Abstract
The understanding of the molecular basis of yeast resistance to ethanol may guide the design of rational strategies to increase process performance in industrial alcoholic fermentations. In this study, the yeast disruptome was screened for mutants with differential susceptibility to stress induced by high ethanol concentrations in minimal growth medium. Over 250 determinants of resistance to ethanol were identified. The most significant gene ontology terms enriched in this data set are those associated with intracellular organization, biogenesis, and transport, in particular, regarding the vacuole, the peroxisome, the endosome, and the cytoskeleton, and those associated with the transcriptional machinery. Clustering the proteins encoded by the identified determinants of ethanol resistance by their known physical and genetic interactions highlighted the importance of the vacuolar protein sorting machinery, the vacuolar H+-ATPase complex, and the peroxisome protein import machinery. Evidence showing that vacuolar acidification and increased resistance to the cell wall lytic enzyme β-glucanase occur in response to ethanol-induced stress was obtained. Based on the genome-wide results, the particular role of the FPS1 gene, encoding a plasma membrane aquaglyceroporin which mediates controlled glycerol efflux, in ethanol stress resistance was further investigated. FPS1 expression contributes to decreased [3H]ethanol accumulation in yeast cells, suggesting that Fps1p may also play a role in maintaining the intracellular ethanol level during active fermentation. The increased expression of FPS1 confirmed the important role of this gene in alcoholic fermentation, leading to increased final ethanol concentration under conditions that lead to high ethanol production.
Saccharomyces cerevisiae and related yeast species have been extensively used in fermentation, wine making, sake making, and brewing processes. Bioethanol production by yeast is also a growing industry due to energy and environmental demands (38). The successful performance of alcoholic fermentations depends on the ability of the yeast strains used to cope with a number of stress factors occurring during the process (45, 48). These include osmotic pressure imposed by initial high sugar concentration and stress induced by fermentation end products or subproducts such as ethanol and acetate. However, the stress induced by increasing amounts of ethanol, accumulated to toxic concentrations during ethanolic fermentation, is the major factor responsible for reduced ethanol production and, eventually, for stuck fermentations (14). Thus, yeast strains that can endure stress imposed by high ethanol concentrations are highly desired.
Throughout the years many efforts to characterize the mechanisms underlying ethanol stress tolerance, aiming to increase ethanol productivity, have been made (3, 16, 45, 53). The successful engineering of yeast transcription machinery for this purpose was recently reported (3). A number of studies based on detailed physiological and molecular analyses have contributed to increasing the understanding of the processes underlying ethanol toxicity and yeast tolerance of stress induced by this metabolite (34-36, 45). These studies indicate that ethanol interferes with membrane lipid organization, affecting its function as a matrix for enzymes, perturbing the conformation and function of membrane transporters, increasing the nonspecific plasma membrane permeability, and leading to the dissipation of transmembrane electrochemical potential (36, 45). Concomitantly, yeast responses to ethanol-induced stress include changes in the levels and composition of membrane phospholipids and ergosterol (1, 6, 53). Through its effect at the level of plasma membrane organization and function, ethanol also produces intracellular acidification (26, 34-36). In response to this effect, yeast exhibits increased plasma membrane H+-ATPase activity, which is important to maintain the intracellular pH and secondary transport mechanisms, which are dependent on the proton gradient across the plasma membrane (1, 29, 31, 34). Ethanol has also been shown to inhibit crucial glycolytic enzymes (5) and to induce the generation of reactive oxygen species (7, 11); yeast tolerance is dependent on the stability of the mitochondrial genome (18) and on the activity of the mitochondrial superoxide dismutase, encoded by SOD2 (7). Another factor contributing to the protection of yeast cells is cell wall composition and structure (31), although the exact changes occurring at this level are not well characterized.
A deeper understanding of the molecular mechanisms underlying yeast tolerance of ethanol stress is expected to increase the ability to delineate the genetic engineering of yeast strains and process strategies aiming at increasing alcoholic fermentation performance. In recent years, genome-wide surveys have been used to identify the genes involved in the yeast response and resistance to ethanol stress (2, 13, 16, 21, 46, 52). Specifically, four of these global studies were focused on the analysis of the yeast disruptome in yeast extract-peptone-dextrose (YPD) rich medium, both in solid (13, 21, 46) and liquid (52) media. In all four cases, only nonessential genes were considered. Each screening used slightly different genetic backgrounds. Nonetheless, the small differences in the approaches pursued in these previous studies led to a low level of overlap in the results, only four genes being observed in all experiments to confer ethanol resistance. This discrepancy seems to suggest that the expression of ethanol tolerance determinants is highly variable depending of the strain and growth condition. Thus, in the present work we decided to screen the yeast disruptome in minimal medium and to extend the previous analyses to include also yeast essential genes. To do this, we tested a collection of BY4741-derived haploid single-deletion mutants in which each nonessential yeast gene was individually deleted and a BY4743-derived collection of heterozygous diploid strains in which one of the two copies of each essential yeast gene was individually deleted. This chemogenomics approach led to the identification of over 250 determinants of yeast resistance to stress induced by high ethanol concentrations, most of them encoding proteins involved in intracellular trafficking, transcription, pH homeostasis, and peroxisome biogenesis. These include 121 new determinants of yeast resistance to ethanol-induced stress. Based on the genome-wide results, the effects of ethanol on peroxisome proliferation, vacuolar acidification, and cell wall resistance were assessed. Furthermore, the role of one of the most interesting determinants of yeast resistance to ethanol stress identified in this study, the FPS1 gene, encoding the plasma membrane aquaglycerolporin (43), was studied in more detail. Aquaglyceroporins facilitate transmembrane transport of small uncharged molecules like polyols and urea, which play important roles in osmoregulation and nutrient uptake (32). Under hyperosmotic shock, Fps1 activity is reduced, leading to glycerol accumulation, whereas upon shifting back to hypo-osmotic conditions the Fps1 channel opens to release glycerol and thus relieve turgor pressure (17, 43). Fps1 has also been shown to facilitate the diffusion of toxic compounds across the yeast plasma membrane, including arsenite and antimonite (51), acetic acid (28), and boron (30). In the present study, the participation of Fps1 in reducing the intracellular concentration of ethanol in yeast cells was evaluated. The manner in which FPS1 expression increases yeast ability to achieve high final concentrations of ethanol during alcoholic fermentation was also analyzed.
MATERIALS AND METHODS
Strains and plasmids.
The parental strains Saccharomyces cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0) and the Euroscarf collection of BY4741-derived haploid mutant strains, with all nonessential open reading frames (ORFs) individually deleted, and BY4743-derived diploid heterozygous mutant strains, in which one of the two copies of each essential gene was individually deleted (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/), were used for the chemogenomics analysis carried out in this study. Strains W303-1A (MATa ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 GAL SUC2 mal0) and the derived Δfps1 deletion mutant were used for studies focused on the FPS1 role in ethanol stress resistance. Plasmid pYEP-FPS1, expressing the FPS1 gene, and the corresponding cloning vector were also used for complementation assays (43).
Genome-wide screening for deletion mutants with differential susceptibility to ethanol-induced stress.
To screen the entire Euroscarf deletion mutant collection for sensitivity to high concentrations of ethanol, the different strains were grown in 96-well plates in MM4 medium, at pH 4.0, 30°C, and 250 rpm, for 24 h under aerobic conditions. MM4 liquid medium contains, per liter, 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco Laboratories, Detroit, MI) and 20 g glucose, 2.65 g (NH4)2SO4, 20 mg methionine, 20 mg histidine, 60 mg leucine, 20 mg uracil, 40 mg tryptophan, and 30 mg lysine, all from Sigma (Spain). A 96-pin replica plater was used to spot these cells onto the surface of MM4 solid medium, which contained, besides the above-indicated ingredients, 20 g/liter agar (Iberagar), supplemented or not with ethanol. To ensure that ethanol concentration was kept constant in all tests, fresh plates were used in every assay. The final concentrations tested were 8% (vol/vol) for the haploid mutant collection and 12% (vol/vol) for the diploid mutant collection. Ethanol concentrations leading to moderate growth inhibition of the wild-type strain were chosen based on preliminary testing. Different concentrations had to be selected for the haploid and diploid strains since the diploid strain is intrinsically more resistant. Susceptibility phenotypes were registered after incubation at 30°C for 2 to 3 days, depending on the severity of growth inhibition. At least two independent replicates were obtained for each set of mutants, and results were confirmed by low-throughput spot assays. The eventual over- or underrepresentation of gene ontology (GO) terms associated with our data set, compared to the yeast genome, was determined using GOToolBox (24). The hypergeometric test was used to analyze the data, and enrichment was considered for P values below 0.01. The interaction networks predicted to be established among the determinants of yeast resistance to ethanol were obtained using OSPREY software, applied to the BIOGRID database (http://www.thebiogrid.org/), which takes into account physical and genetic associations between yeast proteins and genes, respectively.
