Abstract
Here we report the characterization of the Chlamydomonas reinhardtii gene ARG9, encoding the plastid resident N-acetyl ornithine aminotransferase, which is involved in arginine synthesis. Integration of an engineered ARG9 cassette in the plastid chromosome of the nuclear arg9 mutant restores arginine prototrophy. This suggests that ARG9 could be used as a new selectable marker for plastid transformation.
In the green alga Chlamydomonas reinhardtii, the arg9-1 and arg9-2 mutations result in arginine auxotrophy because of a deficiency in N-acetyl ornithine aminotransferase activity (NAOAT) (6). Of the two arg9 mutants originally isolated (6), only the arg9-2 strain was found to be an arginine auxotroph, while the arg9-1 mutant had reverted to wild type. We reasoned that the arg9-2 mutation mapped to the structural gene for NAOAT and identified a candidate ARG9 gene (XP_001698091), based on the similarity of the predicted gene product to the Saccharomyces cerevisiae NAOAT, Arg8p. Three full-length cDNAs were identified and sequenced. Both the ARG9 genomic DNA and full-length cDNAs restored arginine prototrophy when introduced into the nucleus of the arg9-2 mutant (data not shown). Sequencing the ARG9 genomic locus in the arg9-2 strain identified a G-to-A transition at codon 317, resulting in a glycine-to-arginine mutation at a strictly conserved residue in NAOATs.
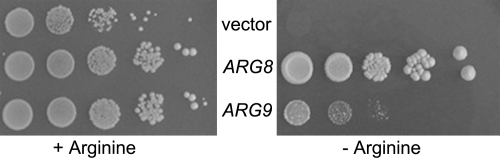
Next, we tested if Chlamydomonas ARG9 could functionally replace the Arg8 protein in yeast by expressing the ARG9 cDNA in an arg8-null mutant that is deficient in NAOAT. Figure 1 shows that expression of the ARG9 cDNA from the plasmid-borne PGK1 promoter is able to partially restore arginine prototrophy. Since the yeast arg8 mutant can be complemented by the Chlamydomonas ARG9 protein, it is likely that the algal protein expressed in yeast is targeted to the mitochondria, where NAOAT typically functions in fungi (10). Indeed, the N-terminal extension of the candidate ARG9 protein exhibits features typical of a plastid- or mitochondrion-targeting sequence, such as the propensity to form an amphiphilic α-helix (7).
FIG. 1.
Heterologous functional complementation of the S. cerevisiae arg8 mutant by the C. reinhardtii ARG9 cDNA encoding NAOAT. The yeast arg8 mutant (NB880) was transformed by the yeast expression vector pFL61, by pFL61/ARG8 (expressing the yeast ARG8 gene), and by pFL61/ARG9 (expressing the Chlamydomonas ARG9 cDNA). Dilution series (10×) of each transformant were plated on synthetic complete medium with or without arginine and incubated at 28°C for 7 or 14 days, respectively.
The sublocalization of the ARG9 protein was examined by immunoblot analysis using an antibody raised against Arg8p, the S. cerevisiae NAOAT that is resident in the mitochondrial matrix. The anti-Arg8p antibody cross-reacted with species in mitochondrial and plastid fractions of Chlamydomonas cells (Fig. 2). We identified the 48-kDa species in the plastid fraction as the ARG9 protein, based on the predicted size of the mature protein. This species was also present in the arg9-2 strain, suggesting that the arg9-2 mutant accumulates a nonfunctional ARG9 protein. The cross-reacting species detected in the mitochondrial fraction have higher electrophoretic mobilities and could correspond to nonspecific bands, splicing variants of the ARG9 transcript that specify a mitochondrial protein or dually targeted ARG9 protein. Complementation experiments described below indicate that the primary site of action of ARG9 is the plastid. Based on our analyses, we concluded that NAOAT is located in the plastid in Chlamydomonas, but we cannot exclude the possibility that it also operates in the mitochondrion. The operation of plastid-localized enzymes involved in arginine biosynthesis in Chlamydomonas is also supported by studies suggesting that argininosuccinate lyase could also be resident in the chloroplast (1).
FIG. 2.
