LC
liquid chromatography
- MAP2
microtubule-associated protein 2
- mGluR5
metabotropic glutamate receptor 5
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NMDA
N-methyl-d-aspartate
- PACS1a
phosphofurin acidic cluster sorting protein 1
- PAS
protein A-sepharose
- PKC
protein kinase C
- PLC
phospholipase C
- PPID
Protein–Protein Interaction Database
- PSD
postsynaptic density
- PVDF
polyvinylidene difluoride
- Q-TOF
Quadrupole Time-of-Flight
- TBS
Tris-buffered saline
- TFA
trifluoroacetic acid
References
- Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem. 1992;267:13 361–13 368. [PubMed] [Google Scholar]
- Alagarsamy S, Rouse ST, Gereau RWT, Heinemann SF, Smith Y, Conn PJ. Activation of N-methyl-d-aspartate receptors reverses desensitization of metabotropic glutamate receptor, mGluR5, in native and recombinant systems. Ann NY Acad Sci. 1999;868:526–530. doi: 10.1111/j.1749-6632.1999.tb11321.x. [DOI] [PubMed] [Google Scholar]
- Ashman K, Moran MF, Sicheri F, Pawson T, Tyers M. Cell signalling – the proteomics of it all. Sci STKE. 2001;2001:E33. doi: 10.1126/stke.2001.103.pe33. [DOI] [PubMed] [Google Scholar]; Signal Transduction Knowledge Environment (STKE) is a resource on cell signaling provided by the journal Science. [30 August 2004]; www.stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/103/pe33.
- Bécamel C, Alonso G, Galéotti N, Demey E, Jouin P, Ullmer C, Dumuis A, Bockaert J, Marin P. Synaptic multiprotein complexes associated with 5-HT (2C) receptors: a proteomic approach. EMBO J. 2002;21:2332–2342. doi: 10.1093/emboj/21.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Ciruela F, Robbins MJ, Willis AC, McIlhinney RA. Interactions of the C terminus of metabotropic glutamate receptor type 1alpha with rat brain proteins: evidence for a direct interaction with tubulin. J Neurochem. 1999;72:346–354. [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate recepors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Croci C, Sticht H, Brandstatter JH, Enz R. Group I metabotropic glutamate receptors bind to protein phosphatase 1C. Mapping and modeling of interacting sequences. J Biol Chem. 2003;278:50 682–50 690. doi: 10.1074/jbc.M305764200. [DOI] [PubMed] [Google Scholar]
- Croucher MJ, Thomas LS, Ahmadi H, Lawrence V, Harris JR. Endogenous sulphur-containing amino acids: potent agonists at presynaptic metabotropic glutamate autoreceptors in the rat central nervous system. Br J Pharmacol. 2001;133:815–824. doi: 10.1038/sj.bjp.0704138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Xiang Y, Thomas L, Gu F, Austin C, Tooze SA, Thomas G. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 2001;20:2191–2201. doi: 10.1093/emboj/20.9.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. Actin and microtubules in neurite initiation: are MAPs the missing link? J Neurobiol. 2004;58:18–33. doi: 10.1002/neu.10284. [DOI] [PubMed] [Google Scholar]
- tom Dieck S, Sanmartí-Vila L, Langnaese K, et al. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Synapse structure: glutamate receptors connected by the shanks. Curr Biol. 1999;9:R848–R850. doi: 10.1016/s0960-9822(00)80043-3. [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Faas GC, Adwanikar H, Gereau RWT, Saggau P. Modulation of presynaptic calcium transients by metabotropic glutamate receptor activation: a differential role in acute depression of synaptic transmission and long-term depression. J Neurosci. 2002;22:6885–6890. doi: 10.1523/JNEUROSCI.22-16-06885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Bessis AS, Rossignol F, Pin JP, Olivo-Marin JC, Hemar A. The metabotropic glutamate receptor mGluR5 is endocytosed by a clathrin-independent pathway. J Biol Chem. 2003;278:12 222–12 230. doi: 10.1074/jbc.M205663200. [DOI] [PubMed] [Google Scholar]
- Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., 3rd Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- Gereau RWT, Conn PJ. Roles of specific metabotropic glutamate receptor subtypes in regulation of hippocampal CA1 pyramidal cell excitability. J Neurophysiol. 1995;74:122–129. doi: 10.1152/jn.1995.74.1.122. [DOI] [PubMed] [Google Scholar]
- Gereau RWT, Heinemann SF. Role of protein kinase C phosphorylation in rapid desensitization of metabotropic glutamate receptor 5. Neuron. 1998;20:143–151. doi: 10.1016/s0896-6273(00)80442-0. [DOI] [PubMed] [Google Scholar]
- Gray NW, Fourgeaud L, Huang B, Chen J, Cao H, Oswald BJ, Hemar A, McNiven MA. Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and Homer. Curr Biol. 2003;13:510–515. doi: 10.1016/s0960-9822(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Hermans E, Challiss RA. Structural, signalling and regulatory properties of the group I metabotropic glutamate receptors: prototypic family C G-protein-coupled receptors. Biochem J. 2001;359:465–484. doi: 10.1042/0264-6021:3590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuss C, Scanziani M, Gahwiler BH, Gerber U. G-protein-independent signaling mediated by metabotropic glutamate receptors. Nat Neurosci. 1999;2:1070–1077. doi: 10.1038/15996. [DOI] [PubMed] [Google Scholar]
- van Hooft JA, Giuffrida R, Blatow M, Monyer H. Differential expression of group I metabotropic glutamate receptors in functionally distinct hippocampal interneurons. J Neurosci. 2000;20:3544–3551. doi: 10.1523/JNEUROSCI.20-10-03544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert GW, Paquet M, Smith Y. Differential subcellular localization of mGluR1a and mGluR5 in the rat and monkey Substantia nigra. J Neurosci. 2001;21:1838–1847. doi: 10.1523/JNEUROSCI.21-06-01838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Nash SR, Nishimune A, Neki A, Kaneko S, Nakanishi S. Competitive interaction of seven in absentia homolog-1A and Ca2+/calmodulin with the cytoplasmic tail of group 1 metabotropic glutamate receptors. Genes Cells. 1999;4:381–390. doi: 10.1046/j.1365-2443.1999.00269.x. [DOI] [PubMed] [Google Scholar]
- Kaech S, Fischer M, Doll T, Matus A. Isoform specificity in the relationship of actin to dendritic spines. J Neurosci. 1997;17:9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR. Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 2000;20:7238–7245. doi: 10.1523/JNEUROSCI.20-19-07238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Jun KS, Lee SB, Kang NG, Min DS, Kim YH, Ryu SH, Suh PG, Shin HS. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature. 1997;389:290–293. doi: 10.1038/38508. [DOI] [PubMed] [Google Scholar]
- Kim M, Jiang LH, Wilson HL, North RA, Surprenant A. Proteomic and functional evidence for a P2X7 receptor signalling complex. EMBO J. 2001;20:6347–6358. doi: 10.1093/emboj/20.22.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Lin HC, Gilman AG. Regulation of dynamin I GTPase activity by G protein betagamma subunits and phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1996;271:27 979–27 982. doi: 10.1074/jbc.271.45.27979. [DOI] [PubMed] [Google Scholar]
- Lin HC, Duncan JA, Kozasa T, Gilman AG. Sequestration of the G protein beta gamma subunit complex inhibits receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1998;95:5057–5060. doi: 10.1073/pnas.95.9.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis Albasanz J, Fernandez M, Martin M. Internalization of metabotropic glutamate receptor in C6 cells through clathrin-coated vesicles. Brain Res Mol Brain Res. 2002;99:54–66. doi: 10.1016/s0169-328x(02)00103-1. [DOI] [PubMed] [Google Scholar]
