Abstract
Development of the amygdala, a central structure of the limbic system, remains poorly understood. Using mouse as a model, our studies reveal that two spatially distinct and early specified telencephalic progenitor pools marked by the homeodomain transcription factor Dbx1 are major sources of neuronal cell diversity in the mature amygdala. We find that Dbx1+ cells of the ventral pallium (VP) generate excitatory neurons of the basolateral complex and cortical amygdala nuclei. Moreover, Dbx1-derived cells comprise a novel migratory stream that emanates from the preoptic area (POA), a ventral telencephalic domain adjacent to the diencephalic border. The Dbx1+ POA-derived population migrates specifically to the amygdala, and as defined by both immunochemical and electrophysiological criteria, generates a unique subclass of inhibitory neurons in the medial amygdala nucleus. Thus, this POA-derived population represents a novel progenitor pool dedicated to the limbic system.
The broad function of the amygdala is to integrate and process information from external stimuli and coordinate appropriate behavioral outputs1–3. These complex functions are differentially regulated by the 11–15 distinct functional subnuclei of the amygdala, which make numerous connections with multiple cortical and subcortical brain regions. These subnuclei can also be grouped together according to a variety of criteria including connectivity, function and neuronal diversity. The most studied of these nuclear groups is the basolateral complex, which comprises the lateral, basolateral and basomedial nuclei. The prime function of the basolateral complex is the processing and storage of information with emotional salience, especially fear. In contrast, the function of the medial nucleus, via connections with the olfactory bulb and hypothalamus, is to integrate chemosensory and hormonal signals to control social, reproductive, feeding and defensive behaviors. The importance of the amygdala in human behavior is evidenced by the fact that amygdala dysfunction is a key component of numerous prevalent human disorders such as autism and autism spectrum disorders as well as stress related psychological disorders such as post - traumatic stress disorder (PTSD)4–6. Thus, unraveling the development of this complex structure is an important goal with potential translational applications.
The unique connectivity and complex nuclear organization of the amygdala sets it apart from all other telencephalic structures, most notably from laminar brain structures such as the cerebral cortex and hippocampus. In addition to classification according to functional criteria, individual amygdala nuclei have also been grouped based on whether their principal output neurons are excitatory (glutamatergic) or inhibitory (GABAergic)1–3. Major excitatory nuclei include the basolateral complex and cortical nuclei and major inhibitory nuclei include the central and medial nuclei. Although differing in their neuronal composition, both excitatory and inhibitory nuclei are also comprised of a variety of functionally diverse interneuron subtypes that are essential for the proper regulation of the firing of the output neurons. The coordinated development of these neuronal cell types during embryogenesis is essential for the formation of complex amygdala circuitry. However, despite its central role in normal and abnormal brain function and behavior, little is currently known regarding how neuronal cell diversity is generated in the amygdala.
In this study we examined the contribution of progenitor cells to excitatory and inhibitory cell diversity in the mature amygdala. Previous work from our laboratory, as well as others, has indicated that the pallial-subpallial boundary (PSB) of the telencephalon, the region of the telencephalon where pallial (e.g. high Pax6, Ngn2) and subpallial (e.g. Gsh2, Dlx1/2) gene expression abuts, is a major source of amygdala neural progenitors7–10. In addition to expressing regional markers, the pallial and subpallial aspects of the PSB express a unique combination of genes that includes the homeodomain transcription factor, Dbx1, which marks the ventral pallial (VP) progenitors of the PSB9,11–13. In addition to the PSB, Dbx1 is expressed in other progenitor regions of the developing telencephalon such as the preoptic area (POA) and septum. While a previous study has revealed that Dbx1-derived progenitors, specifically from the PSB and septum, contribute early born Cajal-Retzius cells to the piriform and cerebral cortices11, whether this progenitor population contributes to amygdala cell diversity remains unexplored.
Using a combination of mouse genetic fate mapping, in vitro migratory assays and electrophysiological approaches, we find that the VP Dbx1+ progenitor pool is a source of excitatory neurons in the basolateral complex and cortical nuclei, the primary excitatory output nuclei of the amygdala. In addition, we uncover a novel migratory route, termed the PAS (POA-amygdala migratory stream), composed of Dbx1-derived cells from the POA to the emerging amygdala. Our electrophysiological analysis indicates that this migratory population gives rise to a remarkably restricted functional subclass of inhibitory neurons specifically in the mature medial amygdala nucleus. These inhibitory neurons are also characterized by their unique morphology and expression of inhibitory neuronal markers, such as neuronal nitric oxide synthase (nNOS), which together with their electrophysiological characteristics, distinguish them from other known telencephalic inhibitory neuron subclasses. Thus, our data reveal that spatially distinct telencephalic Dbx1+ progenitor pools are major sources of neuronal diversity for distinct nuclei of the mature amygdala, and therefore reveal a novel relationship between genetically marked progenitor pools and their limbic system fate.
RESULTS
Expression of Dbx1 and generation of CreERT2 knock-in mice
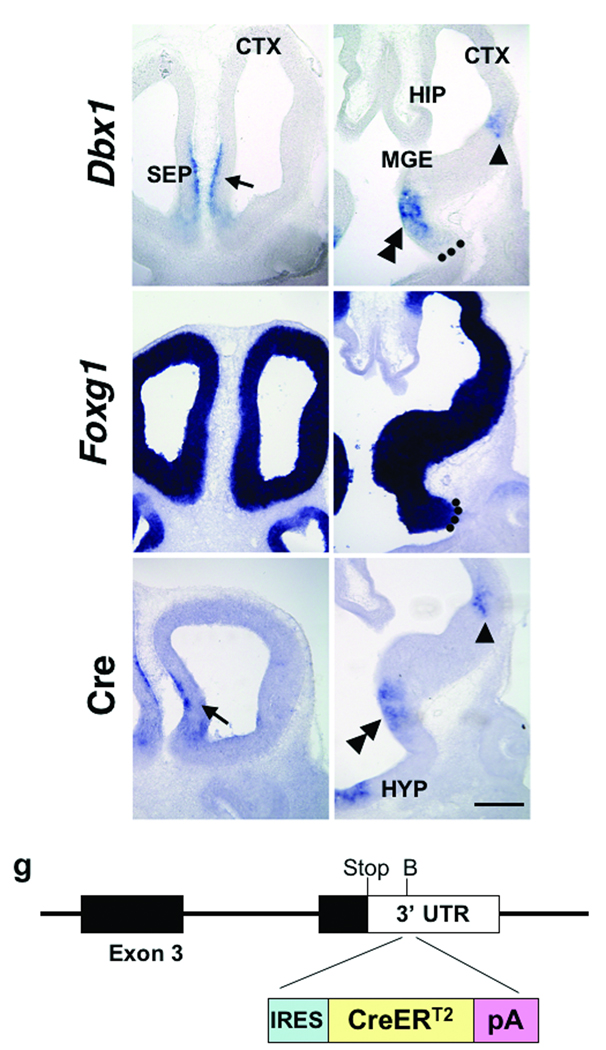
Previous studies have revealed that the pallial-subpallial boundary (PSB) is comprised of progenitors from the ventral pallium (VP) and dorsal LGE (dLGE) and is also the source of lateral cortical stream (LCS) migratory route to the amygdala7,8,13. Expression of the homeodomain transcription factor, Dbx1, marks VP progenitors as well as the developing septum and preoptic area (POA) from embryonic day (E) 9.5 to E14.5 (Fig. 1a,b and data not shown)9,12. The Dbx1 expressing region in the POA is also positive for Foxg114, indicating that this population is of telencephalic rather than the diencephalic origin (Fig. 1c,d).
Figure 1. Expression of Dbx1 and knockin approach.