Ethanol susceptibility assays.
The susceptibility of the parental strain W303-1A and derived Δfps1 deletion mutant, harboring the pYEPmyc-FPS1 plasmid or the corresponding empty vector, to toxic concentrations of ethanol was assessed by comparing curves for growth of the strains in MM4-Leu medium supplemented or not with 6% ethanol. Cell suspensions used to prepare the inocula for growth curves were grown in MM4-Leu medium until mid-exponential phase (optical density at 600 nm [OD600] = 0.4 ± 0.05). Cell growth in liquid media was in Erlenmeyer flasks, at 30°C and 250 rpm, and was followed by measuring culture OD600 during batch cultivation.
pHv assessment.
Vacuolar pH (pHv) values were assessed by fluorescence microscopy using the pH-sensing fluorescent probe 5-(and 6-)carboxy-2,7-dichlorofluorescein diacetate, succinimidyl-ester [5(6)-CDCFDA,SE]. This technique makes use of a membrane-permeable fluorescent probe which accumulates in the yeast vacuole and undergoes pH-dependent changes, in both its absorption and fluorescence emission spectra, over a wide range of pH values (33). This property has been exploited to successfully determine pHv in yeast cells (9, 12, 33) and also to screen for mutants defective in maintenance of a low pHv (9, 33). Briefly, to assess pHv, yeast cells were harvested by filtration, washed twice with CF buffer (50 mM glycine, 10 mM NaCl, 5 mM KCl, and 1 mM MgCl2 in 40 mM Tris-100 mM MES [morpholineethanesulfonic acid] [pH 4.0]) and resuspended in 2 ml of CF buffer (supplemented with 2% glucose to assure proper vacuolar staining) to an OD600 of 10. The 5(6)-CDCFDA,SE probe (9, 47, 49) was added to the cell suspension (final concentration of 45 μM), and the mixture was vortexed in one burst of 10 s and incubated for 20 min at 30°C with orbital agitation (250 rpm). After fluorescent labeling, cells were centrifuged at 5,500 × g for 5 min (at 4°C), washed twice with CF buffer, and finally resuspended in 2 ml of the same buffer. Cells were immediately examined with a Zeiss Axioplan microscope equipped with epifluorescence interference filters (Zeiss; BP450-490 and LP520). Fluorescent emission was collected with a cooled charge-coupled device camera (Cool SNAPFX; Roper Scientific Photometrics). Bright-field images for pHv determination were obtained concurrently and recorded at 1-min intervals, each experiment being completed within 15 min. The images were analyzed using MetaMorph 3.5, and fluorescence images were background corrected using dark-current images. In each experiment at least 500 individual cells were examined and the value of fluorescence intensity emitted by each cell was obtained pixel by pixel in the region of interest to estimate pHv. The average pHv values presented, expressed in terms of fluorescence intensity, are the means of at least three independent experiments.
Peroxisome proliferation analysis.
Peroxisome proliferation was examined by in vivo fluorescence microscopy based on the pPTS2-GFP plasmid expressing a chimera that localizes properly to the peroxisomes, marking their position and relative abundance (42). Wild-type BY4741 yeast cells harboring the pPTS2-GFP plasmid were grown in minimal growth medium in the absence or presence of inhibitory concentrations of ethanol (up to 8% [vol/vol]). Cell samples were collected after 2, 4, 6, 8, and 24 h of cultivation, washed, and visualized by direct fluorescence microscopy using a Zeiss Axioplan microscope coupled to a cooled charge-coupled device camera (Cool SNAPFX; Roper Scientific Photometrics).
Cell wall susceptibility analysis.
To monitor structural changes in the yeast cell wall, a lyticase (a ß-1,3-glucanase from Arthrobacter luteus; Sigma) sensitivity assay was conducted as described by Shimoi et al. (39). Wild-type yeast cells were cultivated in YPD liquid medium, supplemented or not with ethanol (6%), and harvested following 0 h or 3 h of incubation. The harvested cells were washed with distilled water and resuspended in 0.1 mM sodium phosphate buffer (pH 7.5). After the addition of 20 μg/ml of lyticase, cell lysis was followed by measuring the decrease in the OD600 of each cell suspension from its initial value.
Assessment of ethanol, glycerol, and glucose concentrations during alcoholic fermentation.
The parental strain BY4741, harboring the plasmid pYEPmyc-FPS1, expressing FPS1, or the corresponding empty vector, was grown in YPD medium (containing 20 g/liter yeast extract and 10 g/liter peptone [Difco]) supplemented with glucose (Merck) to obtain a final concentration of 300 g/liter and with the amino acids for which the strain is auxotrophic (240 mg/liter leucine, 80 mg/liter histidine, and 80 mg/liter methionine). Cell suspensions used to prepare the inocula were grown in YPD medium until mid-exponential phase (OD600 = 0.5 ± 0.05). Cell growth in liquid media was followed by measuring culture OD600 during batch cultivation. Samples of culture supernatants were harvested by centrifugation and used for the quantification of ethanol, glycerol, and glucose concentrations by high-pressure liquid chromatography. Culture supernatants were analyzed on an Aminex HXP87H ion exchange chromatography column, eluted at room temperature with 0.005 M H2SO4 at a flow rate of 0.6 ml min−1 for 30 min using a refractive-index detector. Under such experimental conditions glucose had a retention time of 8.3 min, ethanol 19.4 min, and glycerol 12.5 min. Reproducibility and linearity of the method were tested, and concentrations were estimated based on appropriate calibration curves.
RESULTS
Identification of genes conferring tolerance of ethanol.
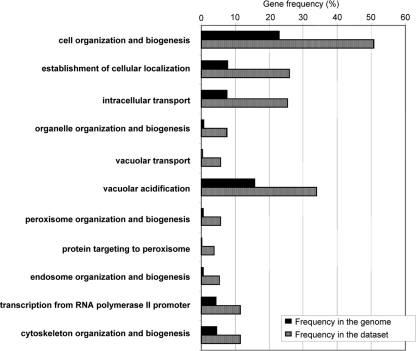
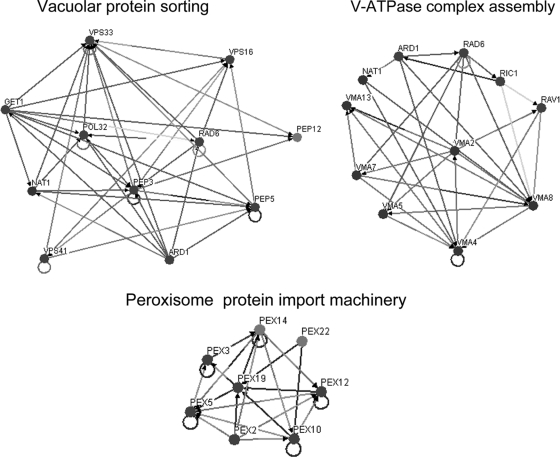
The chemogenomics approach allowed the identification of 254 genes as determinants of yeast resistance to ethanol. These include 121 genes that had never been associated with this phenomenon, 18 of which are essential genes (Table 1). Interestingly, only 3 (CSL4, MED8, and NAT2) of these 18 essential genes were associated with haploinsufficiency in minimal growth medium (10) and none was identified as conferring multidrug resistance in a recent chemical genomic portrait of yeast (15). Thus, their role in ethanol tolerance appears to be more than just a general contribution to overall fitness. Altogether, the 254 genes identified as determinants of ethanol tolerance were first clustered according to their associated GO terms using GOToolBox software (http://crfb.univ-mrs.fr/GOToolBox/index.php). The most significant (P < 0.01) GO terms, enriched in this data set relative to the yeast genome, include mostly those associated with intracellular organization, biogenesis, and transport, in particular, regarding the vacuole, the peroxisome, the endosome, and the cytoskeleton, and those associated with the transcriptional machinery (Fig. 1). Using BioGRID computational tools (http://www.thebiogrid.org/), which allow the clustering of yeast proteins according to the existence of physical or genetic interactions between them, three large protein clusters responsible for yeast tolerance to ethanol are identified (Fig. 2). These include the vacuolar protein sorting machinery, proteins required for assembly of the vacuolar H+-ATPase (V-ATPase) complex, and the peroxisome protein import machinery.
