For more than 30 years, we have understood that genes may be organized within genomic DNA in complex spatial arrangements. In particular, gene-coding sequences can overlap: a given segment of genomic DNA can encode more than one gene product, with the overlapping genes often oriented on opposite strands (3, 22, 44, 55, 67, 69, 71). In some cases, the overlapping genes are organized such that one gene is entirely contained within the chromosomal region occupied by another gene (25, 36). In such instances, the internal gene is referred to as a “nested” gene. Formally, a nested gene is defined as any gene whose entire coding sequence lies within the chromosomal region bounded by the start codon and stop codon of a larger external gene. Nested genes are distinct from alternatively spliced transcripts in that the coding sequence for a nested gene differs greatly from the coding sequence for its external host gene; for example, the nested gene and host do not share transcriptional start sites. This type of nested gene organization is exceptionally interesting, as it holds unique biological implications with respect to gene evolution, function, and regulation.
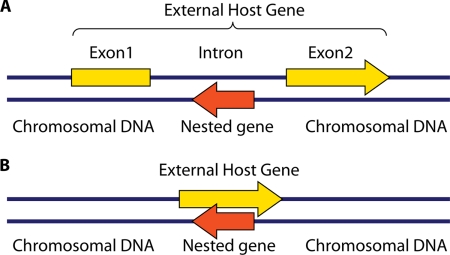
In this review, I provide an overview of nested genes with an emphasis on the occurrence of these genes in eukaryotic genomes. This review describes two principal types of nested genes: (i) genes nested within an intron of the external gene (Fig. 1A) and (ii) genes nested entirely opposite an exon or protein-coding sequence of the external gene (Fig. 1B). The first type of nested gene is fairly common, particularly in the introns of higher eukaryotes; however, the second nested gene type is quite rare, with very few observed examples of nested genes opposite protein-coding DNA in eukaryotic genomes. Each class of nested gene is discussed separately in this article, and examples of each gene type are provided. In particular, my group has been active in identifying nested genes opposite coding sequences in the budding yeast. I present two examples of nested yeast genes as a platform for the consideration of unique functional and regulatory implications associated with this model of gene organization. This review also provides a summary of the evolutionary implications associated with nested genes and a discussion of the broad significance of this gene structure for biological subdisciplines ranging from evolutionary biology to bioinformatics-based genome annotation.
FIG. 1.
Chromosomal context of a nested gene. (A) Diagram of an intronic nested gene. The nested gene is represented by a red arrow, while the external host gene is represented by a yellow arrow interrupted by an intron. (B) Diagram of a nonintronic nested gene. The nested gene and host gene are indicated as above. This review focuses on these two types of nested genes; it does not consider protein pairs that arise by alternative splicing or gene pairs that share a transcriptional start site or termination sequence.
NESTED INTRONIC GENES
A nested intronic gene at the Drosophila Gart locus.
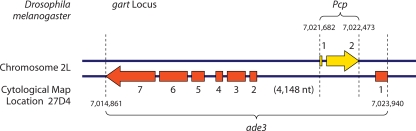
The term “nested gene” was used in the mid-1980s to describe gene organization at the Gart locus in Drosophila melanogaster (Fig. 2) (25). The Gart locus resides on the left arm of chromosome II and encodes three enzymatic activities involved in de novo purine biosynthesis (29). Subsequent to the initial studies by Henikoff and colleagues, the purine pathway gene at this locus has been renamed adenosine 3 (ade3) (65). The ade3 gene is interrupted by six introns; the 5′-most intron contains an open reading frame (ORF) of approximately 200 codons oriented antisense and opposite the ade3 coding strand. This ORF encodes a pupal cuticle protein and has accordingly been named Pupal cuticle protein (Pcp). Pcp and ade3 have no sequences in common, being derived from opposite DNA strands. The nested organization of Pcp within ade3 is not specific to D. melanogaster; Henikoff and Eghtedarzadeh (24) found this nested pattern of gene organization conserved at the orthologous locus in the divergent species Drosophila pseudoobscura.
FIG. 2.
Chromosomal gene organization at the gart locus in D. melanogaster. The arrows represent protein-coding sequences interrupted by introns for both the Pcp and ade3 genes. Chromosomal coordinates are derived from data deposited in FlyBase version FB2009_04 (http://flybase.org/).
In many respects, Pcp is a prototypical nested intronic gene. Relative to ade3, Pcp is encoded on the opposite strand and in an antisense orientation (25); this is typical, although not uniformly characteristic, of nested intronic genes. In addition, the Pcp and ade3 genes encode functionally unrelated proteins. Pcp encodes a fully functional structural constituent of the pupal chitin-based cuticle, while ade3 encodes the purine pathway enzyme activities phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, and phosphoribosylglycinamide transformylase (26). The majority of nested intronic genes are functionally unrelated to their host genes, and no functional inferences can be drawn a priori from this pattern of gene organization (72). Finally, the expression patterns of Pcp and ade3 differ: Pcp expression is restricted to the epidermis during the prepupal stage, while the ade3 gene is expressed throughout development (25). Typically, nested intronic genes are not coexpressed with their external host genes, although, again, this is not an absolute rule (2, 72).
Nested intronic genes in metazoan genomes.
Recent studies indicate that nested intronic genes are widespread in metazoan genomes (Table 1). Assis et al. (2) analyzed NCBI annotation records to identify 792 nested genes in D. melanogaster, 429 nested genes in Caenorhabditis elegans, and 233 nested genes in Caenorhabditis briggsae. In D. melanogaster, nested intronic genes constitute approximately 6% of the organism's total gene complement, and 85% of these nested genes are predicted to encode protein. The remaining D. melanogaster nested genes produce noncoding RNAs (46). Similarly, in C. elegans and in C. briggsae, the majority of nested genes are predicted to encode proteins.
TABLE 1.
Nested intronic genes in metazoan genomes
| Metazoan genome | No. of nested intronic genes | Total no. of genesc | % Nested |
|---|---|---|---|
| D. melanogaster | 792a | ∼14,601 (r5.1) | ∼5.4 |
| C. elegans | 429a | ∼20,061 (WS176) | ∼2.1 |
| C. briggsae | 233a | ∼19,500 | ∼1.2 |
| H. sapiens | 158b | ∼28,755 (r36.2) | ∼0.5 |
Nested gene counts were determined by Assis et al. (2).
Identification of nested protein-coding genes and pseudogenes in the human genome was performed by Yu et al. (70).
The genome release or reference from which each gene count was obtained is presented in parentheses after the gene count if available. These gene tallies represent best current estimates and should be viewed as approximations.