Expression of Dbx1 (a,b) and Foxg1 (c,d) in wild-type embryos, and Cre (e,f) in Dbx1+/CreERT2 knock-in embryos, at E11.5 is shown at rostral and caudal levels in coronal sections. Dbx1 is expressed in the septum (a, arrow), the ventral pallium (b, arrowhead) and the preoptic area (b, double arrowhead). Foxg1 expression delineates the telencephalon from the diencephalon (c, d). Dotted lines in b,d indicate the telencephalic-diencephalic border. Cre expression recapitulates Dbx1 expression in the septum (e, arrow), the ventral pallium (f, arrowhead) and the preoptic area (f, double arrowhead). Schematic of the knockin approach in which an IRES-CreERT2 cassette was inserted into the 3’ UTR of the 4th exon of Dbx1 is shown in (g). Abbr: CGE; caudal ganglionic eminence, CTX; cerebral cortex, HIP; hippocampus, HYP; hypothalamus, MGE; medial ganglionic eminence SEP; septum (Note: Embryonic anatomy is based on37). Scale bar; 250 µm
To examine the fate of Dbx1+ cells, we used a genetic fate mapping approach in which an IRES-CreERT2 cassette was inserted into the 3’ UTR region of Dbx1 gene (Fig. 1g, see material and methods). This strategy allows for the indelible marking of genetically defined progenitor populations from embryogenesis to adulthood, and thus is a powerful tool to track the fate of specific progenitor populations15,16. Our analysis of Dbx1+/CreERT2 embryos reveals that cre is expressed in the same pattern as Dbx1 (Fig. 1e,f and data not shown) and insertion of an IRES-CreERT2 cassette into the 3’ UTR of the Dbx1 locus also importantly does not appear to interfere with normal Dbx1 expression (Supplementary Fig. 1)11.
Dbx1-derived cells contribute to the post-natal amygdala
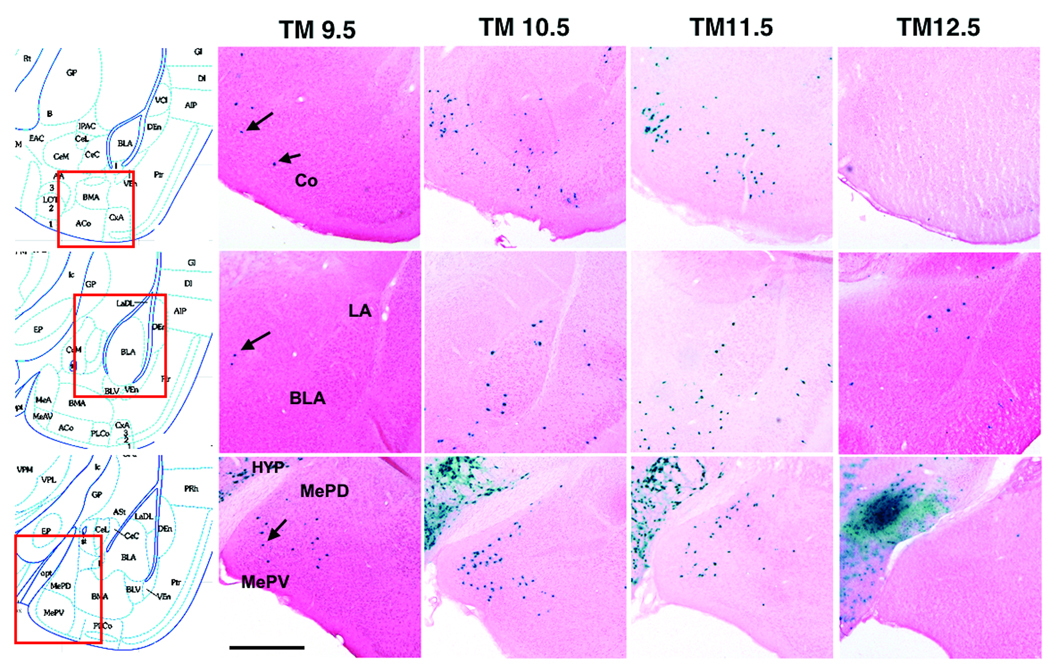
To examine the fate of Dbx1-derived cells in the amygdala, Dbx1+/CreERT2 mice were crossed to R26RLacZ reporter animals that permanently express β-galactosidase upon Cre-induced recombination17. The fusion of Cre to the tamoxifen-sensitive estrogen receptor (ERT2) allows Cre translocation to the nucleus to be controlled by the administration of tamoxifen, with recombination typically occurring 6–12 hours after tamoxifen delivery.18 Pregnant dams were administered tamoxifen at E9.5, E10.5, E11.5, or E12.5 and the distribution of β-galactosidase-labeled cells was examined at post-natal day (P) 21 in both the telencephalon and diencephalon (Fig. 2 and Supplementary Fig. 2). Tamoxifen administration at E9.5 results in recombined cells mainly in the medial amygdala nucleus, with a few recombined cells observed in the cortical nucleus and none in the basolateral complex (lateral, basolateral and basomedial nuclei) (Fig. 2b,g,l). However, tamoxifen administration one day later at E10.5 (Fig. 2c,h,m) or two days later at E11.5 (Fig. 2d,i,n), results in recombined cells in the basolateral complex, cortical and medial nuclei. In contrast, tamoxifen delivery at E12.5 results in recombined cells primarily in the basolateral complex with only a few recombined cells detected in the medial and cortical nuclei (Fig. 2e,j,o) These data indicate that Dbx1+ progenitors generate cells destined for the amygdala in two waves; medial and cortical nuclei cells first (starting at approximately E10.0), then, approximately one day later, cells of the basolateral complex.
Figure 2. Dbx1-derived cells in the post natal amygdala.
β-galactosidase staining of coronal sections from Dbx1+/CreERT2;R26RLacZ brains at P21 reveals numerous recombined LacZ+ cells in the amygdala. Distribution of recombined cells from tamoxifen (TM) administration at E9.5 (b,g,l), E10.5 (c,h,m), E11.5 (d,i n) or E12.5 (e, j o) are shown. Boxed area on schematics (a, f, k) indicate the region of the amygdala shown in each panel. E9.5 TM delivery results in recombined cells in the cortical and medial nuclei (b,g,l, arrowheads), but not the lateral and basolateral nuclei. In addition to the cortical and medial nuclei, TM delivery at E10.5 (c,h,m) or E11.5 (d,i,n) results in recombined cells in the lateral and basolateral nuclei. TM delivery at E12.5 results in recombined cells in the lateral and basolateral nuclei only (e,j,o). Abbr: BLA; basolateral nucleus, Co; cortical nucleus, HYP; hypothalamus, LA; lateral nucleus, MePD; medial nucleus posterior dorsal, MePV; medial nucleus posterior ventral. Scale bar; 500 µm
Origins of amygdala Dbx1-derived cells
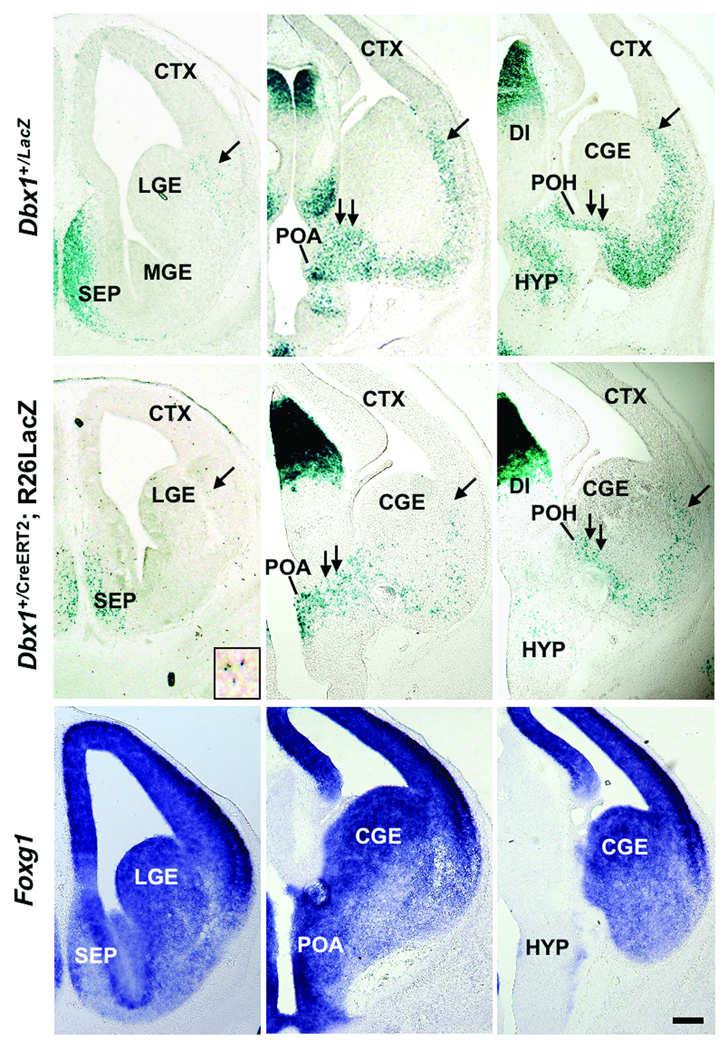
As Dbx1 is expressed in multiple domains in the embryonic telencephalon we examined the origins of Dbx1-derived cells by analyzing patterns of putative migratory pathways in two sets of transgenic mice in which Dbx1+ progenitors and their progeny are marked: previously generated Dbx1+/LacZ mice in which LacZ was knocked into the Dbx1 locus19 and the above described Dbx1+/CreERT2;R26RLacZ mice (Fig. 3). Analysis of Dbx1+/LacZ embryos at E13.5 reveals cohorts of LacZ+ cells along the LCS migratory route from the VP to the basal telencephalon (Fig. 3a,b). In addition, we detect a large novel putative migratory route between the POA and the developing amygdala, which we term the “PAS” (POA-amygdala migratory stream) (Fig. 3b,c).