TABLE 1.
Essential genes identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Description of protein |
|---|---|
| Transcription machinery and RNA processing | |
| BDP1 | Essential subunit of RNA polymerase III transcription factor (TFIIIB); involved in transcription of genes encoding tRNAs, 5S rRNA, U6 snRNA, and other small RNAs |
| CSL4 | Subunit of the exosome, which is an essential complex present in both the nucleus and cytoplasm that mediates RNA processing and degradation |
| CWC25 | Component of a complex containing Cef1p, involved in pre-mRNA splicing |
| HTS1 | Cytoplasmic and mitochondrial histidine tRNA synthetase |
| IRR1 | Subunit of the cohesin complex, which is required for sister chromatid cohesion during mitosis and meiosis and interacts with centromeres and chromosome arms |
| MED8 | Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme |
| MPE1 | Essential conserved subunit of CPF (cleavage and polyadenylation factor); plays a role in 3′ end formation of mRNA via the specific cleavage and polyadenylation of pre-mRNA; contains a putative RNA-binding zinc knuckle motif |
| PRP11 | Subunit of the SF3a splicing factor complex, required for spliceosome assembly |
| RRP3 | Involved in rRNA processing; required for maturation of the 35S primary transcript of pre-rRNA and for cleavage leading to mature 18S rRNA |
| SPP381 | mRNA splicing factor, component of U4/U6/U5 tri-snRNP |
| TFC1 | One of six subunits of the RNA polymerase III transcription initiation factor complex (TFIIIC); part of the TauA globular domain of TFIIIC that binds DNA at the BoxA promoter sites of tRNA and similar genes |
| FHL1 | Putative transcriptional regulator with similarity to DNA-binding domain of Drosophila forkhead; required for rRNA processing |
| Others | |
| ARC35 | Subunit of the ARP2/ARP3 complex, which is required for the motility and integrity of cortical actin patches |
| IDI1 | Isopentenyl diphosphate:dimethylallyl diphosphate isomerase (IPP isomerase); catalyzes an essential activation step in the isoprenoid biosynthetic pathway |
| NAT2 | N-α-Acetyltransferase; transfers acetyl group from acetyl coenzyme A to the N-terminal methionine residues of proteins |
| SIS1 | Type II HSP40 cochaperone that interacts with the HSP70 protein Ssa1p |
| STS1 | Protein that interacts with the karyopherin Srp1p; may have a role with Srp1p in ubiquitin-mediated protein degradation |
| TOM40 | Component of the TOM (translocase of outer membrane) complex, responsible for recognition and initial import steps for all mitochondrially directed proteins |
Descriptions are based on those in the SGD (www.yeastgenome.org).
FIG. 1.
Categorization, based on the biological process taxonomy of GO, of the genes required for ethanol stress resistance. Genes were clustered using GOToolBox, and only classes found to be statistically overrepresented in our data set are displayed (P value below 0.01). Striped bars, gene frequency within each class; black bars, frequency registered for the whole genome.
FIG. 2.
Main interaction network maps of the determinants of ethanol tolerance. The map was created using the OSPREY software and shows protein and genetic interactions within three groups of genes associated with vacuolar protein sorting, V-ATPase complex assembly, and peroxisome protein import machinery. The existence of an interaction between two genes or proteins is represented by a connection between two nodes.
A more detailed analysis of all the determinants of resistance to inhibitory concentrations of ethanol allowed us to construct Table S1 in the supplemental material, which groups these 254 genes according to their intracellular localizations and biological functions, based on their associated GO terms and description in the Saccharomyces Genome Database (SGD; www.yeastgenome.org). The roles of the most representative groups are discussed in more detail in the following sections.
Peroxisomal function is involved in yeast resistance to ethanol-induced stress.
Peroxisomal function seems to be required for ethanol stress resistance, as the expression of the PEX1, PEX2, PEX3, PEX4, PEX5, PEX8, PEX10, PEX12, PEX14, PEX19, and PEX22 genes, encoding proteins of the peroxisome transport machinery and/or of the peroxisomal membrane import machinery that are required for peroxisome organization and biogenesis, was found to be essential for maximal tolerance of ethanol (Table 2). It was surprising to identify this group of genes as ethanol resistance determinants in yeast cells growing in the presence of glucose, which represses peroxisomal function. However, although most genes involved in peroxisomal protein transport were found to be required for yeast resistance to ethanol, the individual deletion of genes encoding fatty acid β-oxidation enzymes or proteins involved in lysine biosynthesis, the major metabolic processes taking place in yeast peroxisomes, did not result in increased susceptibility to ethanol stress. The effect of ethanol shock (from 1 to 8% [vol/vol]) on peroxisome proliferation was assessed in cells harboring a plasmid expressing the chimera Pts2_GFP, Pts2 being a signal recognition marker for peroxisomes (42). However, no changes in peroxisome proliferation in response to ethanol stress were detected (results not shown).
TABLE 2.
Genes required for peroxisome organization and biogenesis identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Protein description |
|---|---|
| PEX1 | AAA family ATPase peroxin required for peroxisome biogenesis |
| PEX10 | RING finger peroxisomal membrane peroxin required for peroxisomal matrix protein import |
| PEX12 | RING finger peroxisomal membrane peroxin that plays an essential role in peroxisome biogenesis and peroxisomal matrix protein import |
| PEX14 | Peroxisomal membrane protein that is a central component of the peroxisomal protein import machinery |
| PEX19 | Chaperone and import receptor for newly synthesized class I peroxisomal membrane proteins |
| PEX2 | RING finger peroxin, peroxisomal membrane protein with a C-terminal zinc-binding RING domain |
| PEX22 | Putative peroxisomal membrane protein required for import of peroxisomal proteins |
| PEX3 | Peroxisomal membrane protein required to recruit Pex19p chaperone to peroxisomes |
| PEX4 | Peroxisomal ubiquitin conjugating enzyme required for peroxisomal matrix protein import and peroxisome biogenesis |
| PEX5 | Peroxisomal membrane signal receptor for C-terminal tripeptide signal sequence (PTS1) of peroxisomal matrix proteins |
| PEX8 | Intraperoxisomal organizer of the peroxisomal import machinery |
Descriptions are based on those in the SGD (www.yeastgenome.org).
V-ATPase function is involved in yeast resistance to ethanol-induced stress.
The important role of V-ATPase in yeast tolerance of ethanol is highlighted in the present study, since mutants devoid of PPA1, VMA2, VMA4, VMA7, VMA8, VMA9, VMA13, VMA21, VMA22, THP2, TFP3, or CUP5, encoding V-ATPase subunits, are much more sensitive to ethanol-induced stress than the wild-type strain (Table 3). Other genome-wide studies had pointed out that some of these genes are required for yeast tolerance of ethanol, but their role in this resistance was only suggested (13, 21, 46, 52). It has long been established that exposure to ethanol stress leads to intracellular acidification (35). In this study, we found that ethanol also induces vacuolar acidification. Indeed, it was possible to demonstrate, using a pH-sensitive fluorescent probe, that pHv decreases in a dose-dependent manner in cells exposed for 1 h to ethanol concentrations leading to mild or moderate growth inhibition (6% and 8%) (Fig. 3). The reduction of pHv in response to these ethanol concentrations is hypothesized to be an adaptive response that helps the cell control ethanol- induced cytosolic acidification at more physiological values, as proposed before for weak acids (4, 25, 27).
TABLE 3.
Genes involved in lipid, fatty acid, and ergosterol biosynthesis identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Protein description |
|---|---|
| ERG2 | C-8 sterol isomerase, catalyzes the isomerization of the delta-8 double bond to the delta-7 position at an intermediate step in ergosterol biosynthesis |
| ERG24 | C-14 sterol reductase, acting in ergosterol biosynthesis; mutants accumulate the abnormal sterol ignosterol (ergosta-8,14-dienol) |
| IDI1 | Isopentenyl diphosphate:dimethylallyl diphosphate isomerase; catalyzes an essential activation step in the isoprenoid biosynthetic pathway |
| KCS1 | Inositol hexaphosphate kinase; phosphorylates inositol hexakisphosphate (InsP6) to diphosphoinositol polyphosphates, required for proper vacuole morphology |
| LIP5 | Protein involved in biosynthesis of the coenzyme lipoic acid |
| OPI3 | Phospholipid methyltransferase (methylene-fatty acyl-phospholipid synthase); catalyzes the last two steps in phosphatidylcholine biosynthesis |
| PDX3 | Pyridoxine (pyridoxamine) phosphate oxidase |
Descriptions are based on those in the SGD (www.yeastgenome.org).