The human genome also contains a significant complement of nested intronic genes (33, 58). Yu et al. (72) identified 158 predicted protein-coding genes nested in introns within human genes, drawn from sequence data retrieved from the NCBI Map Viewer Build 34.3. Specifically, these nested genes match available expressed sequence tags and likely encode protein. Yu and colleagues identified an additional 212 human pseudogenes and three snoRNA genes nested in intronic regions (72). Thus, the human genome contains a greater proportion of nested pseudogenes than do the Drosophila and Caenorhabditis genomes. The human nested intronic genes do not exhibit an obvious bias in chromosomal distribution, and 27 genes contain multiple nested intronic genes.
Human nested intronic genes bear out many characteristics inferred from analysis of the Drosophila Pcp gene. In particular, Gene Ontology (GO) terms associated with nested genes seldom overlap with GO annotations of corresponding host genes (1). Of 106 human host genes and 96 corresponding nested genes with GO annotations, only five parallel nested gene pairs and one antiparallel pair exhibit similar functions (72). In Drosophila, Pcp is oriented antisense and opposite ade3 (25), and similar patterns of gene organization are generally observed for nested intronic genes in the human genome. Approximately 63% of human nested genes are found on the strand opposite the host gene, forming antiparallel pairs; the remaining nested genes are oriented in a parallel manner on the same strand as the host gene (72). Henikoff and colleagues identified Pcp in the largest intron of ade3. Similarly, nested genes in the human genome are typically found in large introns; the median length of an intron containing a nested gene is approximately 10-fold that of another intron from the same host gene (72).
Regulated expression of nested intronic genes.
The organization of functional DNA within an intronic region holds interesting implications with respect to the regulatory mechanisms mediating expression of the nested gene. In particular, a clear question arises as to whether these nested genes are coexpressed with their respective host genes. To consider this point using an appropriately large sample size, Yu et al. (72) analyzed human genome microarray data from the Genomics Institute of the Novartis Research Foundation (63). From these data sets, Yu and colleagues identified expression profiles for 45 protein-coding nested gene pairs but found only 3 exhibiting a positive correlation. Of this gene set, 33 gene pairs exhibited statistically significant negative correlation and 9 gene pairs showed insignificant negative correlation in expression profiles. Since these microarray data sets were generated by using a uniform technology platform, the expression profiles are presumed to be highly reliable for the comparison of expression patterns between tissues (63, 72). In independent studies, negative correlations between the expression patterns of nested noncoding RNA genes and host genes have been reported at the human eIF2A (61), Igf2r/Air (70), and α1(I) collagen loci (18).
For loci where the resulting expression patterns of nested and host genes are inversely related, Gibson and colleagues (21) posit that the transcriptional machinery traversing the host gene intron may be subject to steric hindrance from regulatory proteins interacting with the embedded gene. This interference could result in the release of one or both RNA polymerases, thereby affecting gene expression. In addition, transcriptionally convergent host and nested genes may generate nuclear double-stranded RNA from the primary transcripts, forming a substrate for RNase-mediated degradation (16, 66). The formation of double-stranded RNA by nested-host gene pairs may also contribute to RNA editing and imprinting (68). Gibson et al. (21) proposed to use atomic force microscopy (AFM) as a means of imaging individual RNA polymerases transcribing a nested gene pair. Crampton et al. (12) showed that AFM can be used to image individual RNA polymerase molecules transcribing the nested gene AMELX at the human ARH-GAP6 locus. In a separate study, Crampton and colleagues used AFM to monitor collision events between Escherichia coli RNA polymerases on a linear DNA template containing two convergently aligned promoters (11). By similar approaches, future studies should resolve existing questions regarding the transcriptional mechanisms by which nested gene pairs are expressed.
Evolution of nested intronic genes.
The widespread occurrence of nested intronic genes in eukaryotic genomes underscores the importance of understanding the mechanism by which this pattern of gene organization has arisen. Assis et al. (2) have investigated the gene structure of nested-host pairs relative to that of orthologs from vertebrate sister species as a means of inferring the mechanism of nested gene formation. They find that the vast majority of nested intronic genes emerge through the insertion of a DNA sequence into an intron of a preexisting gene. In these cases, the inserted gene arose through either gene duplication or retrotransposition; Assis and colleagues distinguished between these possibilities by searching for the presence or absence of introns in orthologs of the nested and host genes in sister species. Assis et al. also report a smaller number of cases wherein the nested gene evolved de novo from an intronic sequence of a preexisting gene (2). Analysis of the 12 recently sequenced Drosophila genomes indicates that the majority of de novo genes do originate in introns. In support of this observation, 11 nested intronic genes in D. melanogaster exhibit no sequence similarity to any genes in the closely related species D. yakuba (2).
Interestingly, nested intronic gene structures undergo preferential evolutionary gain. Comparative analyses of four representative genomes from vertebrates, fruit flies, and nematodes with closely related species indicate substantially higher rates of evolutionary gains than losses: 55 nested gene emergence events have been documented in the human genome, 52 in Drosophila, and 22 in Caenorhabditis, compared with no detected losses of nested genes in vertebrates, 17 losses in Drosophila, and 2 in Caenorhabditis (2). The acquisition of nested gene structures is thought to be a neutral process in metazoan genomes, driven by the ready availability of long intronic sequences that collectively provide a niche for gene insertion (40, 41). Thus, the evolution of animal genomes is accompanied by a steady rise in the prevalence of nested intronic gene structures, leading to increasingly complex genomic architectures (2).
GENES NESTED OPPOSITE CODING SEQUENCE
Nested genes are not restricted to introns; a rare, but arguably more interesting, type of nested gene has been identified opposite coding sequence specifying a protein or functional RNA product. This second type of nested gene is evident in prokaryotes and has been identified more recently in the budding yeast genome. Several examples are presented here, along with a discussion of the challenges involved in identifying these genes and an analysis of the functional/regulatory implications inherent in this type of nested gene structure.
Nested genes in prokaryotic genomes.
The E. coli genome contains a number of transposable DNA insertion (IS) elements, including IS1, IS2, IS4, IS5, IS903, and IS102 (4, 17, 20, 35, 36, 60). These transposable elements are noteworthy for their genetically compact gene structure. In particular, gene organization has been elucidated clearly for the mobile element IS5. The 1.195-kbp IS5 sequence contains a 978-bp ORF (gene ins5A) extending over the majority of the transposon length (52, 59). The expression of ins5A is driven from an upstream promoter located close to the right-hand end of the element (57). IS5, however, also carries two divergently transcribed genes (ins5B and ins5C) oriented opposite ins5A (57). The ins5B and ins5C genes form an operon, and this transcript is wholly contained within the boundaries of the ins5A transcript (52, 57). Thus, in IS5, two protein-coding genes are oriented opposite and entirely within a third protein-coding gene, presenting an interesting prokaryotic example of genes nested opposite protein-coding sequence.