Figure 3. Dbx1-derived cells in the developing basal telencephalon.
β-galactosidase staining of coronal sections of E13.5 Dbx1+/LacZ knock-in (a-c) and Dbx1+/CreERT2;R26RLacZ embryos (d-f) reveals putative migrating cells to the region of the developing amygdala. LacZ+ cells are shown along the both LCS (a-f, single arrow, inset in d shows a higher magnification of LacZ+ cells of the LCS) and the putative POA-amygdala migratory stream (PAS) (b,c,e,f, double arrows). Foxg1 expression in corresponding sections in Dbx1+/LacZ knock-in embryos (g-i) indicates that Dbx1-derived cells distributed along PAS are located within the telencephalon. Note the relative paucity of LacZ+ cells in the ganglionic eminences and the developing cerebral cortex. Abbr: LGE; lateral ganglionic eminence, POA; preoptic area, POH; preopto-hypothalamic region, TH; thalamus. Scale bar; 250 µm
Similar patterns of putatively migrating LacZ+ recombined cells are also observed in Dbx1+/CreERT2;R26RLacZ embryos (Fig. 3d–f). As revealed by Foxg1 expression, the PAS appears to originate within the telencephalon as opposed to the diencephalon (Fig. 3g–i). Most strikingly, the overwhelming majority of Dbx1+ and Dbx1-derived cells accumulate in the region of the developing amygdala. Therefore, based on these patterns of LacZ expression in two sets of mice that mark Dbx1-expressing cells and their progeny it appears that two spatially separated Dbx1+ progenitor regions, the VP and POA, are the likely primary sources of the post-natal Dbx1-derived amygdala cells shown in Fig. 2.
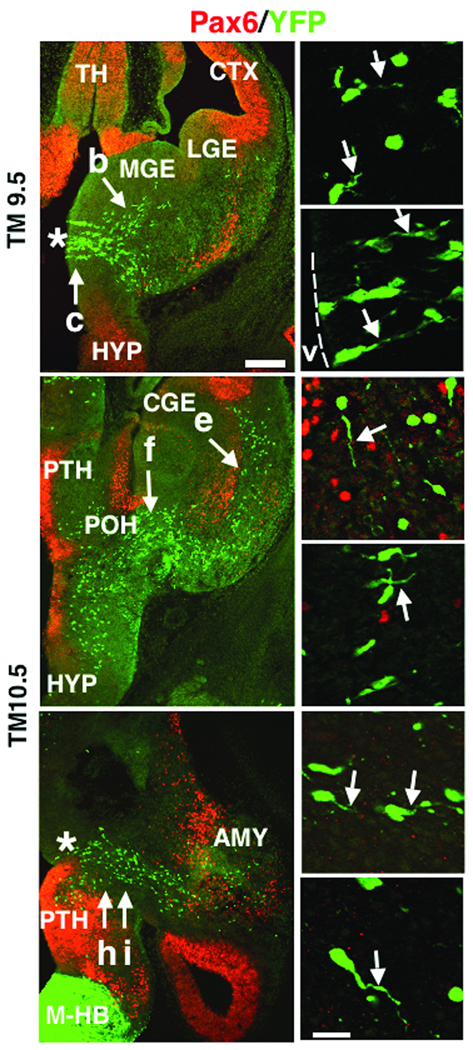
To more closely examine the patterns of putative migration from these sources, Dbx1+/CreERT2 mice were also crossed to R26RYFP reporter mice, which express yellow fluorescent protein (YFP) in the presence of Cre-recombinase20 (Fig. 4). In contrast to β-galactosidase, YFP is typically expressed in the entire cell, which allows for examination of migratory profiles. Tamoxifen treatment at E9.5 results in recombined cells emanating from the POA, with leading processes primarily oriented toward the basal telencephalon (Fig. 4a–c). Interestingly, at this age of tamoxifen delivery only a few YFP+ cells can be detected along the LCS. In contrast, E10.5 tamoxifen treatment results in numerous YFP+ cells along the LCS, as well as a continued putative migration from the region of the POA (Fig. 4d–i). Surprisingly, many of the Dbx1-derived cells of the LCS do not express Pax6, a pallial marker typically expressed in many cells of the LCS8. The leading processes of many of POA-derived and LCS recombined cells were oriented toward the basal telencephalon suggestive of active migration to the developing amygdala. Notably, the orientation of the leading processes of the Dbx1-derived POA population appeared different from that of Dbx1-derived cells along the LCS, indicating that these are two separate migratory populations (Fig. 4d,e).
Figure 4. Distribution of YFP+ recombined cells at embryonic stages.
Tamoxifen was administered to Dbx1+/CreERT2;R26RYFP embryos at E9.5 (a-c) or E10.5 (d-i) and analyzed at E12.5 as shown. E9.5 TM administration results in YFP+ recombined cells (green) primarily in the POA (asterisk), with only a few YFP+ recombined cells observed along the LCS as shown in a coronal section at low power magnification (a). Higher power magnification of areas highlighted by arrows in (a) shows leading processes of recombined cells (arrows,b, c). Numerous recombined cells are observed in the VZ of the POA (c, dotted line marks the edge of the tissue and the location of the lateral ventricle “v”). As shown in a low power coronal section (d), E10.5 TM administration (d, g) results in recombined cells along both the PAS migratory stream (arrow, f) and LCS (arrow, e). Higher power magnification shows Pax6 (red) negative, recombined cells along the LCS with migratory profiles directed toward the basal telencephalon (arrow, e). Recombined PAS cells also display migratory profiles directed toward the developing amygdala (arrow, f). The PAS migratory stream is most evident in horizontal sections (g, asterisk marks the POA), with higher power magnification of YFP+ cells also revealing recombined cells with leading processes oriented toward the developing amygdala (arrows, h, i). Abbr: AMY; developing amygdala, M-HB; midbrain-hindbrain region, PTH; prethalamus. Scale bar in (a) for (a, b, g); 250 µm. Scale bar in (i) for (b, c, e, f, h, i); 20 µm
Thus, our analysis of patterns of LacZ and YFP expression in a series of mice in which Dbx1+ cells and their progeny are marked (Fig. 3 and Fig. 4) indicate that the post-natal Dbx1-derived cells in the amygdala (Fig. 2) primarily originate in the VP and POA. These analyses also reveal that Dbx1-derived cells of the POA are generated earlier in development than the Dbx1-derived cells of the VP. This also correlates with the one-day difference in the timing of the generation of Dbx1-derived medial nuclei cells versus Dbx1-derived basolateral complex cells as shown in Fig. 2.
POA-derived cells are highly migratory
While the above studies suggest that Dbx1-derived cells of the POA migrate to the developing amygdala, we wanted to directly examine and compare their migratory capacity to that of the PSB and septum, the two other telencephalic domains of Dbx1+ expression (Fig. 1). First, the POA, PSB and septum were dissected from E12.5 embryos and cultured for three days in vitro (DIV) in matrigel, a permissive substrate for migration (Supplementary Fig. 3). The majority (17/21) of POA explants exhibit moderate to robust migration (Supplementary Fig 3a,b). This is in contrast to explants from the septum, from which either no or minimal migration is observed (Supplementary Fig. 3c). Consistent with our previous studies8, most (14/16) PSB explants also display robust migration (Supplementary Fig. 3d).