FIG. 3.
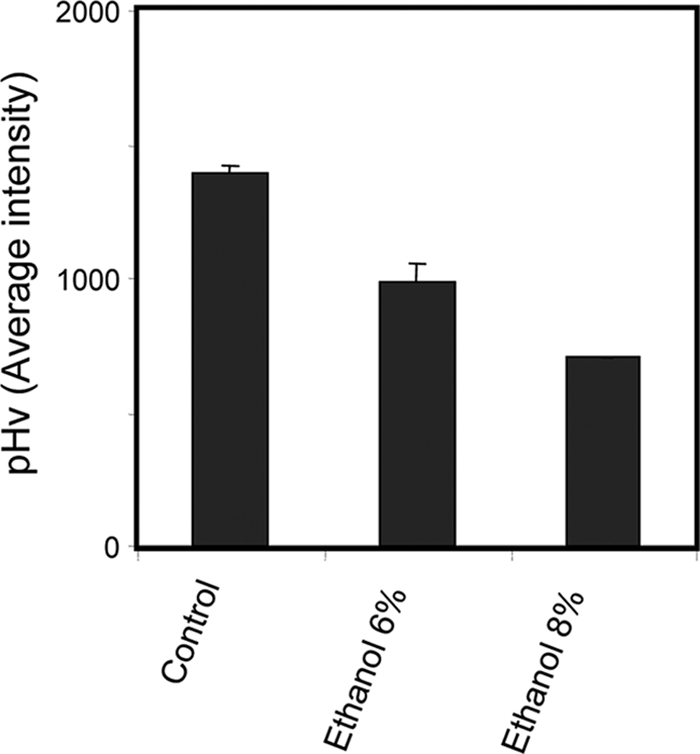
pHv, assessed as fluorescence intensity values as described in Materials and Methods, upon 1 h of BY4741 cell cultivation in growth medium in the absence (control) or presence of inhibitory concentrations of ethanol (6% or 8% [vol/vol]). Values are the averages of at least three independent experiments; standard variations are displayed as error bars.
Genes involved in membrane and cell wall composition are required for yeast resistance to ethanol-induced stress.
Another interesting set of genes required for yeast tolerance of ethanol stress includes those involved in membrane (Table 4) and cell wall (Table 5) biosynthesis. Genes involved in membrane composition identified as conferring resistance to high ethanol concentrations include ERG2 and ERG24, involved in ergosterol biosynthesis, and KCS1, LIP5, OPI3, PDX3, and IDI1, involved in phospholipid biosynthesis (Table 3). Among cell wall-related genes involved in ethanol stress tolerance we found the ANP1, HOC1, and MNN10 genes, encoding three of the five subunits of mannosyl polymerase complex II (M-Pol II). Besides this protein complex, other cell wall-related genes were herein identified as determinants of yeast resistance to high concentrations of ethanol, including KRE6, encoding a β-glucan synthase required for β-1,6-glucan biosynthesis, and SLG1 and SLT2, encoding a sensor and a kinase belonging to the stress-activated cell wall integrity protein kinase C (PKC) signaling pathway.
TABLE 4.
Genes involved in V-ATPase composition and assembly identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Protein description |
|---|---|
| CUP5 | Proteolipid subunit of the V-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit) |
| PPA1 | Subunit c″ of V-ATPase; one of three proteolipid subunits of the V0 domain |
| RAV1 | Subunit of the RAVE complex (Rav1p, Rav2p, Skp1p), which promotes assembly of the V-ATPase holoenzyme |
| TFP3 | V-ATPase V0 domain subunit c′, involved in proton transport activity |
| THP2 | V-ATPase V0 domain subunit c′, involved in proton transport activity; hydrophobic integral membrane protein (proteolipid) containing four transmembrane segments; N and C termini are in the vacuolar lumen |
| VMA2 | Subunit B of the eight-subunit V1 peripheral membrane domain of the V-ATPase |
| VMA4 | Subunit E of the eight-subunit V1 peripheral membrane domain of the V-ATPase |
| VMA5 | Subunit C of the eight-subunit V1 peripheral membrane domain of V-ATPase |
| VMA7 | Subunit F of the eight-subunit V1 peripheral membrane domain of V-ATPase |
| VMA8 | Subunit D of the eight-subunit V1 peripheral membrane domain of the V-ATPase |
| VMA9 | Subunit e of the V-ATPase V0 subcomplex |
| VMA13 | Subunit H of the eight-subunit V1 peripheral membrane domain of the V-ATPase |
| VMA21 | Integral membrane protein that is required for the assembly of V-ATPase |
| VMA22 | Integral membrane protein that is required for the assembly of V-ATPase |
Descriptions are based on those in the SGD (www.yeastgenome.org).
TABLE 5.
Genes involved in cell wall biosynthesis identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Protein description |
|---|---|
| ANP1 | Subunit of the α-1,6-mannosyltransferase complex; type II membrane protein; involved in osmotic sensitivity |
| HOC1 | α-1,6-Mannosyltransferase involved in cell wall mannan biosynthesis |
| KRE6 | Protein required for β-1,6-glucan biosynthesis; putative β-glucan synthase |
| MNN10 | Subunit of a Golgi mannosyltransferase complex, also containing Anp1p, Mnn9p, Mnn11p, and Hoc1p, that mediates elongation of the polysaccharide mannan backbone |
| SLG1 | Sensor-transducer of the stress-activated PKC1-MPK1 kinase pathway involved in maintenance of cell wall integrity |
| SLT2 | Serine/threonine mitogen-activated protein kinase involved in regulating the maintenance of cell wall integrity; regulated by PKC1-mediated signaling pathway |
Descriptions are based on those in the SGD (www.yeastgenome.org).
The identification of genes involved in the control of cell wall or membrane composition as determinants of resistance to ethanol suggests that these structures undergo remodeling in response to ethanol stress, thus leading to increased cell resistance. Changes in membrane composition in response to ethanol-induced stress at the level of phospholipids and ergosterol composition have been described before (1, 6, 53). To check whether cell wall remodeling occurs during the yeast response to ethanol shock, the very simple 1,3-ß-glucanase sensitivity assay, which has been shown to be valuable in monitoring cell wall alterations (40, 41), was used. Yeast cells grown in a culture medium in the absence of ethanol were seen to be more susceptible to lyticase activity than cells exposed for 3 hours to cultivation in the presence of 6% ethanol (Fig. 4). These results support the concept that ethanol induces the remodeling of the yeast cell wall and suggest that this event is important for cell adaptation and resistance to ethanol.
FIG. 4.
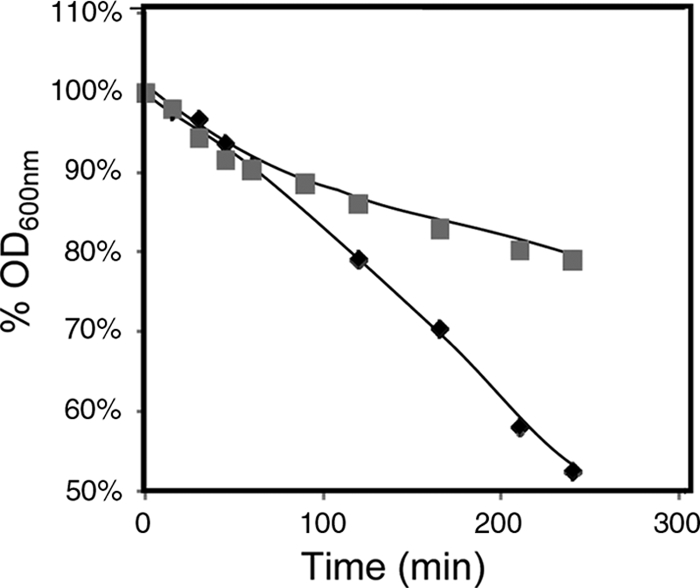
Comparison of the susceptibilities to lyticase of cells of S. cerevisiae parental strain BY4741 grown in the absence of ethanol and then incubated in a growth medium that was either left unsupplemented (⧫) or supplemented with 6% ethanol (▪). Cells were harvested after 3 h of incubation. The different cell populations were washed with water and resuspended in 0.1 M sodium phosphate buffer at pH 7.5. After the addition of 20 μg of lyticase (Sigma) per ml, the decrease in the OD600 of the cell suspension was measured periodically.