As evidenced by this example, nested genes do exist opposite coding sequences in prokaryotic genomes, but the existence of these nested genes is not commonplace. Analysis of the E. coli K-12 genome (6) reveals only a few genes nested entirely opposite protein-coding sequences. A larger study by Pallejà et al. (50) identified gene pairs that overlap by 60 or more bp from a set of 338 sequenced prokaryotic genomes. The authors analyzed 42,055 gene overlaps of this type and found that the vast majority (92%) of these gene pairs partially overlap each other on the same strand. Nested genes were identified very infrequently; only 1% of the gene pairs overlap on opposite strands, and of these identified cases, very few genes are nested or wholly embedded opposite another gene.
Nested genes in metazoan genomes.
Genes nested opposite coding sequences have been identified extremely infrequently, if at all, in metazoan genomes. Sanna et al. (56) examined overlapping genes in the mouse genome and identified 28 gene pairs with overlapping exons; however, these overlaps are partial, in that they do not encompass the entire coding sequence of either gene. Analysis of the human genome reveals 51 exon-exon overlaps on opposite strands, but again the overlaps are partial and do not represent true nested genes. Neither the human nor the mouse genome contains any overlapping genes that share coding sequences on the same strand (56). Nested genes in D. melanogaster and C. elegans have been found exclusively as embedded sequences in introns (2, 46). Thus, at present, there exists an extreme scarcity of reports describing genes nested opposite protein-coding sequences in the genomes of higher eukaryotes.
Nested genes opposite coding sequences in yeast.
At present, two nonintronic nested genes have been identified in the genome of Saccharomyces cerevisiae: NAG1 and TAR1. The NAG1 gene is a protein-coding gene nested antisense to another protein-coding gene. TAR1 is a protein-coding gene nested opposite a tandemly repeated gene encoding a functional RNA product; unlike NAG1 and the prokaryotic nested genes mentioned above, it does not reside opposite protein-coding sequence. Interestingly, neither of these genes was readily apparent upon initial annotation of the yeast genome.
The S. cerevisiae genome was sequenced in 1996, and a first draft of its annotation was completed by using a straightforward set of rules (23, 45, 51). All previously known genes were annotated, and simple gene-finding algorithms were employed to identify putative genes from chromosomal sequence data (19). As part of this process, ORF length was used as an important criterion: ORFs at least 100 codons long were typically annotated as genes. Shorter ORFs were annotated as genes if they corresponded to known genes or exhibited sequence similarity to known proteins upon conceptual translation. Overlapping ORFs were also annotated as genes, provided they satisfied the above criteria; however, genes wholly nested or embedded within longer ORFs were excluded from consideration (19, 23, 45). By implementing these criteria, a nested ORF would only be identified as a gene if its protein-coding potential had been uncovered from previous experimental studies. No nested yeast genes had been discovered experimentally by the late 1990s, and accordingly, the initial release of the S. cerevisiae genome did not indicate any nested genes (23, 45, 51).
A similar case exists in regard to the Candida albicans genome. In annotating the C. albicans genome, nested nonintronic ORFs were not considered, unless the nested ORF exhibited sequence similarity to another gene (47). By this criterion, no nested nonintronic genes have been identified in C. albicans. The C. albicans genome contains 415 introns by recent estimate (7), but nested intronic genes are not readily apparent and have not been annotated as such (47).
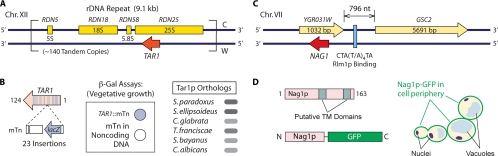
TAR1 nested in the yeast rDNA locus.
In 2002, researchers in Michael Snyder's group identified the gene TAR1 (transcript antisense to ribosomal RNA) nested antisense to the 25S rRNA gene in the rDNA repeat region of yeast chromosome XII (Fig. 3A) (10). In the S. cerevisiae genome, the rDNA cluster consists of approximately 150 tandemly repeated copies of a 9.1-kb rDNA unit located 450 kb from the left end of chromosome XII. Each rDNA unit ultimately gives rise to 5S, 5.8S, 18S, and 25S transcripts; TAR1 resides wholly opposite the 25S rRNA gene within each rDNA unit. The TAR1 gene is 124 codons in length and encodes a protein with a predicted molecular mass of 14.34 kDa. The Tar1p protein sequence is conserved in hemiascomycetous species (Fig. 3B) but does not possess clear orthologs in higher eukaryotes (10).
FIG. 3.
Nested nonintronic genes in the budding yeast genome. (A) Gene organization at the rDNA repeat locus of S. cerevisiae chromosome (Chr.) XII. The TAR1 gene (represented by the red arrow) is nested opposite and within the 25S rRNA gene. For simplicity, a single repeat unit of the rDNA locus is presented here; approximately 140 tandem copies of this unit are present on chromosome XII. (B) TAR1 encodes a protein product and is conserved among related fungi. The TAR1 gene was originally identified by transposon-based gene trapping in S. cerevisiae. Twenty-three different sites of transposon insertion identified within TAR1 are represented here by vertical blue lines along the arrow representing the TAR1 gene. Transposon insertions at each of these sites within TAR1 resulted in detectable levels of β-galactosidase (β-Gal) activity under vegetative growth conditions, as indicated by the blue circle. The protein-coding potential of TAR1 was confirmed by Western blotting, and the Tar1p product was localized to yeast mitochondria. TAR1 is conserved among hemiascomycetous species, and a sampling of these species is shown here. The similarity of the indicated orthologous sequence to that of S. cerevisiae Tar1p is indicated by the intensity of the shaded ellipse; the darkest colors indicate the strongest sequence similarity. (C) Gene organization at the YGR031W locus on S. cerevisiae chromosome VII. Transcription of the nested gene NAG1 is regulated by Rlm1p binding at the indicated site shared with the GSC2 promoter. The consensus sequence for Rlm1p binding is shown here. (D) NAG1 encodes a protein product with two predicted transmembrane (TM) domains. A Nag1p-green fluorescent protein (GFP) chimera localizes to the yeast cell periphery, consistent with its likely role as a plasma membrane protein contributing to yeast cell wall biogenesis.