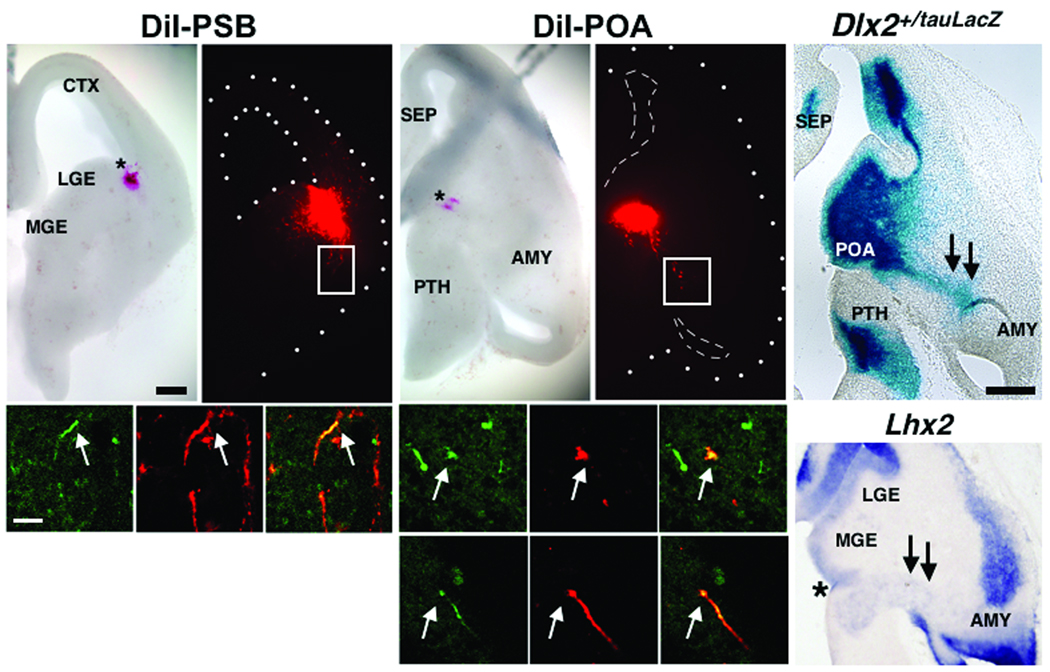
Second, we also carried out a series of DiI experiments in telencephalic slices from Dbx1+/CreERT2;R26RYFP embryos at E13.5 (Fig. 5). After two days in vitro, numerous DiI+ cells from the POA and PSB migrate toward the region of the developing amygdala (Fig. 5a,b,f,g). Many DiI+ cells from both the PSB and POA also express YFP (Fig. 5c–e, h–m), revealing that these migratory streams were comprised, at least in part, of Dbx1-derived cells. Interestingly, the PAS is also marked by expression of Dlx2+/tauLacZ and Lhx2, suggesting that this novel migratory route is comprised of a heterogeneously marked population of cells (Fig. 5n–o). Although their mode of migration remains to be fully elucidated, Dbx1-derived cells of the POA also intermingle with RC2+ radial glia, suggesting that these cells may migrate along radial glia (Supplementary Fig. 4).
Figure 5. Migration from the PSB and POA.
DiI crystals (red) were placed in either the PSB in coronal slices (a, b, n=2) or in the POA (f, g, n=4) in horizontal slices at E13.5 in Dbx1+/CreERT2;R26RYFP embryos treated with tamoxifen at E10.5. After two days in vitro (DIV), DiI labeled cells (red) from both the PSB and the POA migrate to the basal telencephalon (b, g). Boxed regions show that migratory streams from both the PSB (arrows, c-e) and POA (arrows, h-j) comprise Dbx1-derived cells as revealed by co-expression of DiI (red) and YFP (green). DiI and YFP co-expressing cells are also shown from another brain slice (arrows, k-m). β-galactosidase stained horizontal sections from E12.5 Dlx2+/tauLacZ embryos (n) and Lhx2 mRNA expression (o) also mark the PAS migratory route (double arrows, asterisk marks the POA in (o)). Scale bar in (a) for (a, b, f, g); 250 µm. Scale bar in (c) for (c-e, h-m); 50 µm. Scale bar in (n) for (n, o); 250 µm.
Fate of Dbx1-derived cells in the post-natal amygdala
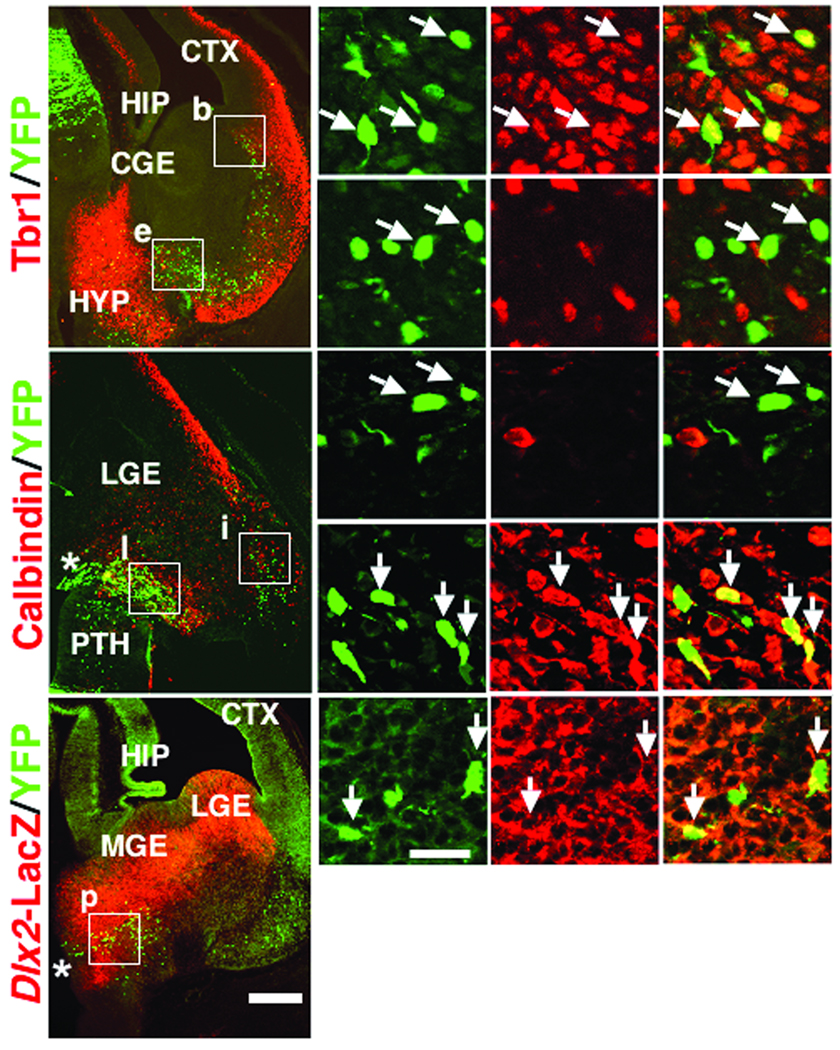
Based on their location in the pallium and subpallium respectively, we predicted that VP Dbx1-derived cells give rise to amygdala excitatory neurons and the POA Dbx1-derived cells give rise to amygdala inhibitory neurons. To test this, we examined if Dbx1-derived cells from these locations express regional neuronal subtype markers. In these experiments, sections from E12.5 Dbx1+/CreERT2;R26RYFP embryos in which tamoxifen was administered at E10.5 were double immunolabeled with a variety of well-characterized cell specific markers including Tbr1, a marker of developing and mature telencephalic excitatory neurons21, and calbindin and Dlx2, broad markers of developing inhibitory neurons22,23 (Fig. 6). Consistent with their pallial origin, numerous Dbx1-derived cells of the LCS express Tbr1 (90.2%), but do not express calbindin (Fig. 6a–n). In contrast, Dbx1-derived cells of the PAS do not express Tbr1 (Fig. 6a–g), but instead many express the subpallial markers calbindin (41.4%) or Dlx2 (80.8%) (Fig. 6h–r). Thus, it appears that Dbx1-derived cells of the VP generate amygdala excitatory neurons and Dbx1-derived cells of the PAS differentiate into inhibitory neurons.
Figure 6. Dbx1-derived cells express regional cell fate markers during embryogenesis.