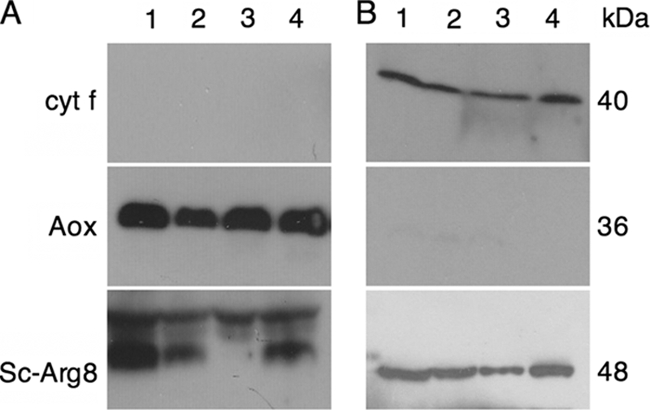
ARG9 localizes to the plastid in Chlamydomonas. Proteins from mitochondrion-enriched (A) and chloroplast-enriched (B) fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immobilized on a nylon membrane. The membrane was immunodecorated using antibodies against Arg8p, Aox, or cytochrome f. The purity of each fraction was verified by using an antibody against a known protein resident in the mitochondria (Aox) or in the chloroplast (cyt f). Lanes 1 to 3, arg9-2 transformants complemented by genomic ARG9; lane 4, arg9-2 mutant.
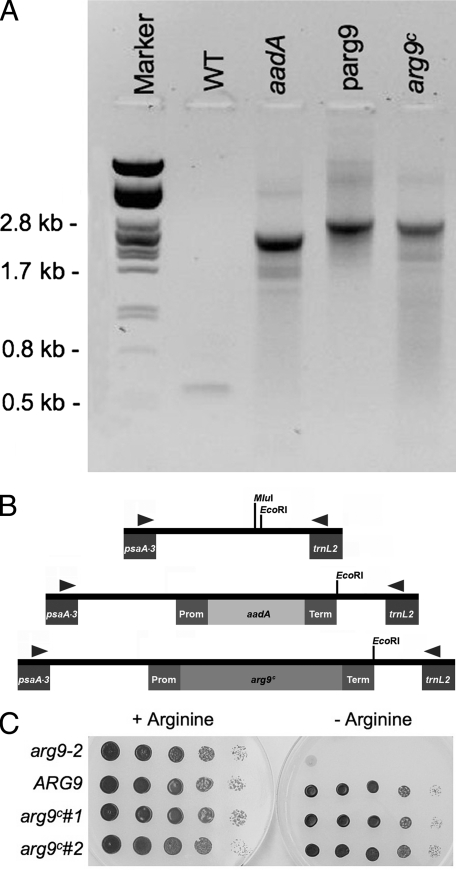
We reasoned that the ARG9 gene relocated to the plastid chromosome of an arg9-2 strain should be able to restore arginine prototrophy if the ARG9 protein is successfully expressed and active in the organelle. For this experiment, we chose the ARG9 cDNA from Arabidopsis thaliana, whose nuclear genome has a codon bias (44% GC) closer to that of the plastid genome than the nuclear genome of Chlamydomonas. Moreover, the frequency of codons in the Chlamydomonas plastid genes made it likely that the Arabidopsis cDNA would be translated by the set of plastid tRNAs. Using an existing spectinomycin resistance cassette (aadA) for plastid transformation, we designed an arg9c (c for chloroplast) cassette for expression of the Arabidopsis NAOAT in the chloroplast of Chlamydomonas (3). As a proof of concept, we first targeted the aadA cassette to an integration site on the plastid chromosome that is neutral with respect to chloroplast function (4). A spectinomycin-resistant (Specr) transformant was generated by biolistic transformation of the mt+ arg9-2 mutant (4). The presence of the aadA gene at the expected location in the plastid chromosome was detected by diagnostic PCR (Fig. 3A and B). Next, we replaced the aadA cassette with the arg9c cassette, which expresses the A. thaliana ARG9 protein from the same promoter and terminator as the aadA cassette (plasmid KS/arg9c) (Fig. 3B). Based on our alignments of NAOATs, we reasoned that the first 48 residues of ARG9 might include the plastid-targeting sequence and were therefore not necessary if ARG9 is synthesized on plastid ribosomes. Replacement of the aadA marker by the arg9c cassette is expected to occur by homologous recombination between the atpA promoter and rbcL terminator sequences, which are common to the aadA and arg9c cassettes (Fig. 3B). The mt+ arg9-2 Specr strain was bombarded with the pKS/arg9c plasmid and transformants were selected on medium lacking arginine. A few transformants were able to grow on the selective medium (Arg+ transformants), suggesting that ARG9 was successfully expressed from the plastid genome. Molecular analysis of two independent Arg+ transformants showed that they were heteroplasmic for the presence of the aadA and arg9c cassettes and were also Specr, as expected (data not shown). In a previous study, we showed that the segregation of plastid markers is facilitated if heteroplasmic transformants are converted to gametes, presumably because gametes undergo a reduction of the copy number of their chloroplast genomes (4). To achieve homoplasmy for the arg9c marker, we induced gametogenesis of two arg9-2 Specr Arg+ plastid transformants and retrieved vegetative clones by plating the gametes on medium without arginine. Two independent clones were further analyzed by using diagnostic primers lying outside the regions of homology between the aadA and arg9c cassettes and were found to contain the arg9c cassette (Fig. 3A and B). As expected, the clones were still arginine prototrophs but had lost the spectinomycin resistance (data not shown), indicating that they had become homoplasmic for the arg9c cassette. This suggests that the arg9c cassette was able to replace the aadA cassette in the plastid chromosome and could be successfully expressed to complement the arg9-2 mutation. Interestingly, we found that the level of growth on medium without arginine of the plastid transformants expressing arg9c was comparable to that of nuclear transformants complemented with ARG9 genomic DNA (Fig. 3C).