The protein-coding potential of TAR1 was evident from its initial identification; TAR1 was discovered by large-scale transposon tagging by using a modified transposable element bearing a 5′-truncated lacZ reporter as a simple gene trap (Fig. 3B) (37, 38, 54). By virtue of this transposon, in-frame insertions in protein-coding genes generate a β-galactosidase translational fusion. More than 20 independent transposon insertions in TAR1 yielded detectable levels of β-galactosidase under conditions of vegetative growth, indicating that the TAR1 gene likely encodes a protein (Fig. 3B). Paulo Coelho and colleagues (10) detected a 13myc-tagged form of Tar1p by Western blotting and subsequently localized this protein to the mitochondria. Thus, TAR1 is a protein-coding gene nested opposite a structural RNA gene.
The function of TAR1 is also interesting and is consistent with the mitochondrial localization of the protein it encodes. In a set of experiments independent from the transposon-based screen described above, researchers in Gerald Shadel's group found that a low-copy plasmid bearing a partial rDNA repeat unit is capable of suppressing the petite phenotype resulting from a point mutation in the mitochondrial RNA polymerase gene RPO41 (53). Since this partial rDNA repeat unit encompasses the region encoding Tar1p, researchers in the Shadel and Snyder laboratories hypothesized that this surprising result could stem from the presence of TAR1 opposite the 25S rRNA gene. As reported by Coelho et al. (10), these groups identified TAR1 as a multicopy suppressor of the rpo41 point mutant; TAR1 rescued the growth defect of the RNA polymerase mutant strain at both 30°C and 37°C. While the exact function of TAR1 is unknown, its ability to suppress a mutation in a domain of mitochondrial RNA polymerase involved in maintaining mitochondrial DNA stability and posttranscriptional gene expression suggests that it may contribute to both of these processes. Further studies will no doubt define the functional role of TAR1 more completely.
NAG1 gene in budding yeast.
The gene NAG1 (nested antisense gene) was also identified through large-scale transposon tagging in yeast; however, unlike TAR1, the NAG1 gene represents an extremely rare eukaryotic example of a protein-coding gene wholly nested opposite another protein-coding gene. Analysis of random transposon insertions generated with the lacZ gene trap described above revealed a putative protein-coding ORF (NAG1) nested opposite the gene YGR031W on yeast chromosome VII (Fig. 3C) (42). YGR031W consists of a single uninterrupted ORF encoding a mitochondrial protein 343 amino acids in length. The NAG1 gene is 163 codons in length and encodes a 19-kDa protein, as detected by Western blotting of a hemagglutinin-tagged NAG1 allele (42). Nag1p contains two putative transmembrane segments, and a Nag1p-green fluorescent protein chimera localizes to the yeast plasma membrane (Fig. 3D). To investigate NAG1 function, we constructed a point mutation (nag1-1) that introduces a premature stop codon into the NAG1 coding sequence but is silent with respect to the opposite gene, YGR031W (42). The nag1-1 point mutant is hypersensitive to the cell wall-perturbing agent calcofluor white at 30°C and 37°C. Consistent with its apparent role in cell wall biogenesis, the phenotype caused by nag1-1 is exacerbated in the W303 strain of yeast; W303 is defective for the cell wall integrity gene SSD1 (31), and accordingly, genuine cell wall defects are often exaggerated in the W303 genetic background. Thus, the nested NAG1 gene encodes a plasma membrane protein contributing to the regulated synthesis and/or maintenance of the yeast cell wall.
NAG1 is conserved as a unit with YGR031W in several bacterial species and numerous fungi, while YGR031W is itself highly conserved in organisms ranging from bacteria to humans (42). Protein-based BLAST searches using a conceptual six-frame translation of genomic sequences do not indicate orthologs of NAG1 in higher eukaryotes, possibly owing to the function of NAG1 in cell wall biogenesis. Several fungal orthologs of NAG1 may be nonfunctional, as nonsense mutations are present in some of the identified coding sequences. As a result, additional analysis is required to determine the potential functionality of NAG1 in eukaryotes other than S. cerevisiae.
The regulated expression of NAG1 is interesting with respect to both its function and its chromosomal context. In response to calcofluor white, Nag1p levels are elevated 1.4-fold, as is typical of proteins encoded by cell wall-related genes under conditions of cell wall stress. In the budding yeast, cell wall integrity is regulated through the Slt2p mitogen-activated protein kinase cascade and its downstream transcription factor Rlm1p (14, 15, 34, 39, 73). Interestingly, the Rlm1p-regulated gene GSC2 is positioned 796 nucleotides (nt) upstream of YGR031W on the Watson strand of chromosome VII. GSC2 encodes a subunit of 1,3-β-glucan synthase that is upregulated by cell wall stress (27, 28). The GSC2 promoter contains a palindromic Rlm1p binding site that is shared with the putative promoter of NAG1 (30). Deletion of RLM1 causes a significant decrease in the production of Nag1p under conditions of vegetative growth and cell wall stress (42). It is highly likely, therefore, that both NAG1 and GSC2 are coordinately regulated from the same Rlm1 binding site. The spatial relationship between NAG1 and GSC2 is conserved in many fungi (e.g., in Kluyveromyces lactis, Vanderwaltozyma polyspora, Candida glabrata, and Debaryomyces hansenii), and the coordinated regulation of these genes may be conserved as well. The YGR031W gene, however, is regulated from a separate promoter, and its expression pattern does not correlate with the expression of NAG1.
Nested gene pair in Tetrahymena thermophila.
Recently, Zweifel et al. (74) implemented an antisense RNA-based screen for cell division defects in the ciliate T. thermophila and, from this work, identified a pair of nested protein-coding genes, CDA12 and CDA13. The 552-nt CDA12 gene consists of a single continuous ORF nested within and opposite the 591-nt CDA13 gene; CDA13 also consists of a single continuous ORF. Although neither predicted gene product exhibits strong sequence similarity to a previously annotated protein, both coding sequences specify putative transmembrane domains and may have related functions in membrane trafficking. Cda12p localizes to diverse membrane-bound compartments, including perinuclear vesicles and recycling endosomes. Cda13p localizes to vesicular structures adjacent to both the endoplasmic reticulum-Golgi body system and the associated cortical mitochondria. Consistent with these diverse localizations, antisense RNA-mediated inhibition of CDA12 and CDA13 expression yields defects in cytokinesis, macronuclear segregation, pinosome processing, and conjugation (74). Interestingly, both genes can be actively transcribed during conjugation, although only CDA12 produces detectable transcript under conditions of vegetative growth. In total, the CDA12/CDA13 locus presents a strong eukaryotic example of a nested gene pair exhibiting both similar expression patterns and broadly related cellular functions.
FINDING NESTED GENES: CHALLENGES AND PROSPECTS
Obstacles to the identification of nested genes.