The expression of Tbr1 (a-g) and caldindin (h-n) in Dbx1+/CreERT2;R26RYFP embryos, and Dlx2 (o-r) in Dbx1+/CreERT2;R26RYFP; Dlx2tauLacZembryos at E12.5 in which TM was administered at E10.5 is shown. As shown in a coronal section (a), expression of Tbr1 (red) is detected in 90.2% (46/51) of YFP+ recombined cells (green) along the LCS (boxed region “b” and arrows in b-d show double labeled cells). In contrast, Tbr1 is not detected in any (0/33) YFP+ recombined cells along the POA-amygdala migratory stream (PAS) (boxed region “e” and arrows in e-g show lack of double labeling). As shown in a horizontal section (h), no (0/88) YFP+ recombined cells (green) of the LCS co-express calbindin (red) (boxed region “i” and arrows in i-k show lack of double labeling). However, 41.4% (46/111) of YFP+ recombined cells of the PAS (green) co-express calbindin (red) (boxed region “i” and arrows in l-n show double labeled cells). As shown in a coronal section (o), 80.8% (21/26) of YFP+ recombined cells of the PAS (green) also co-express Dlx2 (red) (boxed region “p” and arrows in p-r show double labeled cells). Asterisks in h and o mark the POA. Scale bar in (o) for (a, h, o); 250 µm. Scale bar in (p) for (b-g, i-n, p-r); 20 µm
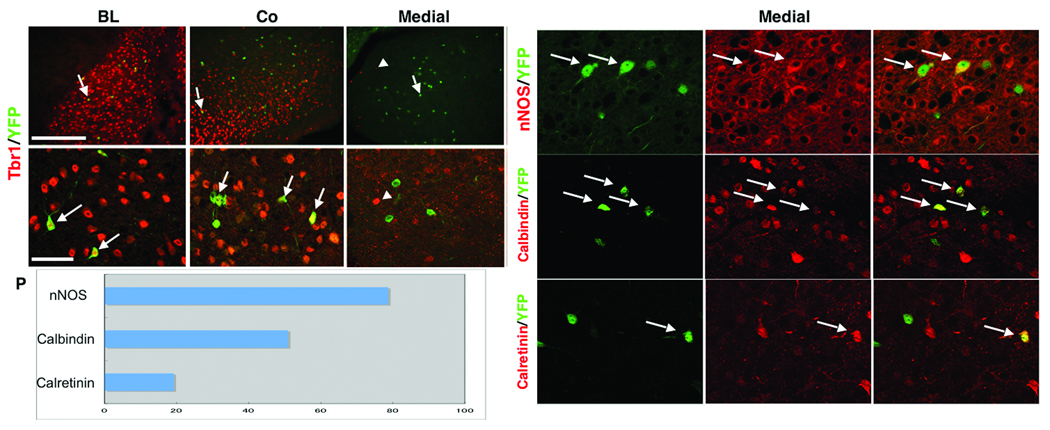
We next examined the post-natal cellular fate of Dbx1-derived cells in the amygdala. Tbr1 was used to mark excitatory cells, and based on previous studies of amygdala neuronal cell diversity, neuronal nitric oxide synthase (nNOS), calretinin and calbindin were used to mark mature inhibitory neurons24,25. Our analyses reveals that all Dbx1-derived cells in the basolateral complex and most (95.3%) of the cells in the cortical nucleus are Tbr1+ (Fig. 7a,b,d,e). In contrast, none of the recombined cells in the medial nucleus, a region only sparsely populated by excitatory neurons, are Tbr1+ (Fig. 7c,f). Instead, most (78.6%) of the recombined cells of the medial amygdala express nNOS, a marker of medial nuclei projection neurons26 (Fig. 7g–i,p). In addition, 50.8% of YFP+ cells in the medial nucleus express calbindin and 19.1% express calretinin (Fig. 7j–p), consistent with an inhibitory neuronal fate. Thus, Dbx1-derived cells give rise to both excitatory and inhibitory cell diversity in the post-natal amygdala.
Figure 7. Dbx1+ progenitors generate amygdala excitatory and inhibitory neurons.
The cellular phenotype of recombined cells in the amygdala was examined at P21 in Dbx1+/CreERT2;R26RYFP mice that were administered tamoxifen at E10.5. All (32/32) YFP+ recombined cells (green) in the basolateral complex (a,d) and 95.3% (41/43) of recombined cells in the cortical nucleus (b,e) are positive for Tbr1 (red) as shown at low and high power magnification (arrows show double labeled cells). In contrast, no (0/41) YFP+ recombined cells (green, arrow) in the medial nucleus express Tbr1 (arrowhead) (c,f). Higher power magnification of the medial nucleus reveals YFP+ recombined cells co-expressing nNOS (arrows, g-i), calbindin (arrows, j-l), or calretinin (arrows, m-o). Percentages of YFP+ cells co-expressing each inhibitory marker in the medial nucleus is shown in (p), with 78.6% (114/145) of recombined YFP+ cells in the medial nucleus are positive for nNOS, 50.8% (30/59) are positive for calbindin, and 19.1% (18/94) are positive for calretinin. Scale bar in (a) for (a-c); 250 µm. Scale bar in (d) for (d-o); 20 µm.
Collectively, these results indicate that the Dbx1-derived cells of the VP and POA are major differential sources of this excitatory and inhibitory cell diversity in the basolateral complex/cortical nuclei and medial nuclei, respectively, in the post-natal amygdala. The evidence in support of this is three-fold: 1) the Dbx1-derived cells of the POA are generated one day earlier than those of the VP (shown in Fig. 4). This correlates with the one day earlier generation of Dbx1-derived cells in the mature medial nucleus as compared to the basolateral complex (shown in Fig. 2), 2) the leading processes of VP and POA Dbx1-derived cells are oriented toward the developing amygdala indicating that these two regions are separate sources of migrating cells (shown in Fig. 4), and 3) during their migration to the developing amygdala, VP and POA Dbx1-derived cells express excitatory and inhibitory neuronal markers, respectively (shown in Fig. 6).
Physiological analysis of Dbx1-derived medial nuclei cells
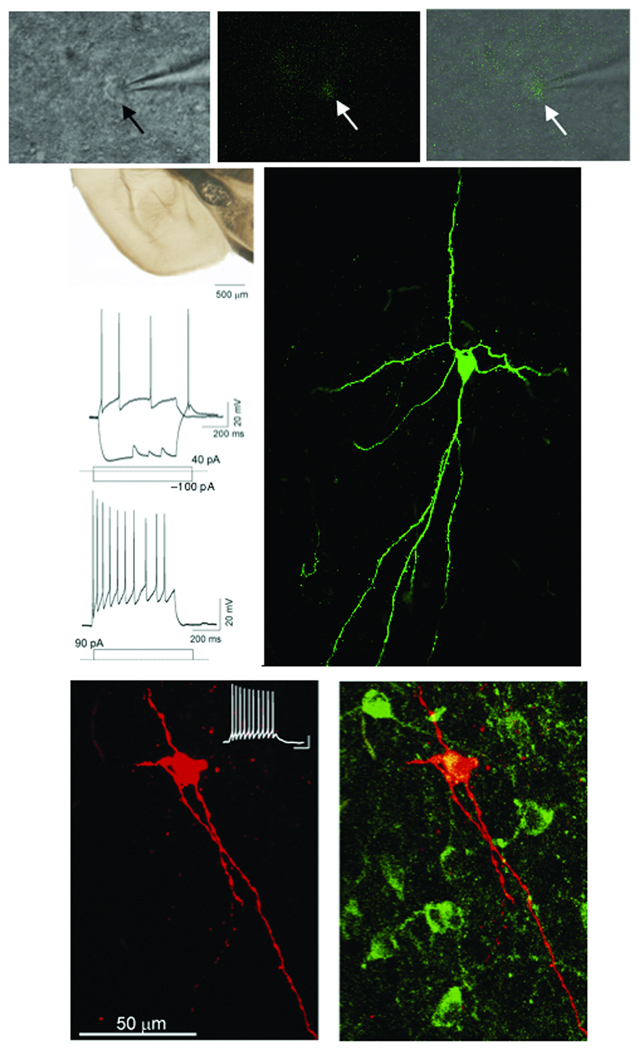
In addition to expressing unique combinations of immunomarkers, subtypes of telencephalic inhibitory neurons also have distinct electrophysiological signatures34. To determine if Dbx1-derived neurons possess functional features of inhibitory neurons, we carried out whole cell patch clamp recordings of recombined YFP+ cells in the medial nucleus from Dbx1+/CreERT2;R26RYFP mice at P17–27 (Fig. 8 and Supplementary Table 1). YFP+ neurons in the medial amygdala were identified under fluorescent and infrared differential interference contrast (IR-DIC) video microscopy (Fig.8a–c). The average Dbx1-derived neuron has a membrane potential of −58.21 ± 1.1 mV (n=35) with an input resistance at 491.34 ± 26.7 MΩ (n=35). Action potential discharge patterns were obtained by injecting current pulses in current clamp mode and monitoring discharge responses. A remarkable feature of Dbx1-derived neurons is the lack of diversity in action potential firing patterns as all (n=35) recorded cells exhibit accommodating firing patterns (accommodation ratio = 0.568 ±0.029) at threshold and all have sharp rising after hyperpolarizing potentials (Fig. 8d–f). In addition, Dbx1-derived neurons share some biochemical and physiological similarities with inhibitory neurons in other brain regions. For example, although there are differences in many characteristics, the firing pattern is similar to the recently identified nNOS+ “Ivy cells” of the hippocampus27 and the calretinin+ classic-accommodating “Martinotti cells” of the cerebral cortex28. Additionally, at hyperpolarizing current pulses in the −100pA range, Dbx1-derived neurons reveal a small sag (4.1 ± 0.55 mV n=27) indicative of an Ih current, and at the conclusion of the 600 ms hyperpolarizing pulse, cause a rebound burst indicative of low threshold calcium currents in the inhibitory low threshold spiking cells of the cerebral cortex29–31. Current pulses at more negative levels are deleterious to the cells and result in rapid depolarization in membrane potential. As revealed by biocytin filling, Dbx1-derived cells possess large sparsely spiny proximal dendrites with numerous long typically bipolar oriented projections (Fig. 8g–i and Supplementary Fig. 5). This morphology is similar to recently characterized medial amygdala neurons, some of which displayed inhibitory characteristics32. Therefore, a combination of immunomarker, electrophysiological and morphological analyses (Fig. 7 and Fig. 8) indicate that Dbx1-derived neurons, while sharing similarities with a variety of classic inhibitory features outside of the amygdala, appear to be a distinct subclass of neurons.