Genes involved in carbohydrate and amino acid metabolism are involved in yeast resistance to ethanol-induced stress.
A number of genes involved in the control of carbohydrate metabolism were identified in this work as determinants of yeast resistance to ethanol stress (Table 6). These include four genes (HAP2, MIG1, RTG3, and GCR1) encoding transcription factors that act in carbohydrate metabolism control. Moreover, 30 genes encoding mitochondrial proteins (see Table S1 in the supplemental material) are determinants of yeast resistance to high concentrations of ethanol, suggesting that, even in the presence of glucose, mitochondrial functions are essential for ethanol tolerance. These results are in agreement with the notion that ethanol tolerance depends on the stability of the mitochondrial genome (18). A high proportion of the mitochondrion-related genes required for ethanol stress resistance are involved in mitochondrial protein synthesis, respiration, and mitochondrial DNA maintenance (see Table S1 in the supplemental material). Among the carbohydrate metabolism-related genes, TPS1 and GPH1, encoding enzymes required for the synthesis of trehalose and degradation of glycogen, respectively, were also identified as ethanol resistance determinants. Trehalose and glycogen are two molecules that are synthesized and that accumulate under stress, with roles in carbon storage and as compatible solutes. Moreover, trehalose protects membranes from desiccation and prevents protein denaturation (50).
TABLE 6.
Genes involved in carbohydrate and amino acid metabolism identified in this study as determinants of yeast resistance to inhibitory ethanol concentrationsa
| Gene/ORF name | Protein description |
|---|---|
| Carbohydrate metabolism | |
| FPS1 | Plasma membrane glycerol channel, involved in efflux of glycerol and in uptake of the trivalent metalloids arsenite and antimonite |
| HAP2 | Subunit of the heme-activated, glucose-repressed Hap2p/-3p/-4p/-5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression |
| MIG1 | Transcription factor involved in glucose repression; sequence-specific DNA binding protein containing two Cys2His2 zinc finger motifs |
| TPS1 | Synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage of carbohydrate trehalose |
| REG1 | Regulatory subunit of type 1 protein phosphatase Glc7p, involved in negative regulation of glucose-repressible genes |
| PFK26 | 6-Phosphofructo-2-kinase, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate |
| GCR1 | Transcriptional activator of genes involved in glycolysis; interacts and functions with the transcriptional activator Gcr2p |
| GPH1 | Nonessential glycogen phosphorylase required for the mobilization of glycogen |
| RTG3 | Basic helix-loop-helix leucine zipper (bHLH/Zip) transcription factor that forms a complex with another bHLH/Zip protein, Rtg1p, to activate the retrograde and TOR pathways |
| Amino acid metabolism | |
| SLM4 | Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p |
| GTR1 | Cytoplasmic GTP binding protein and negative regulator, with homolog Gtr2p, of the Ran/Tc4 GTPase cycle; component of GSE complex, which is required for sorting of Gap1p |
| AGP2 | High-affinity polyamine permease, preferentially uses spermidine over putrescine; plasma membrane carnitine and low-affinity amino acid permease |
| BNA1 | 3-Hydroxyanthranilic acid dioxygenase, required for biosynthesis of nicotinic acid from tryptophan via kynurenine pathway |
| LST8 | Protein required for the transport of amino acid permease Gap1p from the Golgi complex to the cell surface |
| PTR3 | Component of the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p) |
| GCN4 | Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation |
| GLY1 | Threonine aldolase, catalyzes the cleavage of l-allo-threonine and l-threonine to glycine; involved in glycine biosynthesis |
Descriptions are based on those in the SGD (www.yeastgenome.org).
Genes related to amino acid metabolism identified in this study as required for yeast resistance to ethanol stress include mostly those involved in sensing or regulatory processes (Table 6). Among them are the transcription factor-encoding gene GCN4, the master regulator of amino acid biosynthesis; SLM4, GTR1, and LST8, encoding three proteins involved in protein trafficking required for the proper localization of the major amino acid permease Gap1p; PTR3, encoding a component of the SPS plasma membrane amino acid sensor system; and AGP2, encoding a low-affinity amino acid permease with a high affinity for polyamines. The importance of amino acid permeases and sensors in ethanol tolerance may be related to the effect of this metabolite on yeast plasma membrane organization and on the activity of embedded transport systems (26, 34, 36, 45).
The important role of FPS1 in yeast resistance to ethanol-induced stress.
One of the genes found to play a role in yeast resistance to ethanol is FPS1. This gene encodes an aquaglyceroprotein involved in the control of the intracellular level of glycerol that fortuitously facilitates the entrance of a number of toxic compounds (17, 28, 30, 32, 43, 51). In the absence of FPS1, yeast cells experience a period of lag phase when 6% (vol/vol) ethanol is suddenly added to an exponentially growing cell culture in MM4 medium. This period of latency, which was not observed in parental cells, was followed by exponential growth resumption with reduced growth rate compared to that of parental cells expressing the FPS1 gene (Fig. 5). The expression of the FPS1 gene in Δfps1 cells allows the population to behave similarly to parental cells (Fig. 5). Moreover, wild-type cells expressing FPS1 from the multicopy plasmid pYEPmyc-FPS1 exhibit slightly increased tolerance of ethanol-induced stress (Fig. 5). The effect of FPS1 expression on the intracellular accumulation of [3H]ethanol was assessed during yeast cultivation in the presence of a moderately inhibitory concentration of 6% (vol/vol) ethanol (Fig. 6), under conditions identical to those used in the growth experiment shown in Fig. 5. The accumulation of [3H]ethanol in yeast cells that had not been previously adapted to this compound when suddenly exposed to 6% ethanol is higher when the cells are devoid of the FPS1 gene (Fig. 6). The expression of FPS1 from pYEPmyc-FPS1 clearly decreases the intracellular accumulation of radiolabeled ethanol exhibited by parental cells and further reduces ethanol accumulation in the mutant strain, back to levels close to those detected in the parental strain (Fig. 6).
FIG. 5.
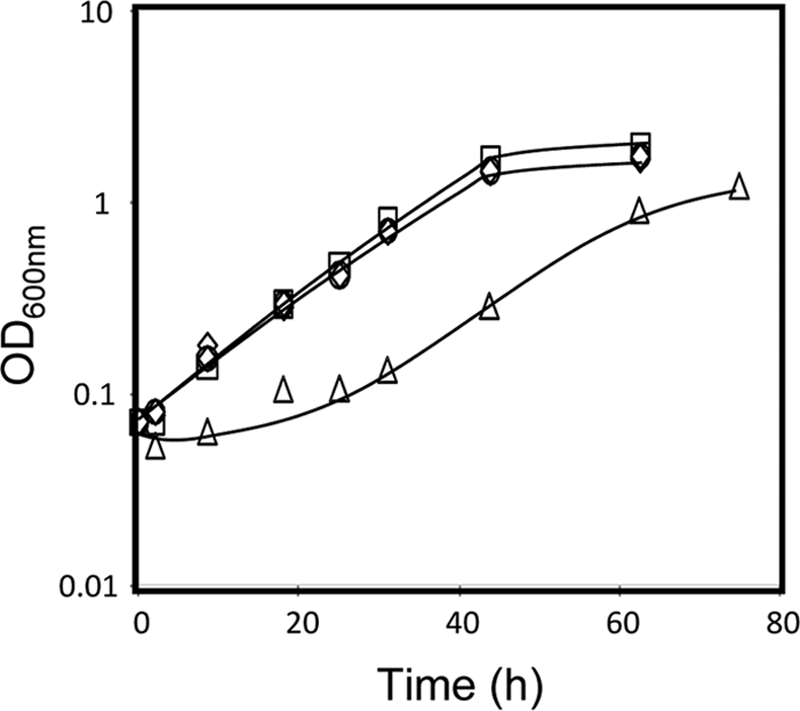
Comparison of the susceptibilities to ethanol-induced stress of S. cerevisiae parental strain W303-1A, harboring the FPS1 expression plasmid (□) or the corresponding empty vector (⋄), to those of the derived deletion mutant Δfps1, harboring the FPS1 expression plasmid (○) or the corresponding empty vector (▵). Cells were grown in the absence of ethanol and then incubated in MM4 liquid medium supplemented with 6% ethanol. Growth curves are representative of at least three independent experiments.
FIG. 6.