Nested genes, and in particular nonintronic nested genes, have largely escaped the attention of many researchers—a fact that principally stems from the difficulties encountered in identifying these genes and their resulting underrepresentation in annotated genome sequences. At present, gene annotation is a predominantly computational process wherein gene-finding algorithms and simple criteria are used to discriminate between chance ORFs and genuine protein-coding genes. These algorithms and criteria are relatively ineffective in identifying nonintronic nested genes. For example, neither the NAG1 nor the TAR1 gene was identified as a protein-coding ORF upon annotation of the yeast genome. The decision to exclude such nested and nonintronic ORFs from consideration during the gene identification process exacerbates the situation; however, this decision is understandable. Because of biases in nucleotide usage associated with codons, ORF sequences are statistically likely to contain antisense ORFs on the opposite strand (43). Thus, even the simplest eukaryotic genomes contain an extremely large number of antisense ORFs, the vast majority of which do not encode proteins. It is not practical at present to consider all nested antisense ORFs for annotation, and decisions to exclude nested genes are cost-effective but nonetheless compromise the ultimate accuracy of gene identification during genome annotation.
Without a large training set of nested antisense gene sequences for the development of new computational gene-finding methods, in silico approaches are unlikely to be effective as a means of discriminating between true nested genes and chance antisense ORFs; instead, experimental approaches provide a significantly better means to identify nested, nonintronic genes. Transposon-based gene traps have been used successfully to identify previously nonannotated genes such as TAR1 and NAG1 in yeast (10, 42). Experimental approaches using modified forms of antisense RNA-based technologies may be effective in higher eukaryotes for this purpose (64, 74). Transcriptional profiling studies using tiling microarrays and deep sequencing technologies have proven extremely useful in identifying transcribed regions of eukaryotic genomes (5, 8, 9, 13, 32, 48, 49, 62); however, since many antisense sequences may be transcribed as regulatory RNAs, this approach would presumably be less effective for the identification of protein-coding genes. Mass spectrometric methods offer a promising avenue toward the experimental identification of proteins, and hence protein-coding genes, for a given organism. For this to be achieved, technical obstacles limiting both the scale and the quality of protein detection need to be overcome, but proteomic methods are potentially exciting as a means of achieving more comprehensive levels of gene identification.
Do additional nested genes exist in eukaryotic genomes?
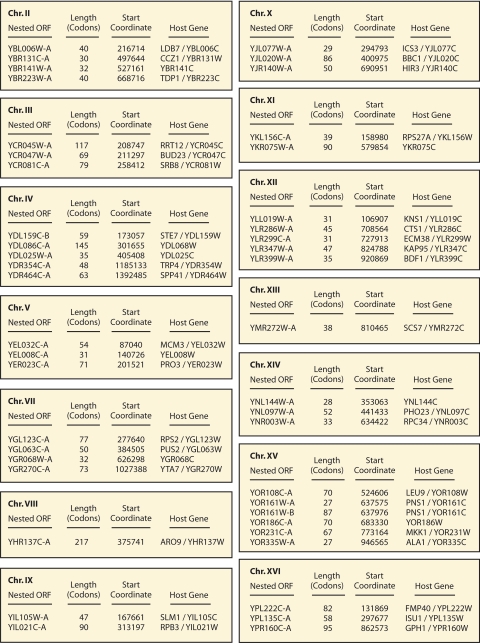
In the budding yeast, a substantial body of evidence suggests that additional nested antisense genes may be present. The gene-trapping study that ultimately resulted in the identification of NAG1 and TAR1 (38, 42, 54) also indicated at least 45 additional nested ORFs producing detectable levels of β-galactosidase activity upon in-frame insertion of a modified transposon bearing a truncated form of lacZ (37). The chromosomal location and systematic name of each nested ORF are indicated in Fig. 4. For these putative genes, β-galactosidase levels were detected under vegetative growth conditions from transposon-mediated lacZ fusions. Each nested ORF was identified by multiple independent transposon insertions and/or by strong levels of β-galactosidase activity. In addition, transcripts corresponding to each of these nested ORFs were detected by large-scale RNA sequencing, as presented by Nagalakshmi et al. (49); the likelihood that these sequences are expressed is therefore very high. These nested and nonintronic ORFs range from 27 to 215 codons in length, with moderate-to-strong expression levels as detected by RNA array analysis using strand-specific oligonucleotide probes (37). The putative nested genes, however, are not uniformly well conserved in related yeast species. Thus, additional studies are needed to verify the protein-coding potential of these nested ORFs, but the outlook remains promising for the confirmation of these and/or other putative nonintronic nested genes in S. cerevisiae.
FIG. 4.
ORFs in S. cerevisiae that may encode nonintronic nested genes were identified by large-scale gene-trapping and strand-specific expression analyses. The putative nested gene is indicated, along with the length of the ORF in codons, its chromosomal start coordinate, and the host gene name. Chromosomal coordinates were determined from sequence deposited in the Saccharomyces Genome Database as of May 2009 (http://www.yeastgenome.org/). This gene list is not comprehensive but rather presents a starting point for the continued identification of nonintronic nested genes in S. cerevisiae. Chr., chromosome.
The experimental approaches implemented by Zweifel et al. (74) for the identification of the nested gene pair CDA12/CDA13 also suggest the presence of additional nonintronic nested genes in Tetrahymena; in fact, Zweifel and colleagues speculate that the occurrence of these nested genes may be relatively common. The identification of nonintronic nested genes in metazoan genomes, however, may not be quite as imminent, as some of the technical obstacles limiting the throughput of mutational studies will complicate attempts to identify nested genes in these organisms. In this regard, antisense RNA-based gene knockdowns and mass spectrometric approaches are viable means to identify genes without prior in silico annotation in metazoan genomes. In total, nonintronic nested genes are likely present in many, if not most, eukaryotes.
Functional benefits of nested genes.
In prokaryotes, it is generally assumed that a compact genome is advantageous, conferring a selective advantage due to the speed with which DNA replication can be carried out. In eukaryotes, no such size constraints are in place. Thus, an open question remains why nested genes exist in eukaryotic genomes.
We can only speculate regarding the functional benefits of nested eukaryotic genes. As evidenced in some of the examples presented here, the expression of nested genes may be correlated with host genes—both positively and negatively. The nested-host gene structure presents a mechanism for the coordinated regulation of functionally related gene pairs. However, many nested genes exhibit functions and expression patterns distinct from those of host genes; thus, this cannot be an overriding principle governing nested gene structures. Most likely, the acquisition of a nested gene is an evolutionarily neutral process facilitated by the presence of numerous introns in eukaryotic genes that present a niche for gene insertion (2). Accordingly, in many instances, the nested gene is an evolutionary remnant, and collectively, it is difficult to derive a generally applicable functional benefit associated with the nested gene structure.