Figure 8. Electrophysiological properties of Dbx1-derived medial amygdala neurons.
DIC (a), fluorescent (b) and overlay (c) images of a typical YFP+ recombined medial amygdala neuron (arrows) with attached recording electrode. Live coronal slice indicating region of physiological recordings (d, asterisk). Firing patterns of a typical medial amygdala YFP+ recombined cell at threshold and sub-threshold (e) and supra-threshold (f) firing. Injected level of current for each trace is indicted below. Confocal image of biocytin filled cell (g) represented in d and e following physiological characterization. Note the fusiform-like dendritic arborization and the presence of sparsely spiny dendrites. Confocal image of biocytin filled cell conjugated to texas red following whole cell patch clamp recording is shown in (h). The current clamp trace of this cell is located in the upper right corner (scale bar: 200 ms horizontal, 20 mV vertical.) Fluorescent immunostaining for nNOS (green) reveals that this cell is also nNOS+ (yellow) (i).
DISCUSSION
By a combination of in vivo and in vitro approaches, including genetic fate mapping, cell migration assays and electrophysiology, we examined the link between the embryonic origin of progenitor cells and their fate in the mature amygdala. Our primary finding is the identification of a novel neural progenitor pool specifically dedicated to the generation of a single subtype of inhibitory neuron in the mature medial amygdala nucleus, a major inhibitory output nucleus of the amygdala. Our results reveal that this population, which is marked by the expression of the homeodomain encoding gene Dbx1, migrates along a newly identified migratory stream, the PAS, which arises from the preoptic area (POA) of the subpallial telencephalon, a region previously not recognized as a source of migratory cells. Moreover, our analyses indicate that the ventral pallial (VP) Dbx1+ population is a source of excitatory neurons in the basolateral complex and cortical nuclei, the two major excitatory nuclei of the amygdala (Schematic shown in Supplementary Fig. 6). These findings also provide strong support for an emerging model in which aspects of the program for the development of the amygdala are distinct from that of other telencephalic structures such as the cerebral cortex.
The POA is a novel source of amygdala inhibitory neurons
The embryonic subpallium is the origin of most, if not all, inhibitory neuronal cell populations in the cerebral cortex and striatum33– 35. It also appears that the vast inhibitory interneuronal diversity that is characteristic of the telencephalon is derived from spatially distinct progenitor pools within the subpallial ganglionic eminences, most prominently the medial (MGE) and caudal (CGE) ganglionic eminences. Emerging evidence also suggests that the genetic mechanisms for establishment of inhibitory neuronal cellular diversity in the telencephalon is strikingly similar to that in the spinal cord in which the combinatorial expression of specific transcription factor genes (typically of the homeodomain and bHLH classes) intrinsically instruct spatially separate progenitor populations to give rise to mature neurons with differential functional fates34,36. Based on this idea of ‘combinatorial genetic codes’, recent studies have revealed that the subpallium can be subdivided into at least 18 distinct progenitor zones37. One of these unique regions is the POA, which is distinguished in part by its focal expression of Dbx1. By use of a Cre-based fate mapping strategy combined with in vitro migratory assays, we identify a novel POA migratory stream termed the “POA-amygdala stream (PAS)” that is comprised, at least in large part, of Dbx1-derived progenitors. As the POA and PAS is also combinatorially marked by a number of other transcription factors, most notably Nkx6.2, Lhx2 and Dlx1/2(37 and data shown here), it is likely that similar to the lateral cortical stream (LCS) migratory route from the PSB, the PAS may be comprised of a diverse set of progenitor cells.
The primary function of the medial amygdala is to process sensory information from the olfactory bulb and relay this information the hypothalamus, a brain region involved in hormonal regulation. As such, the medial amygdala has reciprocal connections with multiple hypothalamic nuclei, which in turn project to numerous regions of the brainstem. These pathways are involved in the regulation of social, feeding and reproductive behaviors3,38. Along with the central nucleus, the medial amygdala comprises the major inhibitory output of the amygdala. The predominant neuronal subtype of the medial amygdala is inhibitory projection neurons. Based on their electrophysiological properties, cellular morphology and expression of nNOS, the Dbx1-derived medial amygdala neurons resemble a previously characterized population of inhibitory projection neurons of the medial nucleus26,32. Although their function remains unknown, these nNOS+ neurons project to the paraventricular nucleus of the hypothalamus, and therefore may be involved in the modulation of hormone release and autonomic responses. Thus, in addition to identification of a novel Dbx1-derived POA-amygdala migratory stream, our results indicate that these cells generate a specific subtype of inhibitory neurons in the medial amygdala, and therefore uncover a novel relationship between gene expression, developmental origin and electrophysiological fate in the amygdala.
Ventral pallial source of excitatory neurons
The PSB comprises progenitors from the VP and dLGE aspects of the developing telencephalon. These progenitors migrate to the basal telencephalic limbic system via the lateral cortical stream (LCS)8,39. A series of studies has revealed that VP and dLGE progenitors express a combination of markers that define this region as distinct from other telencephalic progenitor domains9–11,40,41. Based on their expression of pallial markers, such as Pax6, the VP population is predicted to give rise to excitatory neurons. In contrast, dLGE progenitors, which express subpallial markers such as Gsh2 and Dlx1/2, may generate subtypes of inhibitory neurons of the amygdala. However, to date a direct link between gene expression domains of the PSB and neuronal cell fate in the amygdala has not been demonstrated. Our data indicate that Dbx1+ progenitors derived from the VP are a source of excitatory neurons of basolateral complex and cortical amygdala nuclei, major excitatory outputs of the amygdala. During their migration along the LCS, a subpopulation of these basally-migrating Dbx1-derived cells express Tbr1, a marker of excitatory neurons, but, interestingly, do not express Pax6. As Pax6 expression marks a large number of LCS cells8,12,13,41,42 this result was somewhat surprising, and indicates that the VP generates a heterogeneous pool of progenitors destined for the amygdala. In addition to the VP as a source of excitatory cells of the amygdala, fate mapping studies of Emx1-derived cells, whose expression marks more dorsally located progenitors in the lateral pallium (LP), are also a presumptive source of amygdala excitatory neurons43,44. Whether these progenitor populations that are differentially marked by Dbx1, Pax6 and/or Emx1 give rise to distinct functional and or spatially located excitatory neurons in the mature amygdala remains to be explored.
Comparison of amygdala development to other brain regions
A number of recent studies have revealed there are notable similarities between development of the amygdala and other major telencephalic structures such as the cerebral cortex and striatum. The main commonality is a sharing of progenitor populations for the generation of neuronal diversity. For example, in utero cell transplantation fate mapping and gene expression studies have revealed that the medial (MGE) and caudal (CGE) ventral telencephalic ganglionic eminences are sources of inhibitory interneuronal cell diversity for the amygdala, cerebral cortex and striatum45,46. In addition, recent genetic fate mapping studies have revealed that subpallial progenitors expressing Nkx2.1 generate parvalbumin interneuronal subtypes in both the amygdala and cerebral cortex47. Moreover, the lateral olfactory tract nucleus (nLOT) of the amygdala appears to share a common developmental origin with the cerebral cortex48.