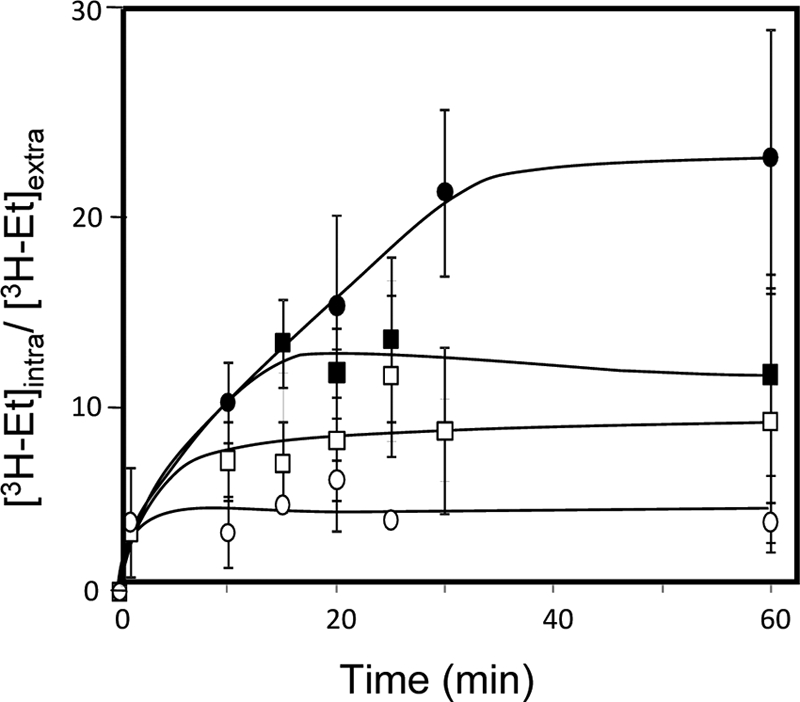
Comparison of levels of [3H]ethanol accumulation in nonadapted cells of S. cerevisiae parental strain W303-1A, harboring the FPS1 expression plasmid (○) or the corresponding empty vector (□), and the derived Δfps1 deletion mutant, harboring the FPS1 expression plasmid (▪) or the corresponding empty vector (•), during cultivation in MM4 liquid medium suddenly supplemented with 6% ethanol under conditions identical to those used for Fig. 5. The accumulation values are representative of at least three independent experiments. [3H-Et]intra and [3H-Et]extra, intracellular and extracellular [3H]ethanol concentrations, respectively.
To assess whether or not the role of Fps1 in ethanol homeostasis could have an impact on the performance of alcoholic fermentation, BY4741 cells were grown in liquid YPD medium containing 30% glucose and an additional supply of amino acids for which the strain is auxotrophic. This laboratory medium is expected to mimic fermentation conditions leading to high ethanol production. Figure 7 shows that the final concentration of ethanol produced by the wild-type population expressing the FPS1 gene from a multicopy plasmid is increased by more than 15% (Fig. 7A) compared to that produced by the same cells harboring the cloning vector, although the growth curves of both strains are undistinguishable (Fig. 7B). Consistently, glucose consumption was higher in the cell culture producing higher ethanol levels (Fig. 7B). It is interesting to observe that, until 12% (vol/vol) ethanol is reached, the ethanol production rates in cells expressing one or two copies of the FPS1 gene are very similar. However, once higher ethanol concentrations are reached, stationary-phase cells expressing only a single copy of the FPS1 gene become unable to proceed with alcoholic fermentation, while those expressing the FPS1 gene from a multicopy plasmid are able to continue to produce ethanol up to 14% (vol/vol).
FIG. 7.
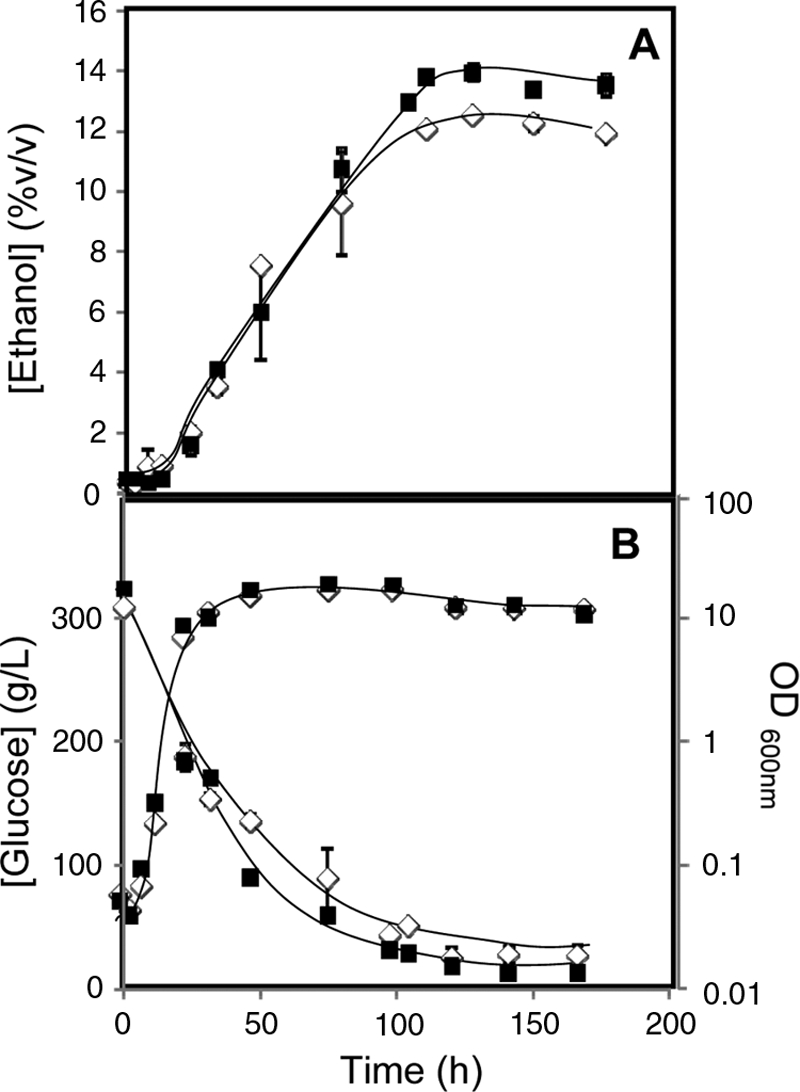
Comparison of extracellular concentrations of ethanol and glucose, accumulated during cultivation in fermentation medium of BY4741 yeast cells harboring the pYEP-FPS1 plasmid (▪) or the corresponding cloning vector (⋄). Growth was followed by measuring culture OD600.
DISCUSSION
The chemogenomics analysis described in this work reveals new information on ethanol resistance mechanisms in yeast. Although other labs have also screened the yeast disruptome for its susceptibility to ethanol stress (13, 21, 46, 52), the observed differences between the five disruptome analyses conducted so far under ethanol stress seem to suggest that the genetic background, growth media, and conditions have a deep impact on the expression of yeast determinants of resistance to ethanol stress. In agreement with this notion, the present genome-wide survey carried out with minimal growth medium uncovered 121 new determinants of resistance to ethanol- induced stress. Among them, 18 essential genes were shown for the first time to be required for increased ethanol tolerance. These essential genes are mostly involved in transcriptional control and in RNA processing, which is consistent with the recent report on the successful engineering of yeast transcription machinery to increase yeast cell ethanol tolerance and production (3).
Among the nonessential genes required for ethanol tolerance in yeast, many are related to intracellular trafficking, including vacuolar protein targeting, endosome transport, and transport mediated by the endosomal sorting complexes required for transport (ESCRT-I, -II, and -III), related to ubiquitin-dependent protein sorting to the vacuole. Endocytosis and vacuolar targeting and degradation are required to regulate transmembrane protein location and turnover, including that of transporters and signaling receptors, which may explain why changes in intracellular trafficking are correlated with multiple-drug resistance (15). In particular, the targeting of membrane transporters is expected to be vital to overcome stress imposed by lipophilic agents such as ethanol, which is known to, at least, affect glucose, ammonium, and amino acid transport (45). Significantly, some of the identified determinants of ethanol resistance are specifically involved in the targeting of the major amino acid permease, Gap1p. Corroborating the idea that intracellular protein trafficking through the endosome and vacuole is important for ethanol stress tolerance, vacuolar morphology and endocytosis have been shown to be altered in yeast cells exposed to ethanol stress (26). The results presented in this work showing that yeast cells under ethanol stress exhibit increased vacuolar acidification also reinforce the notion that a functional vacuolar system is required for yeast resistance to ethanol. Furthermore, since vacuolar acidification has been proposed as one of the mechanisms contributing to the recovery from stress-induced cytosolic acidification (12, 19, 25), the obtained results suggest that V-ATPase plays a role in the recovery of cytosolic pH, known to be acidified under ethanol stress (34).