The need to identify nested genes.
Genome sequences are available in draft or finished form for an expanding set of at least 5,400 species (http://www.ncbi.nlm.nih.gov). This knowledge base of sequence data is extremely valuable, but its practical utility depends, in part, upon the accuracy with which the encompassed genome sequences are annotated. In order to accurately annotate a genome, we must, of course, accurately annotate nested genes. By overlooking a nested gene, we raise the likelihood of incorrectly assigning gene functions. For example, any observed phenotypes resulting from the deletion of a genetic locus containing a previously overlooked nested gene would be incorrectly assumed to identify functions strictly for the external host gene. Also, the presence of a nested gene holds potential evolutionary implications for the host gene, and this information may be useful in interpreting comparative genomic results for a given locus. Finally, it is difficult to accurately determine the regulatory control of a given locus without accurately identifying the constituent genes; as indicated by studies of the yeast NAG1 locus, the regulatory mechanisms controlling a nested gene may be coordinated with regulatory events affecting a neighboring gene. Nested genes potentially represent a wealth of overlooked biology, and the identification of these genes and their encoded functions promises to clarify underlying biological processes across a diverse spectrum of eukaryotes.
SUMMARY
Nested genes fall into two broad categories: (i) genes nested within the intron of another gene and (ii) nonintronic genes nested opposite coding sequence for the host gene. Nested intronic genes have been identified in many eukaryotes and are fairly common, with at least 158 protein-coding genes nested within introns in the human genome. These nested intronic genes are not necessarily functionally related to their host genes; also, expression patterns of nested intronic genes are most frequently noncorrelated or inversely correlated with respect to host genes, at least in the human genome. Nested intronic genes have emerged predominantly through insertion of a DNA sequence into the intron of a preexisting gene. Nonintronic nested genes are rare but have been identified in prokaryotes, yeast, and Tetrahymena. The sample size is extremely small, but as with intronic nested genes, nonintronic nested genes do not necessarily share functions or expression patterns with host genes. Nested genes are difficult to identify in silico without prior experimental evidence, and even experimental approaches must be designed carefully to distinguish a nested gene from its opposite host gene. Ultimately, however, it is critical to identify nested genes as a means of ensuring accurate genome annotation for subsequent experimental analysis.
Acknowledgments
Research in the Kumar laboratory is supported by grants RSG-06-179-01-MBC from the American Cancer Society and DBI 0543017 from the National Science Foundation.
I thank Craig J. Dobry and Randy Bo-Bandy for critically reading the manuscript.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assis, R., A. S. Kondrashov, E. V. Koonin, and F. A. Kondrashov. 2008. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 24:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrell, B. G., G. M. Air, and C. A. Hutchison III. 1976. Overlapping genes in bacteriophage phiX174. Nature 264:34-41. [DOI] [PubMed] [Google Scholar]
- 4.Bernardi, A., and F. Bernardi. 1981. Complete sequence of an IS element present in pSC101. Nucleic Acids Res. 9:2905-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertone, P., V. Stolc, T. E. Royce, J. S. Rozowsky, A. E. Urban, X. Zhu, J. L. Rinn, W. Tongprasit, M. Samanta, S. Weissman, M. Gerstein, and M. Snyder. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306:2242-2246. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. R., M. van Het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morschhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V. B. Bajic, S. E. Brenner, S. Batalov, A. R. Forrest, M. Zavolan, M. J. Davis, L. G. Wilming, V. Aidinis, J. E. Allen, A. Ambesi-Impiombato, R. Apweiler, R. N. Aturaliya, T. L. Bailey, M. Bansal, L. Baxter, K. W. Beisel, T. Bersano, H. Bono, A. M. Chalk, K. P. Chiu, V. Choudhary, A. Christoffels, D. R. Clutterbuck, M. L. Crowe, E. Dalla, B. P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C. F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T. R. Gingeras, T. Gojobori, R. E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T. K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S. P. Krishnan, A. Kruger, S. K. Kummerfeld, I. V. Kurochkin, L. F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, F. Nori, O. Ohara, Y. Okazaki, V. Orlando, K. C. Pang, W. J. Pavan, G. Pavesi, G. Pesole, N. Petrovsky, S. Piazza, J. Reed, J. F. Reid, B. Z. Ring, M. Ringwald, B. Rost, Y. Ruan, S. L. Salzberg, A. Sandelin, C. Schneider, C. Schonbach, K. Sekiguchi, C. A. Semple, S. Seno, L. Sessa, Y. Sheng, Y. Shibata, H. Shimada, K. Shimada, D. Silva, B. Sinclair, S. Sperling, E. Stupka, K. Sugiura, R. Sultana, Y. Takenaka, K. Taki, K. Tammoja, S. L. Tan, S. Tang, M. S. Taylor, J. Tegner, S. A. Teichmann, H. R. Ueda, E. van Nimwegen, R. Verardo, C. L. Wei, K. Yagi, H. Yamanishi, E. Zabarovsky, S. Zhu, A. Zimmer, W. Hide, C. Bult, S. M. Grimmond, R. D. Teasdale, E. T. Liu, V. Brusic, J. Quackenbush, C. Wahlestedt, J. S. Mattick, D. A. Hume, C. Kai, D. Sasaki, Y. Tomaru, S. Fukuda, M. Kanamori-Katayama, M. Suzuki, J. Aoki, T. Arakawa, J. Iida, K. Imamura, M. Itoh, T. Kato, H. Kawaji, N. Kawagashira, T. Kawashima, M. Kojima, S. Kondo, H. Konno, K. Nakano, N. Ninomiya, T. Nishio, M. Okada, C. Plessy, K. Shibata, T. Shiraki, S. Suzuki, M. Tagami, K. Waki, A. Watahiki, Y. Okamura-Oho, H. Suzuki, J. Kawai, and Y. Hayashizaki. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, J., P. Kapranov, J. Drenkow, S. Dike, S. Brubaker, S. Patel, J. Long, D. Stern, H. Tammana, G. Helt, V. Sementchenko, A. Piccolboni, S. Bekiranov, D. K. Bailey, M. Ganesh, S. Ghosh, I. Bell, D. S. Gerhard, and T. R. Gingeras. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149-1154. [DOI] [PubMed] [Google Scholar]