However, our finding of a point-to-point correlation between the Dbx1-derived POA source and the medial nucleus of the amygdala suggests a putative additional and novel mechanism for the generation of neuronal cell diversity in the amygdala in which there also exists progenitor pools set aside exclusively for the limbic system. This is interestingly consistent with and predicted by previous molecular anatomical studies, which have identified combinations of genes whose expression marks both embryonic progenitor domains and individual emerging amygdala nuclei, including the medial and central nuclei, suggesting a developmental relationship between the two9,49. A major prediction of these gene expression studies is that progenitor pools that express unique combinations of genes will generate distinct functional cell types. As revealed by our studies, at least in the case of the Dbx1-derived POA population, this progenitor pool gives rise almost exclusively to a single functional subtype of inhibitory neurons of the medial amygdala. Although possessing some characteristics in common with other telencephalic inhibitory cell types, by a combination of criteria including neuronal subtype marker expression, cell morphology and electrophysiological properties, the POA Dbx1-derived neurons of the medial amygdala appear to be distinct from other telencephalic inhibitory cell types. Therefore, our data, in conjunction with previous studies, suggest a model in which there are both overlapping and distinct mechanisms for amygdala development in comparison to other telencephalic structures. Thus, for the generation of cell types that are found in other brain regions (such as parvalbumin interneurons which are generated in the MGE), there are shared progenitor pools. In contrast, the generation of cell types that may be exclusive to the amygdala, such as the medial amygdala Dbx1-derived inhibitory neurons, appears to require the dedication of progenitor pools solely for the production of that cell type.
In summary, our data provide novel and important new insight into the developmental mechanisms underlying the generation of neuronal diversity in the amygdala, a brain structure essential for major aspects of complex mammalian behavior. First, we identify a new major migratory stream (the PAS) in the developing brain that is comprised of Dbx1+ progenitors derived from the POA. As this population generates a specific cell type in a distinct functional nucleus of the amygdala, we reveal a novel and remarkable direct correlation between an embryonic progenitor population and a functional neuronal cell type in the mature telencephalon. Second, our data reveal major differences between the logic for the development of the amygdala and the cerebral cortex, and thus present a revised model of brain development. In addition, as altered function of the amygdala is a hallmark characteristic of neuro-developmental disorders such as autism and autism spectrum disorders, our findings may provide a framework for a deeper understanding of the etiology of these disorders.
METHODS
Generation of CreERT2 knock-in mice
All procedures involving the use of animals were carried out in accordance with approved procedures established by the Children’s National Medical Center and Georgetown University Animal Care and Use Committees. The Dbx1+/CreERT2 targeting construct was generated by targeting an IRES-CreERT2-FRT-NEO-FRT into the BamHI site at the 3’ UTR of the Dbx1 locus (obtained from mouse 129sv strain RCPI 21 PAC library from Invitrogen Inc.), the same site previously used to knock Cre into the Dbx1 locus11. Correctly targeted R1 ES cells were injected into C57BL/6 blastulas, and chimeric offspring were crossed to C57BL/6 mice. The Neomycin resistance gene was removed by crossing heterozygous Dbx1+/CreERT2 mice to FLPeR (“flipper”) mice (Jackson Labs).
Reporter mouse lines and tamoxifen delivery
R26RLacZ and R26RYFP reporter mice were obtained from Jackson Labs. Tamoxifen (Sigma T5648) was dissolved at a concentration of 20mg/ml in corn oil (Sigma C8267) and administered by oral gavage to pregnant dams at a concentration of 4–8mg/40g mouse body weight.
Immunohistochemistry and LacZ staining
Embryos were fixed with 4% paraformaldehide for 2hrs at 4□, dehydrated in 30% sucrose, embedded in OCT and sectioned at 30 µm. For detection of LacZ expression in Dbx1+/LacZ knock-in, Dbx1+/CreERT2;R26RLacZ and Dlx2+/tauLacZ embryos, sections were incubated in the X-gal staining solution overnight at 37□. For detection of Dlx2 expression, Dbx1+/CreERT2;R26RYFP mice were crossed to previously generated Dlx2+/tauLacZ mice in which LacZ expression faithfully recapitulates Dlx2 expression50. For immunohistochemistry, sections were incubated with the primary antibody at 4□ overnight, washed and incubated with the corresponding fluorescent secondary antibodies. Primary antibodies used were goat anti-β-galactosidase (1:400; Biogen), rabbit anti-Tbr1 (1:2000; kind gift from R. Hevner), rabbit anti-Pax6 (1:500; Covance Research Products), rabbit anti-nNOS (1:1000; Sigma), goat anti-GFP (to detect YFP expression, 1:500; Novus), rabbit anti-calbindin (1:500; Calbiochem), rabbit anti-calretinin (1:1000; Millipore) and mouse-anti RC2 (1:5; Developmental Hybridoma Studies Bank, University of Iowa, Iowa City, IA).
In situ hybridization
Embryos were processed and sectioned as described above. Sections were refixed with 4% PFA and subsequently treated with proteinase K, refixed with 4% PFA, treated with triethanolamine containing acetic anhydride and then hybridized with a digoxigenin-UTP-labeled RNA probes overnight. Next day, the probes were washed with 50% formamide, 2xSSC at 65 °C, treated with 20mg RNase and washed in 50% formamide, 2xSSC at 65 °C. Signals were detected with an anti-digoxigenin antibody (Roche) and BM purple (Roche). The probes used in this study were Dbx19 (probe used Fig. 1), Foxg114, Lhx237 and Cre recombinase (5’ coding region was subcloned into pBlusescript and RNA probe was generated from NcoI digested plasmid using T3 RNA polymerase). For analysis of Dbx1 expression in Dbx1+/CreERT2 embryos (Supplementary Fig. 1), a PCR amplified fragment of the 5’ coding region was subcloned into pBluescript, and RNA probe was generated from BamHI digested plasmid using T7 RNA polymerase.
Migration assays
For matrigel assays, tissue was dissected from E12.5 embryos using Lumsden Bioscissors. The explants were placed in matrigel (BD Biosciences) mixed with L-15 media and cultured in Neurobasal medium (Invitrogen), supplemented with penicillin/streptomycin (1:100; Invitrogen), B27 supplement (1:50; Invitrogen) and glutamine (1:100; Invitrogen). After 3 DIV, explants were fixed in 4% PFA for 30 min, rinsed briefly in PBS, covered with 50% glycerol and photographed. For DiI migration assays, the 300µm horizontal or coronal slices were made from E13.5 embryos with a vibratome and DiI crystals were placed in the POA or VP, and cultured in Neurobasal medium (Invitrogen), supplemented with penicillin/streptomycin (1:100; Invitrogen), B27 supplement (1:50; Invitrogen) and glutamine (1:100; Invitrogen). After taking low power pictures of both light field and fluorescence, specimens were fixed in 4% PFA for 2 hrs, re-sectioned on a vibratome at 100 µm and processed for immunofluorescence.
Electrophysiology and biocytin
After sacrifice, brains of animals were immediately immersed in ice-cold - oxygenated (95% O2/ 5% CO2) sucrose solution (mM): 234 sucrose, 11 glucose, 24 NaHCO3, 2.5 KCl, 1.25 NaH2PO4*H2O, 10 MgSO4 and 0.5 CaCl2. Coronal slices were cut on a vibratome (Leica) at 250 ⌠m and placed in preheated (32°C), oxygen equilibrated, artificial cerebral spinal fluid (ACSF) for 1 hour. Slices were then placed in a recording chamber and visualized with a fixed staged, upright microscope (Nikon, E600 FN) equipped with infrared (IR) illumination, Nomarski optics, an IR-sensitive video camera (COHU) and fluorescent lamp (Nikon) with a 450–490λ filter. Intracellular pipettes were pulled with a Flaming/Brown Micropipette Puller (Sutter Instruments) to a resistance of 3–5 MΩ when filled with a solution containing (mM): 130 K-gluconate, 10 KCl, 10 HEPES, 10 EGTA, and 2 MgCl2. YFP+ fluorescent cells were recorded at room temperature (21–23°C) with continuous perfusion of ACSF in current clamp mode (Multiclamp 700A, DigiDATA, Axon). After recording membrane potential and input resistance values, cells were categorized based on their response to depolarizing and hyperpolarizing current pulses. The response properties were analyzed off-line using pClamp software (Axon) and graphing software (Origin).