Genes involved in peroxisome protein import machinery were also shown to be required for yeast tolerance of ethanol in minimal medium, corroborating a previous study using YPD (52). However, no increase in peroxisome proliferation appears to occur in yeast cells exposed to ethanol stress, consistent with the lack of ethanol sensitivity found in strains devoid of genes required for the regulation of peroxisome size and number. The role of peroxisomes in ethanol stress response can only be hypothesized. Since yeast cells deficient in peroxisomal functions are unable to effectively control the fatty acid composition of membrane phospholipids (23) and the composition of membrane phospholipids is a key feature of the ethanol stress response (53), peroxisomal function may be required for the synthesis or degradation of membrane phospholipids for remodeling cell membranes. Moreover, the subgroup of peroxisomal genes identified in this work as involved in ethanol stress tolerance were identified previously as conferring resistance to a number of oxidative stress inducing agents in yeast (15). The peroxisomes are known to be involved in the metabolism of peroxides and other reactive oxygen species (37), which are known to be generated in cells challenged with ethanol stress (11). Thus, peroxisomal function may also be required to confer resistance to ethanol-induced oxidative stress.
Our results point out an important role of some genes involved in phospholipid and ergosterol biosynthesis as part of this adaptive response, consistent with previous reports indicating that there are changes at the level of plasma membrane composition in response to ethanol stress (1, 6, 53). Decreased cell envelope permeability to weak organic acids was found to depend on cell wall remodeling, leading to increased lyticase resistance (40, 41). The importance of the cell wall composition in yeast resistance to ethanol-induced stress has been previously pointed out, based on the fact that sake yeasts, which tolerate a very high ethanol concentration (up to 20%), are more resistant to the cell wall-targeted drugs K1 toxin and zymolase (31). In this study, we show that ethanol stress induces cell wall remodeling, leading to an increase in cell wall resistance to the lytic enzyme β-1,3-glucanase. Given this result, we were surprised to see that two genes, SPI1 and YGP1, previously related to cell wall remodeling conferring lyticase resistance, were not found among the ethanol resistance determinants obtained through the disruptome analysis. However, comparison of the growth curves of wild-type cells and of the derived Δspi1 and Δygp1 deletion mutants shows that indeed these two genes are also required for ethanol stress resistance (our unpublished results). Interestingly, microarray analysis of yeast cells subjected to ethanol stress further shows that SPI1 and, particularly, YGP1 are upregulated in response to ethanol-induced stress (2). It is possible to hypothesize that, similar to what has been observed for weak acids, the cell wall remodeling found in this study to occur upon ethanol exposure is possibly dependent on Spi1p or Ygp1p (40, 41). Furthermore, the emergence of ANP1, HOC1, and MNN10 genes, encoding three of the five subunits of the M-Pol II complex, as determinants of ethanol stress resistance seems to indicate that the composition and size of the mannan external layer of the yeast cell wall are crucial for allowing yeast cells to adapt to toxic concentrations of ethanol. Indeed, in Saccharomyces cerevisiae, cell wall proteins and secretory proteins are often modified with mannan, a long branched polymer of approximately 200 mannoses. The activity of α-1,6-mannosyltransferase is provided by Och1p and two enzyme complexes, M-Pol I and M-Pol II. Och1p catalyzes the addition of the first mannose, while M-Pol I is required for the addition of approximately 10 mannoses to the initial mannose residue and M-Pol II is required for the elongation of the mannose backbone (22). This mannan modification is very extensive: mannoproteins constitute up to 40% of the yeast cell wall dry weight. Mannans provide an external layer to the cell wall, which is believed to contribute to its structural integrity and to determine the permeability of the cell wall to macromolecules (20). This work further suggests that the composition and extension of this mannan backbone may be essential to increase yeast tolerance of ethanol-induced stress.
The screening of the yeast disruptome carried out in this work for mutants exhibiting increased susceptibility to ethanol stress also led to the identification of FPS1 as a determinant of resistance to ethanol-induced stress in Saccharomyces cerevisiae. This result is corroborated by one of the previous disruptome analyses that included this gene in the list of genes required for ethanol resistance without any further reference to its involvement in the underlying mechanism (13). The current study further explores this phenotype and provides evidence showing that FPS1 gene expression decreases the intracellular accumulation of radiolabeled ethanol against a concentration gradient in stressed yeast cells. Although it is surprising to see that a lipophilic molecule such as ethanol can accumulate in yeast cells, as found here to occur in the absence of the FPS1 gene, a previous report had shown that ethanol accumulates intracellularly up to 20-fold in the first hours of alcoholic fermentation in media containing 20% glucose (8). Interestingly, 20% glucose induces osmotic stress, a condition in which the Fps1p channel is closed to prevent the export of glycerol (43), thus contributing to osmotic stress tolerance. It is possible to hypothesize that, under osmotic stress, in which yeast cells keep Fps1p in an inactive state (43), as in Δfps1 mutant cells (this work), ethanol accumulation takes place, suggesting a role for Fps1p in ethanol export. The obtained results led us to hypothesize that Fps1 expression could also increase yeast capability to produce higher ethanol concentrations, an advantage for industrial processes. Indeed, in conditions that lead to high ethanol production (medium containing 30% glucose and no other nutrient limiting growth), the increased expression of FPS1 increases ethanol productivity in wild-type cells. In a previous study, FPS1 deletion had been shown to slightly increase ethanol production upon fermentation of 8% glucose, mainly due to a reduced glycerol production and the simultaneous redirection of yeast metabolism to ethanol production (54). However, in the work by Zhang and coworkers, final ethanol concentration did not go beyond 5% (vol/vol) (54). In this study, wild-type BY4741 cells reaching a high final concentration of ethanol (above 15%) were found to produce slightly higher levels of ethanol than the derived Δfps1 mutant (results not shown). Thus, under conditions leading to high ethanol production, closer to those desired in, for example, bioethanol production processes, Fps1 expression is proved here to be beneficial. The exact effect of FPS1 overexpression in this context remains to be clarified. Its role in increasing ethanol tolerance by reducing intracellular ethanol accumulation through Fps1-mediated ethanol export is suggested here. However, the described effect of FPS1 on membrane ergosterol content (44) might also play a role in this context. Indeed, increased ergosterol concentrations have been previously related to higher ethanol tolerance in yeast (6). Increased overexpression of the FPS1 gene from its chromosomal locus in industrial yeast strains is expected to be a promising strategy to increase the capacity to produce higher ethanol concentrations in industrial processes.
Supplementary Material
Acknowledgments
Research described in this article was financially supported by FEDER and Fundação para a Ciência e a Tecnologia (contract PTDC/BIO/66151/2006 and postdoctoral and Ph.D. grants to N.P.M. and A.B.L., respectively).