- 10.Coelho, P. S., A. C. Bryan, A. Kumar, G. S. Shadel, and M. Snyder. 2002. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 16:2755-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crampton, N., W. A. Bonass, J. Kirkham, C. Rivetti, and N. H. Thomson. 2006. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 34:5416-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crampton, N., N. H. Thomson, J. Kirkham, C. W. Gibson, and W. A. Bonass. 2006. Imaging RNA polymerase-amelogenin gene complexes with single molecule resolution using atomic force microscopy. Eur. J. Oral Sci. 114(Suppl. 1):133-138; discussion, 164-165, 380-381. [DOI] [PubMed] [Google Scholar]
- 13.David, L., W. Huber, M. Granovskaia, J. Toedling, C. J. Palm, L. Bofkin, T. Jones, R. W. Davis, and L. M. Steinmetz. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 103:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146(Pt. 9):2121-2132. [DOI] [PubMed] [Google Scholar]
- 15.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 17.Engler, J. A., and M. P. van Bree. 1981. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene 14:155-163. [DOI] [PubMed] [Google Scholar]
- 18.Farrell, C. M., and L. N. Lukens. 1995. Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the α1(I) collagen gene. J. Biol. Chem. 270:3400-3408. [DOI] [PubMed] [Google Scholar]
- 19.Fisk, D. G., C. A. Ball, K. Dolinski, S. R. Engel, E. L. Hong, L. Issel-Tarver, K. Schwartz, A. Sethuraman, D. Botstein, and J. M. Cherry. 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23:857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosal, D., H. Sommer, and H. Saedler. 1979. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 6:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson, C. W., N. H. Thomson, W. R. Abrams, and J. Kirkham. 2005. Nested genes: biological implications and use of AFM for analysis. Gene 350:15-23. [DOI] [PubMed] [Google Scholar]
- 22.Godson, G. N., B. G. Barrell, R. Staden, and J. C. Fiddes. 1978. Nucleotide sequence of bacteriophage G4 DNA. Nature 276:236-247. [DOI] [PubMed] [Google Scholar]
- 23.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 24.Henikoff, S., and M. K. Eghtedarzadeh. 1987. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics 117:711-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff, S., M. A. Keene, K. Fechtel, and J. W. Fristrom. 1986. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell 44:33-42. [DOI] [PubMed] [Google Scholar]
- 26.Henikoff, S., M. A. Keene, J. S. Sloan, J. Bleskan, R. Hards, and D. Patterson. 1986. Multiple purine pathway enzyme activities are encoded at a single genetic locus in Drosophila. Proc. Natl. Acad. Sci. USA 83:720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue, S. B., N. Takewaki, T. Takasuka, T. Mio, M. Adachi, Y. Fujii, C. Miyamoto, M. Arisawa, Y. Furuichi, and T. Watanabe. 1995. Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231:845-854. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone, M. E., D. Nash, and F. N. Naguib. 1985. Three purine auxotrophic loci on the second chromosome of Drosophila melanogaster. Biochem. Genet. 23:539-555. [DOI] [PubMed] [Google Scholar]
- 30.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781-789. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein, M., and L. Guarente. 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160:83-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapranov, P., A. T. Willingham, and T. R. Gingeras. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8:413-423. [DOI] [PubMed] [Google Scholar]
- 33.Karlin, S., C. Chen, A. J. Gentles, and M. Cleary. 2002. Associations between human disease genes and overlapping gene groups and multiple amino acid runs. Proc. Natl. Acad. Sci. USA 99:17008-17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaer, R., S. Kuhn, E. Tillmann, H. J. Fritz, and P. Starlinger. 1981. The sequence of IS4. Mol. Gen. Genet. 181:169-175. [DOI] [PubMed] [Google Scholar]
- 36.Kröger, M., and G. Hobom. 1982. Structural analysis of insertion sequence IS5. Nature 297:159-162. [DOI] [PubMed] [Google Scholar]
- 37.Kumar, A., P. M. Harrison, K. H. Cheung, N. Lan, N. Echols, P. Bertone, P. Miller, M. B. Gerstein, and M. Snyder. 2002. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 20:58-63. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, A., M. Seringhaus, M. Biery, R. J. Sarnovsky, L. Umansky, S. Piccirillo, M. Heidtman, K.-H. Cheung, C. J. Dobry, M. Gerstein, N. Craig, and M. Snyder. 2004. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon. Genome Res. 14:1975-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 13:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch, M. 2002. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. USA 99:6118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch, M., and J. S. Conery. 2003. The origins of genome complexity. Science 302:1401-1404. [DOI] [PubMed] [Google Scholar]
- 42.Ma, J., C. J. Dobry, D. J. Krysan, and A. Kumar. 2008. Unconventional genomic architecture in the budding yeast Saccharomyces cerevisiae masks the nested antisense gene NAG1. Eukaryot. Cell 7:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackiewicz, P., M. Kowalczuk, A. Gierlik, M. R. Dudek, and S. Cebrat. 1999. Origin and properties of non-coding ORFs in the yeast genome. Nucleic Acids Res. 27:3503-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makalowska, I., C. F. Lin, and W. Makalowski. 2005. Overlapping genes in vertebrate genomes. Comput. Biol. Chem. 29:1-12. [DOI] [PubMed] [Google Scholar]
- 45.Mewes, H. W., K. Albermann, M. Bähr, D. Frishman, A. Gleissner, J. Hani, K. Heumann, K. Kleine, A. Maierl, S. G. Oliver, F. Pfeiffer, and A. Zollner. 1997. Overview of the yeast genome. Nature 387:7-65. [DOI] [PubMed] [Google Scholar]