FIG. 3.
Plastid transformation of the arg9-2 strain with the arg9c cassette restores arginine prototrophy. (A) Molecular analysis of the plastid transformants was performed by PCR using diagnostic primers (F and R) lying outside the region of homology in the transforming DNA (see below). PCR amplification products were separated by electrophoresis in agarose gel and ethidium bromide stained. The gel was imaged using an imaging system (Kodak Image Station 2000R). PstI-digested λ phage DNA was used as a size marker. DNAs extracted from strain CC125 (wild type [WT]), an arg9-2 Specr transformant (aadA), an arg9-2 Arg+ transformant (arg9c), and plasmid pmp-arg9c (parg9) were used as templates in the PCR. The plasmid pmp-arg9c contains the arg9c cassette and 5 kb of chloroplast DNA flanking the region of homology between the aadA and arg9c cassettes. (B) Illustration of expected PCR products using the diagnostic primers F (left arrowhead) and R (right arrowhead) and plastid DNAs from the wild-type strain (WT), Specr transformant (aadA), and Arg+ transformant (arg9c) as templates. The neutral site lies between the psaA-3 and trnL2 genes on the chloroplast genome. The MluI site is lost in the cloning of the aadA cassette. The expected sizes for the PCR products are 600 bp (WT), 2,533 bp (aadA), and 2,971 bp (arg9c). (C) Growth of the untransformed arg9 mutant (arg9-2), arg9-2 transformed by pARG9g (containing a 7-kb genomic DNA fragment with the ARG9 gene), and two independent arg9-2 plastid transformants expressing the arg9c cassette (arg9c#1 and arg9c#2) on unsupplemented and arginine-supplemented Tris-acetate-phosphate medium. Cells (4 × 106 for each strain) were serially diluted (10 times for each dilution), plated, and incubated for 4 days.
To provide additional proof of the successful relocation of the arg9c gene to the plastid chromosome, we tested for non-Mendelian segregation of the arginine prototrophy phenotype in the plastid transformants. In genetic crosses of C. reinhardtii, nuclear markers segregate according to Mendelian genetics, but chloroplast genes are inherited uniparentally from the mt+ parent (5). Hence, it is expected that the Arg+ trait can be transmitted to the progeny only if the plastid transformant is of the mt+ sexual type. We crossed an Arg+ plastid transformant (mt+ arg9-2 arg9c) to an mt− arg9-2 mutant and performed bulk segregation of the progeny. As expected, all 40 spores examined in the progeny were arginine prototrophs because they inherited the arg9c cassette from the mt+ parent. An Arg+ (mt− arg9-2 arg9c) spore from this progeny was selected, and diagnostic PCR confirmed the presence of the arg9c cassette in the chloroplast genome (data not shown). This spore was then used to do the reciprocal cross with the mt+ arg9-2 mutant. We examined 42 spores, and all of them were arginine auxotrophs, as expected, because the plastid trait cannot be transmitted by the mt− parent.
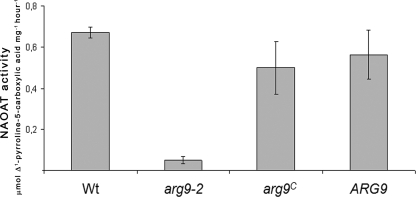
In order to show functional complementation of the arg9-2 mutation, we prepared cellular extracts and measured NAOAT activity in both plastid transformants and nuclear transformants complemented with Chlamydomonas ARG9 genomic DNA. The arg9-2 mutation results in very little detectable NAOAT activity, while the enzymatic activity was significantly restored in both nuclear and plastid transformants (Fig. 4). We concluded that integration of the A. thaliana ARG9 cDNA into the plastid chromosome of C. reinhardtii resulted in the successful expression of an enzymatically active NAOAT. Our results indicate that the arg9c cassette could be developed as a marker for chloroplast transformation in Chlamydomonas. The manipulation of the chloroplast genome of microalgae for the commercial production of recombinant molecules is a recent and promising advance in biotechnology (8). For obvious reasons, the use of arginine as selection for plastid transformation will be of significant value, considering that all markers employed so far are derived from bacterial antibiotic resistance genes (8).