- Lujan R, Roberts JD, Shigemoto R, Ohishi H, Somogyi P. Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J Chem Neuroanat. 1997;13:219–241. doi: 10.1016/s0891-0618(97)00051-3. [DOI] [PubMed] [Google Scholar]
- Matus A, Shepherd GM. The millennium of the dendrite? Neuron. 2000;27:431–434. doi: 10.1016/s0896-6273(00)00054-4. [DOI] [PubMed] [Google Scholar]
- McCool BA, Pin JP, Harpold MM, Brust PF, Stauderman KA, Lovinger DM. Rat group I metabotropic glutamate receptors inhibit neuronal Ca2+ channels via multiple signal transduction pathways in HEK 293 cells. J Neurophysiol. 1998;79:379–391. doi: 10.1152/jn.1998.79.1.379. [DOI] [PubMed] [Google Scholar]
- Minakami R, Jinnai N, Sugiyama H. Phosphorylation and calmodulin binding of the metabotropic glutamate receptor subtype 5 (mGluR5) are antagonistic in vitro. J Biol Chem. 1997;272:20 291–20 298. doi: 10.1074/jbc.272.32.20291. [DOI] [PubMed] [Google Scholar]
- Molloy SS, Thomas L, Kamibayashi C, Mumby MC, Thomas G. Regulation of endosome sorting by a specific PP2A isoform. J Cell Biol. 1998;142:1399–1411. doi: 10.1083/jcb.142.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundell SJ, Matharu AL, Pula G, Roberts PJ, Kelly E. Agonist-induced internalization of the metabotropic glutamate receptor 1a is arrestin- and dynamin-dependent. J Neurochem. 2001;78:546–551. doi: 10.1046/j.1471-4159.2001.00421.x. [DOI] [PubMed] [Google Scholar]
- O'Malley KL, Jong YJI, Gonchar Y, Burkhalter A, Romano C. Activation of metabotropic glutamate receptor mGlu5 on nuclear membranes mediates intranuclear Ca2+ changes in heterologous cell types and neurons. J Biol Chem. 2003;278:28 210–28 219. doi: 10.1074/jbc.M300792200. [DOI] [PubMed] [Google Scholar]
- Okamoto PM, Herskovits JS, Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11 629–11 635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- Okamoto PM, Gamby C, Wells D, Fallon J, Vallee RB. Dynamin isoform-specific interaction with the shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton. J Biol Chem. 2001;276:48 458–48 465. doi: 10.1074/jbc.M104927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Lee FJ, Moszczynska A, Vukusic B, Liu F. Regulation of dopamine D1 receptor function by physical interaction with the NMDA receptors. J Neurosci. 2004;24:1149–1158. doi: 10.1523/JNEUROSCI.3922-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Giampietro DE. The distinct role of mGlu1 receptors in post-ischemic neuronal death. Trends Pharmacol Sci. 2003;24:461–470. doi: 10.1016/S0165-6147(03)00231-1. [DOI] [PubMed] [Google Scholar]
- Pin JP, Joly C, Heinemann SF, Bockaert J. Domains involved in the specificity of G protein activation in phospholipase C-coupled metabotropic glutamate receptors. EMBO J. 1994;13:342–348. doi: 10.1002/j.1460-2075.1994.tb06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN, Romano C, Ghosh P. Metabotropic glutamate receptor mGluR5 subcellular distribution and developmental expression in hypothalamus. J Comp Neurol. 1995;362:134–150. doi: 10.1002/cne.903620108. [DOI] [PubMed] [Google Scholar]
- Popova JS, Rasenick MM. Muscarinic receptor activation promotes the membrane association of tubulin for the regulation of Gq-mediated phospholipase C1 signaling. J Neurosci. 2000;20:2774–2782. doi: 10.1523/JNEUROSCI.20-08-02774.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan EM, Halpain S. Postsynaptic mechanisms for bidirectional control of MAP2 phosphorylation by glutamate receptors. Neuron. 1996;16:357–368. doi: 10.1016/s0896-6273(00)80053-7. [DOI] [PubMed] [Google Scholar]
- Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF. Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem. 1999;274:25 953–25 957. doi: 10.1074/jbc.274.36.25953. [DOI] [PubMed] [Google Scholar]
- Romano C, Sesma MA, McDonald CT, O'Malley K, Van den Pol AN, Olney JW. Distribution of metabotropic glutamate receptor mGluR5 immunoreactivity in rat brain. J Comp Neurol. 1995;355:455–469. doi: 10.1002/cne.903550310. [DOI] [PubMed] [Google Scholar]
- Romano C, Miller JK, Hyrc K, Dikranian S, Mennerick S, Takeuchi Y, Goldberg MP, O'Malley KL. Covalent and noncovalent interactions mediate metabotropic glutamate receptor mGlu5 dimerization. Mol Pharmacol. 2001;59:46–53. [PubMed] [Google Scholar]
- Saugstad JA, Yang S, Pohl J, Hall RA, Conn PJ. Interaction between metabotropic glutamate receptor 7 and alpha tubulin. J Neurochem. 2002;80:980–988. doi: 10.1046/j.0022-3042.2002.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GK, Gu F, Crump CM, Thomas L, Wan L, Xiang Y, Thomas G. The phosphorylation state of an autoregulatory domain controls PACS-1Directed protein traffic. EMBO J. 2003;22:6234–6244. doi: 10.1093/emboj/cdg596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf K, Kostka G, Lammers R, Bashkin P, Daly R, Burgess WH, van der Bliek AM, Schlessinger J, Ullrich A. Dynamin binds to SH3 domains of phospholipase C gamma and GRB-2. J Biol Chem. 1994;269:16 009–16 014. [PubMed] [Google Scholar]
- Sérge A, Fourgeaud L, Hémar A, Choquet D. Active surface transport of metabotropic glutamate receptors through binding to microtubules and actin flow. J Cell Sci. 2003;116:5015–5022. doi: 10.1242/jcs.00822. [DOI] [PubMed] [Google Scholar]
- Sever S. Dynamin and endocytosis. Curr Opin Cell Biol. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shigemoto R, Kinoshita A, Wada E, et al. Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J Neurosci. 1997;17:7503–7522. doi: 10.1523/JNEUROSCI.17-19-07503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Nagayama T, Jin KL, Zhu L, Loeffert JE, Watkins SC, Graham SH, Simon RP. bcl-2 Antisense treatment prevents induction of tolerance to focal ischemia in the rat brain. J Cereb Blood Flow Metab. 2001;21:233–243. doi: 10.1097/00004647-200103000-00007. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Mizutani A, Bito H, Fujisawa K, Narumiya S, Mikoshiba K, Furuichi T. Cupidin, an isoform of Homer/Vesl, interacts with the actin cytoskeleton and activated rho family small GTPases and is expressed in developing mouse cerebellar granule cells. J Neurosci. 1999;19:8389–8400. doi: 10.1523/JNEUROSCI.19-19-08389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev VI, Ochoa GC, Butler MH, Grabs D, Camilli PD. Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science. 1998;281:821–824. doi: 10.1126/science.281.5378.821. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Valenti O, Conn PJ, Marino MJ. Distinct physiological roles of the Gq-coupled metabotropic glutamate receptors co-expressed in the same neuronal populations. J Cell Physiol. 2002;191:125–137. doi: 10.1002/jcp.10081. [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mattson MP, Furukawa K. Endoplasmic reticulum calcium release is modulated by actin polymerization. J Neurochem. 2002;82:945–952. doi: 10.1046/j.1471-4159.2002.01059.x. [DOI] [PubMed] [Google Scholar]
- Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis. 1995;16:1090–1094. doi: 10.1002/elps.11501601185. [DOI] [PubMed] [Google Scholar]
- Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]