- 46.Misra, S., M. A. Crosby, C. J. Mungall, B. B. Matthews, K. S. Campbell, P. Hradecky, Y. Huang, J. S. Kaminker, G. H. Millburn, S. E. Prochnik, C. D. Smith, J. L. Tupy, E. J. Whitfied, L. Bayraktaroglu, B. P. Berman, B. R. Bettencourt, S. E. Celniker, A. D. de Grey, R. A. Drysdale, N. L. Harris, J. Richter, S. Russo, A. J. Schroeder, S. Q. Shu, M. Stapleton, C. Yamada, M. Ashburner, W. M. Gelbart, G. M. Rubin, and S. E. Lewis. 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3:RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitrovich, Q. M., B. B. Tuch, C. Guthrie, and A. D. Johnson. 2007. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 17:492-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura, F., N. Kawaguchi, J. Sese, A. Toyoda, M. Hattori, S. Morishita, and T. Ito. 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA 103:17846-17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagalakshmi, U., Z. Wang, K. Waern, C. Shou, D. Raha, M. Gerstein, and M. Snyder. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pallejà, A., E. D. Harrington, and P. Bork. 2008. Large gene overlaps in prokaryotic genomes: result of functional constraints or mispredictions? BMC Genomics 9:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Philippsen, P., K. Kleine, R. Pohlmann, A. Dusterhoft, K. Hamberg, J. H. Hegemann, B. Obermaier, L. A. Urrestarazu, R. Aert, K. Albermann, R. Altmann, B. Andre, V. Baladron, J. P. Ballesta, A. M. Becam, J. Beinhauer, J. Boskovic, M. J. Buitrago, F. Bussereau, F. Coster, M. Crouzet, M. D'Angelo, F. Dal Pero, A. De Antoni, J. Hani, et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature 387:93-98. [PubMed] [Google Scholar]
- 52.Rak, B., M. Lusky, and M. Hable. 1982. Expression of two proteins from overlapping and oppositely oriented genes on transposable DNA insertion element IS5. Nature 297:124-128. [DOI] [PubMed] [Google Scholar]
- 53.Rodeheffer, M. S., B. E. Boone, A. C. Bryan, and G. S. Shadel. 2001. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 276:8616-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 55.Sanger, F., G. M. Air, B. G. Barrell, N. L. Brown, A. R. Coulson, C. A. Fiddes, C. A. Hutchison, P. M. Slocombe, and M. Smith. 1977. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265:687-695. [DOI] [PubMed] [Google Scholar]
- 56.Sanna, C. R., W. H. Li, and L. Zhang. 2008. Overlapping genes in the human and mouse genomes. BMC Genomics 9:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawers, R. G. 2005. Transcript analysis of Escherichia coli K-12 insertion element IS5. FEMS Microbiol. Lett. 244:397-401. [DOI] [PubMed] [Google Scholar]
- 58.Scherer, S. W., J. Cheung, J. R. MacDonald, L. R. Osborne, K. Nakabayashi, J. A. Herbrick, A. R. Carson, L. Parker-Katiraee, J. Skaug, R. Khaja, J. Zhang, A. K. Hudek, M. Li, M. Haddad, G. E. Duggan, B. A. Fernandez, E. Kanematsu, S. Gentles, C. C. Christopoulos, S. Choufani, D. Kwasnicka, X. H. Zheng, Z. Lai, D. Nusskern, Q. Zhang, Z. Gu, F. Lu, S. Zeesman, M. J. Nowaczyk, I. Teshima, D. Chitayat, C. Shuman, R. Weksberg, E. H. Zackai, T. A. Grebe, S. R. Cox, S. J. Kirkpatrick, N. Rahman, J. M. Friedman, H. H. Heng, P. G. Pelicci, F. Lo-Coco, E. Belloni, L. G. Shaffer, B. Pober, C. C. Morton, J. F. Gusella, G. A. Bruns, B. R. Korf, B. J. Quade, A. H. Ligon, H. Ferguson, A. W. Higgins, N. T. Leach, S. R. Herrick, E. Lemyre, C. G. Farra, H. G. Kim, A. M. Summers, K. W. Gripp, W. Roberts, P. Szatmari, E. J. Winsor, K. H. Grzeschik, A. Teebi, B. A. Minassian, J. Kere, L. Armengol, M. A. Pujana, X. Estivill, M. D. Wilson, B. F. Koop, S. Tosi, G. E. Moore, A. P. Boright, E. Zlotorynski, B. Kerem, P. M. Kroisel, E. Petek, D. G. Oscier, S. J. Mould, H. Dohner, K. Dohner, J. M. Rommens, J. B. Vincent, J. C. Venter, P. W. Li, R. J. Mural, M. D. Adams, and L. C. Tsui. 2003. Human chromosome 7: DNA sequence and biology. Science 300:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnetz, K., and B. Rak. 1992. IS5: a mobile enhancer of transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoner, B., and M. Kahn. 1981. The nucleotide sequence of IS5 from Escherichia coli. Gene 14:165-174. [DOI] [PubMed] [Google Scholar]
- 61.Silverman, T. A., M. Noguchi, and B. Safer. 1992. Role of sequences within the first intron in the regulation of expression of eukaryotic initiation factor 2 alpha. J. Biol. Chem. 267:9738-9742. [PubMed] [Google Scholar]
- 62.Stolc, V., Z. Gauhar, C. Mason, G. Halasz, M. F. van Batenburg, S. A. Rifkin, S. Hua, T. Herreman, W. Tongprasit, P. E. Barbano, H. J. Bussemaker, and K. P. White. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306:655-660. [DOI] [PubMed] [Google Scholar]
- 63.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweeney, R., Q. Fan, and M. C. Yao. 1996. Antisense ribosomes: rRNA as a vehicle for antisense RNAs. Proc. Natl. Acad. Sci. USA 93:8518-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiong, S. Y., and D. Nash. 1990. Genetic analysis of the adenosine3 (Gart) region of the second chromosome of Drosophila melanogaster. Genetics 124:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanhée-Brossollet, C., and C. Vaquero. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211:1-9. [DOI] [PubMed] [Google Scholar]
- 67.Veeramachaneni, V., W. Makalowski, M. Galdzicki, R. Sood, and I. Makalowska. 2004. Mammalian overlapping genes: the comparative perspective. Genome Res. 14:280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, Q., and G. G. Carmichael. 2004. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 68:432-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams, B. A., C. H. Slamovits, N. J. Patron, N. M. Fast, and P. J. Keeling. 2005. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc. Natl. Acad. Sci. USA 102:10936-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wutz, A., O. W. Smrzka, N. Schweifer, K. Schellander, E. F. Wagner, and D. P. Barlow. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745-749. [DOI] [PubMed] [Google Scholar]
- 71.Yelin, R., D. Dahary, R. Sorek, E. Y. Levanon, O. Goldstein, A. Shoshan, A. Diber, S. Biton, Y. Tamir, R. Khosravi, S. Nemzer, E. Pinner, S. Walach, J. Bernstein, K. Savitsky, and G. Rotman. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21:379-386. [DOI] [PubMed] [Google Scholar]
- 72.Yu, P., D. Ma, and M. Xu. 2005. Nested genes in the human genome. Genomics 86:414-422. [DOI] [PubMed] [Google Scholar]
- 73.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15:83-91. [PMC free article] [PubMed] [Google Scholar]
- 74.Zweifel, E., J. Smith, D. Romero, T. H. Giddings, Jr., M. Winey, J. Honts, J. Dahlseid, B. Schneider, and E. S. Cole. 2009. Nested genes CDA12 and CDA13 encode proteins associated with membrane trafficking in the ciliate Tetrahymena thermophila. Eukaryot. Cell 8:899-912. [DOI] [PMC free article] [PubMed] [Google Scholar]