FIG. 4.
Restoration of NAOAT activity in nuclear and plastid transformants. Soluble extracts of the cw15 mt− strain (wild type [Wt]), the cw− arg9-2 strain, the arg9-2 strain transformed with pARG9g (ARG9), and the arg9-2 strain transformed with the arg9c cassette (arg9c) were assayed. Results for only one representative of each plastid or nuclear transformant are shown. For each strain, at least three biological replicates were included in the experiments. Error bars indicate standard deviations.
The arg9-2 mutation is a nonreverting mutation (<10−10). We took advantage of this property and tested the effectiveness of the ARG9 gene as a marker for insertional mutagenesis in the nucleus. In Chlamydomonas, integration of transforming DNA into the nuclear genome occurs via nonhomologous recombination events that are presumed to occur at random loci (2). Thus, nuclear markers such as the ARG7 gene that encodes argininosuccinate lyase are routinely used as tool to generate insertional mutants (2). About 3,000 arginine prototrophic transformants were generated by using the arg9-2 mutant as a recipient strain and the ARG9 gene as a transforming DNA. Since we are interested in a separate project, isolating mutants deficient for mitochondrial function, we screened for candidate mutants on the basis of their slow-growth phenotype in the dark, a phenotypic trait of complex I-deficient mutants (9). Out of 3,000 insertional transformants, two arginine prototrophs displayed a slow-growth phenotype in the dark. Cosegregation of the slow-growth phenotype with the Arg+ trait was observed, suggesting that insertion of the ARG9 gene interrupts a gene controlling respiration. However, neither mutant was deficient for complex I activity, as determined by enzymatic measurement or in-gel staining (data not shown). We concluded that the slow-growth phenotype is probably due to a defect in other respiratory enzymes, but this was not investigated further. In conclusion, our results show that ARG9 could also be used efficiently as an insertional marker to generate nuclear mutants of interest.
Nucleotide sequence accession number.
The cDNAs sequenced in this study have been deposited in the GenBank database under accession number EU711276.
Acknowledgments
V.L. is supported by FRIA (Fonds pour la Recherche Industrielle et Appliquée). Research projects in the laboratories of P.H. and C.R. are funded by a United Mitochondrial Disease Foundation research grant, by grants 2.4638.05, 2.4601.08, and 1.5.255.08 from Fonds de la Recherche Scientifique (FRS-FNRS), and by R.CFRA.0931 (C.R). C.R. was supported by a sabbatical grant from FRS-FNRS during her stay at Ohio State University (summers 2006 and 2007).
We are grateful to J. Schrager and A. Grossman (Stanford) for providing the Chlamydomonas ARG9 cDNAs and the Arabidopsis Biological Resource Center for the Arabidopsis ARG9 cDNA.
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Auchincloss, A. H., A. I. Loroch, and J. D. Rochaix. 1999. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: cloning of the cDNA and its characterization as a selectable shuttle marker. Mol. Gen. Genet. 261:21-30. [DOI] [PubMed] [Google Scholar]
- 2.Galvan, A., D. Gonzalez-Ballester, and E. Fernandez. 2007. Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv. Exp. Med. Biol. 616:77-89. [DOI] [PubMed] [Google Scholar]
- 3.Goldschmidt-Clermont, M. 1991. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of chlamydomonas. Nucleic Acids Res. 19:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamel, P. P., B. W. Dreyfuss, Z. Xie, S. T. Gabilly, and S. Merchant. 2003. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J. Biol. Chem. 278:2593-2603. [DOI] [PubMed] [Google Scholar]
- 5.Harris, E. H. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 6.Loppes, R., and R. Heindricks. 1986. New arginine-requiring mutants in Chlamydomonas reinhardtii. Arch. Microbiol. 143:348-352. [Google Scholar]
- 7.Neupert, W., and J. M. Herrmann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723-749. [DOI] [PubMed] [Google Scholar]
- 8.Purton, S. 2007. Tools and techniques for chloroplast transformation of Chlamydomonas. Adv. Exp. Med. Biol. 616:34-45. [DOI] [PubMed] [Google Scholar]
- 9.Remacle, C., M. R. Barbieri, P. Cardol, and P. P. Hamel. 2008. Eukaryotic complex I: functional diversity and experimental systems to unravel the assembly process. Mol. Genet. Genomics 280:93-110. [DOI] [PubMed] [Google Scholar]
- 10.Slocum, R. D. 2005. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 43:729-745. [DOI] [PubMed] [Google Scholar]