In some cases post recording biocytin (1%) was injected with depolarizing current pulses (1nA). Pictures were taken with an image capturing (Scion) system in order to relocate the cells post-processing. A subset of tissue with biocytin filled cells was also processed for nNOS immunohistochemistry. In these cases, slices the were removed from the recording chamber and fixed overnight in 4% paraformaldehyde and processed for immunohistochemistry.
Microscopy
Light microscopic photographs were taken using a Olympus BX51 microscope and a Olympus CKX41 inverted microscope. Fluorescent photographs were taken using Olympus BX61 (for low power fluorescence and low power DiI images), Zeiss Apotome (for high power fluorescence), a Zeiss LSM510 confocal microscope (for high power DiI/YFP double fluorescence) or an Olympus Fluoview Laser Scanning Confocal Microscope (for biocytin filled cells). Any brightness and contrast adjustments were done equally across pictures using Adobe Photoshop.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank members of the Corbin, Haydar and Zohn labs for input during various stages of this project, with a special acknowledgement to J. L. Olmos-Serrano for his expert insight and advice on amygdala anatomy and development. We also gratefully acknowledge T. Haydar, J. L. Olmos-Serrano, I. Zohn and V. Gallo for critical reading of the manuscript. We also thank the following for reagents: R. Hevner, University of Washington, USA (Tbr1 antibody), S. Aizawa, RIKEN, Japan (Foxg1 and Lhx2 probe) and M. Matise, UMDNJ/Robert Wood Johnson Medical School, USA (Dbxl probe). The RC2 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Science, Iowa City, IA 52242. We also thank the CNMC and Georgetown University Transgenic Cores for the generation of mice. This work was supported by grants from the National Institutes of Health (JGC and MMH). The CNMC microscope core facility is supported by an NIH Intellectual and Developmental Disabilities Research Center (IDDRC) grant.
Footnotes
COMPETING INTEREST STATEMENT
The authors declare they have no competing financial interests
References
- 1.Alheid GF. Extended amygdala and basal forebrain. Ann N Y Acad Sci. 2003;985:185–205. doi: 10.1111/j.1749-6632.2003.tb07082.x. [DOI] [PubMed] [Google Scholar]
- 2.Sah P, Faber ES, Lopez De Armentia M, Power J. The amygdaloid complex: anatomy and physiology. Physiol Rev. 2003;83:803–834. doi: 10.1152/physrev.00002.2003. [DOI] [PubMed] [Google Scholar]
- 3.Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 4.Amaral DG, Bauman MD, Schumann CM. The amygdala and autism: implications from non-human primate studies. Genes Brain Behav. 2003;2:295–302. doi: 10.1034/j.1601-183x.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 5.Baron-Cohen S, et al. The amygdala theory of autism. Neurosci Biobehav Rev. 2000;24:355–364. doi: 10.1016/s0149-7634(00)00011-7. [DOI] [PubMed] [Google Scholar]
- 6.Rauch SL, Shin LM, Phelps EA. Neurocircuitry models of posttraumatic stress disorder and extinction: human neuroimaging research--past, present, and future. Biol Psychiatry. 2006;60:376–382. doi: 10.1016/j.biopsych.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Bai J, et al. The role of DCX and LIS1 in migration through the lateral cortical stream of developing forebrain. Dev Neurosci. 2008;30:144–156. doi: 10.1159/000109859. [DOI] [PubMed] [Google Scholar]
- 8.Carney RS, et al. Cell migration along the lateral cortical stream to the developing basal telencephalic limbic system. J Neurosci. 2006;26:11562–11574. doi: 10.1523/JNEUROSCI.3092-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina L, et al. Expression of Dbx1, Neurogenin 2, Semaphorin 5A, Cadherin 8, and Emx1 distinguish ventral and lateral pallial histogenetic divisions in the developing mouse claustroamygdaloid complex. J Comp Neurol. 2004;474:504–523. doi: 10.1002/cne.20141. [DOI] [PubMed] [Google Scholar]
- 10.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielle F, et al. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 12.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 13.Carney RSE, Cocas LA, Hirata T, Mansfield K, Corbin JG. Differential regulation of telencephalic pallial-subpallial boundary patterning by Pax6 and Gsh2. Cerebral Cortex. 2008 doi: 10.1093/cercor/bhn123. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 15.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 16.Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 19.Pierani A, et al. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29:367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 20.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 22.Porteus MH, Bulfone A, Ciaranello RD, Rubenstein JL. Isolation and characterization of a novel cDNA clone encoding a homeodomain that is developmentally regulated in the ventral forebrain. Neuron. 1991;7:221–229. doi: 10.1016/0896-6273(91)90260-7. [DOI] [PubMed] [Google Scholar]
- 23.del Rio MR, DeFelipe J. Colocalization of calbindin D-28k, calretinin, and GABA immunoreactivities in neurons of the human temporal cortex. J Comp Neurol. 1996;369:472–482. doi: 10.1002/(SICI)1096-9861(19960603)369:3<472::AID-CNE11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.McDonald AJ, Mascagni F. Colocalization of calcium-binding proteins and GABA in neurons of the rat basolateral amygdala. Neuroscience. 2001;105:681–693. doi: 10.1016/s0306-4522(01)00214-7. [DOI] [PubMed] [Google Scholar]
- 25.Olmos JL, Real MA, Medina L, Guirado S, Davila JC. Distribution of nitric oxide-producing neurons in the developing and adult mouse amygdalar basolateral complex. Brain Res Bull. 2005;66:465–469. doi: 10.1016/j.brainresbull.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, et al. Nitrergic neurons in the medial amygdala project to the hypothalamic paraventricular nucleus of the rat. Brain Res. 1997;777:13–21. doi: 10.1016/s0006-8993(97)00948-7. [DOI] [PubMed] [Google Scholar]
- 27.Fuentealba P, et al. Ivy cells: a population of nitric-oxide-producing, slow-spiking GABAergic neurons and their involvement in hippocampal network activity. Neuron. 2008;57:917–929. doi: 10.1016/j.neuron.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beierlein M, Gibson JR, Connors BW. Two dynamically distinct inhibitory networks in layer 4 of the neocortex. J Neurophysiol. 2003;90:2987–3000. doi: 10.1152/jn.00283.2003. [DOI] [PubMed] [Google Scholar]
- 30.Bacci A, Rudolph U, Huguenard JR, Prince DA. Major differences in inhibitory synaptic transmission onto two neocortical interneuron subclasses. J Neurosci. 2003;23:9664–9674. doi: 10.1523/JNEUROSCI.23-29-09664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bacci A, Huguenard JR, Prince DA. Long-lasting self-inhibition of neocortical interneurons mediated by endocannabinoids. Nature. 2004;431:312–316. doi: 10.1038/nature02913. [DOI] [PubMed] [Google Scholar]
- 32.Bian X, Yanagawa Y, Chen WR, Luo M. Cortical-like functional organization of the pheromone-processing circuits in the medial amygdala. J Neurophysiol. 2008;99:77–86. doi: 10.1152/jn.00902.2007. [DOI] [PubMed] [Google Scholar]
- 33.Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4(Suppl):1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 34.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 35.Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 36.Corbin JG, et al. Regulation of Neural Progenitor Cell Development in the Nervous System. Journal of Neurochemistry. 2008 doi: 10.1111/j.1471-4159.2008.05522.x. DOI: 10.1111/j.1471–4159.2008.05522.x, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi GB, et al. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46:647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Molnar Z, Butler AB. The corticostriatal junction: a crucial region for forebrain development and evolution. Bioessays. 2002;24:530–541. doi: 10.1002/bies.10100. [DOI] [PubMed] [Google Scholar]
- 40.Waclaw RR, et al. The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Stoykova A, Fritsch R, Walther C, Gruss P. Forebrain patterning defects in Small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- 42.Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- 43.Tole S, Remedios R, Saha B, Stoykova A. Selective requirement of Pax6, but not Emx2, in the specification and development of several nuclei of the amygdaloid complex. J Neurosci. 2005;25:2753–2760. doi: 10.1523/JNEUROSCI.3014-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 46.Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 48.Remedios R, et al. A stream of cells migrating from the caudal telencephalon reveals a link between the amygdala and neocortex. Nat Neurosci. 2007;10:1141–1150. doi: 10.1038/nn1955. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Lopez M, et al. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506:46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin JG, Gaiano N, Machold RP, Langston A, Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127:5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.