We acknowledge Markus Támas, from the Department of Cellular and Molecular Biology, University of Gothenburg, Sweden, for kindly providing yeast strains W303-1A and the derived Δfps1 deletion mutant, together with the plasmid expressing FPS1, and John D. Aitchison, from the Institute for Systems Biology, Seattle, WA, for the pPTS2-GFP plasmid.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aguilera, F., R. A. Peinado, C. Millan, J. M. Ortega, and J. C. Mauricio. 2006. Relationship between ethanol tolerance, H+-ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int. J. Food Microbiol. 110:34-42. [DOI] [PubMed] [Google Scholar]
- 2.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 3.Alper, H., J. Moxley, E. Nevoigt, G. R. Fink, and G. Stephanopoulos. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565-1568. [DOI] [PubMed] [Google Scholar]
- 4.Carmelo, V., H. Santos, and I. Sá-Correia. 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1325:63-70. [DOI] [PubMed] [Google Scholar]
- 5.Casey, G. P., and W. M. Ingledew. 1986. Ethanol tolerance in yeasts. Crit. Rev. Microbiol. 13:219-280. [DOI] [PubMed] [Google Scholar]
- 6.Chi, Z., and N. Arneborg. 1999. Relationship between lipid composition, frequency of ethanol-induced respiratory deficient mutants, and ethanol tolerance in Saccharomyces cerevisiae. J. Appl. Microbiol. 86:1047-1052. [DOI] [PubMed] [Google Scholar]
- 7.Costa, V., E. Reis, A. Quintanilha, and P. Moradas-Ferreira. 1993. Acquisition of ethanol tolerance in Saccharomyces cerevisiae: the key role of the mitochondrial superoxide dismutase. Arch. Biochem. Biophys. 300:608-614. [DOI] [PubMed] [Google Scholar]
- 8.D'Amore, T., C. J. Panchal, and G. G. Stewart. 1988. Intracellular ethanol accumulation in Saccharomyces cerevisiae during fermentation. Appl. Environ. Microbiol. 54:110-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N. U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chisholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496-503. [DOI] [PubMed] [Google Scholar]
- 10.Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer, R. W. Davis, C. Nislow, and G. Giaever. 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169:1915-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, X., and H. Takagi. 2007. N-Acetyltransferase Mpr1 confers ethanol tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. Appl. Microbiol. Biotechnol. 75:1343-1351. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes, A. R., P. J. Durão, P. M. Santos, and I. Sá-Correia. 2003. Activation and significance of vacuolar H+-ATPase in Saccharomyces cerevisiae adaptation and resistance to the herbicide 2,4-dichlorophenoxyacetic acid. Biochem. Biophys. Res. Commun. 312:1317-1324. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, K., A. Matsuyama, Y. Kobayashi, and H. Iwahashi. 2006. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res. 6:744-750. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, B. R., S. J. Lawrence, J. P. Leclaire, C. D. Powell, and K. A. Smart. 2007. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 31:535-569. [DOI] [PubMed] [Google Scholar]
- 15.Hillenmeyer, M. E., E. Fung, J. Wildenhain, S. E. Pierce, S. Hoon, W. Lee, M. Proctor, R. P. St. Onge, M. Tyers, D. Koller, R. B. Altman, R. W. Davis, C. Nislow, and G. Giaever. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirasawa, T., K. Yoshikawa, Y. Nakakura, K. Nagahisa, C. Furusawa, Y. Katakura, H. Shimizu, and S. Shioya. 2007. Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J. Biotechnol. 131:34-44. [DOI] [PubMed] [Google Scholar]
- 17.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez, J., and T. Benitez. 1988. Yeast cell viability under conditions of high temperature and ethanol concentrations depends on the mitochondrial genome. Curr. Genet. 13:461-469. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 21.Kubota, S., I. Takeo, K. Kume, M. Kanai, A. Shitamukai, M. Mizunuma, T. Miyakawa, H. Shimoi, H. Iefuji, and D. Hirata. 2004. Effect of ethanol on cell growth of budding yeast: genes that are important for cell growth in the presence of ethanol. Biosci. Biotechnol. Biochem. 68:968-972. [DOI] [PubMed] [Google Scholar]
- 22.Lesage, G., and H. Bussey. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockshon, D., L. E. Surface, E. O. Kerr, M. Kaeberlein, and B. K. Kennedy. 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175:77-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, D., C. Brun, E. Remy, P. Mouren, D. Thieffry, and B. Jacq. 2004. GoToolBox: functional investigation of gene datasets based on Gene Ontology. Genome Biol. 5:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Munoz, G. A., and P. Kane. 2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309-20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meaden, P. G., N. Arneborg, L. U. Guldfeldt, H. Siegumfeldt, and M. Jakobsen. 1999. Endocytosis and vacuolar morphology in Saccharomyces cerevisiae are altered in response to ethanol stress or heat shock. Yeast 15:1211-1222. [DOI] [PubMed] [Google Scholar]
- 27.Mira, N. P., A. B. Lourenco, A. R. Fernandes, J. D. Becker, and I. Sá-Correia. 2009. The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res. 9:202-216. [DOI] [PubMed] [Google Scholar]
- 28.Mollapour, M., and P. W. Piper. 2007. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell. Biol. 27:6446-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteiro, G. A., and I. Sá-Correia. 1998. In vivo activation of yeast plasma membrane H+-ATPase by ethanol: effect on the kinetic parameters and involvement of the carboxyl-terminus regulatory domain. Biochim. Biophys. Acta 1370:310-316. [DOI] [PubMed] [Google Scholar]
- 30.Nozawa, A., J. Takano, M. Kobayashi, N. von Wiren, and T. Fujiwara. 2006. Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 262:216-222. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, Y., A. Nitta, H. Uchiyama, T. Imamura, H. Shimoi, and K. Ito. 2000. Tolerance mechanism of the ethanol-tolerant mutant of sake yeast. J. Biosci. Bioeng. 90:313-320. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson, N., C. Filipsson, E. Becit, L. Brive, and S. Hohmann. 2005. Aquaporins in yeasts and filamentous fungi. Biol. Cell 97:487-500. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, C. J., C. K. Raymond, K. T. Yamashiro, and T. H. Stevens. 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194:644-661. [DOI] [PubMed] [Google Scholar]
- 34.Rosa, M. F., and I. Sá-Correia. 1991. In vivo activation by ethanol of plasma membrane ATPase of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 57:830-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosa, M. F., and I. Sá-Correia. 1996. Intracellular acidification does not account for inhibition of Saccharomyces cerevisiae growth in the presence of ethanol. FEMS Microbiol. Lett. 135:271-274. [DOI] [PubMed] [Google Scholar]
- 36.Salgueiro, S. P., I. Sá-Correia, and J. M. Novais. 1988. Ethanol-induced leakage in Saccharomyces cerevisiae: kinetics and relationship to yeast ethanol tolerance and alcohol fermentation productivity. Appl. Environ. Microbiol. 54:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrader, M., and H. D. Fahimi. 2004. Mammalian peroxisomes and reactive oxygen species. Histochem. Cell Biol. 122:383-393. [DOI] [PubMed] [Google Scholar]
- 38.Schubert, C. 2006. Can. biofuels finally take center stage? Nat. Biotechnol. 24:777-784. [DOI] [PubMed] [Google Scholar]
- 39.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simões, T., N. P. Mira, A. R. Fernandes, and I. Sá-Correia. 2006. The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl. Environ. Microbiol. 72:7168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simões, T., M. C. Teixeira, A. R. Fernandes, and I. Sá-Correia. 2003. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: important role of SPI1. Appl. Environ. Microbiol. 69:4019-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Támas, M. J., K. Luyten, F. C. Sutherland, A. Hernandez, J. Albertyn, H. Valadi, H. Li, B. A. Prior, S. G. Kilian, J. Ramos, L. Gustafsson, J. M. Thevelein, and S. Hohmann. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087-1104. [DOI] [PubMed] [Google Scholar]
- 44.Toh, T. H., G. Kayingo, M. J. van der Merwe, S. G. Kilian, J. E. Hallsworth, S. Hohmann, and B. A. Prior. 2001. Implications of FPS1 deletion and membrane ergosterol content for glycerol efflux from Saccharomyces cerevisiae. FEMS Yeast Res. 1:205-211. [DOI] [PubMed] [Google Scholar]
- 45.Van Uden, N. 1985. Ethanol toxicity and ethanol tolerance in yeasts. Ann. Rep. Ferment. Process. 8:11-58. [Google Scholar]
- 46.van Voorst, F., J. Houghton-Larsen, L. Jonson, M. C. Kielland-Brandt, and A. Brandt. 2006. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23:351-359. [DOI] [PubMed] [Google Scholar]
- 47.Viegas, C. A., P. F. Almeida, M. Cavaco, and I. Sá-Correia. 1998. The H+-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl. Environ. Microbiol. 64:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viegas, C. A., M. F. Rosa, I. Sá-Correia, and J. M. Novais. 1989. Inhibition of yeast growth by octanoic and decanoic acids produced during ethanolic fermentation. Appl. Environ. Microbiol. 55:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vindelov, J., and N. Arneborg. 2002. Saccharomyces cerevisiae and Zygosaccharomyces mellis exhibit different hyperosmotic shock responses. Yeast 19:429-439. [DOI] [PubMed] [Google Scholar]
- 50.Wiemken, A. 1990. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie van Leeuwenhoek 58:209-217. [DOI] [PubMed] [Google Scholar]
- 51.Wysocki, R., C. C. Chery, D. Wawrzycka, M. Van Hulle, R. Cornelis, J. M. Thevelein, and M. J. Tamas. 2001. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol. Microbiol. 40:1391-1401. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa, K., T. Tanaka, C. Furusawa, K. Nagahisa, T. Hirasawa, and H. Shimizu. 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9:32-44. [DOI] [PubMed] [Google Scholar]
- 53.You, K. M., C. L. Rosenfield, and D. C. Knipple. 2003. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl. Environ. Microbiol. 69:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, A., Q. Kong, L. Cao, and X. Chen. 2007. Effect of FPS1 deletion on the fermentation properties of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 44:212-217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.