Abstract
Many pathogens have the ability to survive and multiply in abiotic environments, representing a possible reservoir and source of human and animal exposure. Our objective was to develop a methodological framework to study spatially explicit environmental and meteorological factors affecting the probability of pathogen isolation from a location. Isolation of Listeria spp. from the natural environment was used as a model system. Logistic regression and classification tree methods were applied, and their predictive performances were compared. Analyses revealed that precipitation and occurrence of alternating freezing and thawing temperatures prior to sample collection, loam soil, water storage to a soil depth of 50 cm, slope gradient, and cardinal direction to the north are key predictors for isolation of Listeria spp. from a spatial location. Different combinations of factors affected the probability of isolation of Listeria spp. from the soil, vegetation, and water layers of a location, indicating that the three layers represent different ecological niches for Listeria spp. The predictive power of classification trees was comparable to that of logistic regression. However, the former were easier to interpret, making them more appealing for field applications. Our study demonstrates how the analysis of a pathogen's spatial distribution improves understanding of the predictors of the pathogen's presence in a particular location and could be used to propose novel control strategies to reduce human and animal environmental exposure.
The transmission cycle of many pathogens involves biotic hosts and abiotic environments. After infection of a host with a pathogen like Listeria monocytogenes, Bacillus anthracis, enterohemorrhagic Escherichia coli, Salmonella spp., or Toxoplasma gondii, large numbers of the pathogen may be shed into the environment where, under favorable conditions, they may survive, multiply, and infect new hosts, including humans (6, 11, 13, 30, 37). It is important to identify spatially explicit environmental and meteorological factors that favor a pathogen's presence in a particular environmental location. That information could be used to design novel measures to reduce the presence of the pathogen in the environment and prevent exposure and infection of animal and human hosts. For analysis of pathogens' spatial distribution in the environment, geographic information systems (GIS) integrated with standard statistical and epidemiological methods provide tremendous opportunities (5).
Detection of pathogens in environmental samples is usually based on culturing methods without enumeration, resulting in presence/absence data. For such data, a standard statistical approach to predict microbial presence as influenced by covariates would be logistic regression (LR). However, classification trees (CT) have recently been suggested as a powerful yet simple alternative to LR in ecological studies (7, 48). It is therefore of interest to contrast the performance of the CT with that of the standard LR approach in predicting pathogen isolation from a spatial location.
The objective of this study was to develop a methodological framework to study spatially explicit determinants affecting the local probability of pathogen isolation by using Listeria spp. as a model system. Specifically, our goals were (i) to examine the effect of environmental and meteorological factors on isolation of Listeria spp. from a spatial location and from soil, vegetation, and water layers of a location and (ii) to compare the predictive performance of LR and CT models. The genus Listeria was chosen as a model system because of the convenience of gathering data (Listeria bacteria are relatively prevalent in the environment; they have been isolated from 28% of sampled locations in the natural environment [38]) and because the genus Listeria includes the human-pathogenic species L. monocytogenes. There are currently six species in the genus Listeria, including two known pathogens (L. monocytogenes and Listeria ivanovii) and four nonpathogens (Listeria innocua, Listeria seeligeri, Listeria welshimeri, and Listeria grayi). L. ivanovii is predominantly an animal pathogen, infecting ruminants, while L. monocytogenes can cause disease (listeriosis) in a wide range of animal species, including humans. In addition, a new Listeria species, “Listeria marthii” sp. nov., has recently been identified (L. M. Graves, L. O. Helsel, A. G. Steigerwalt, R. E. Morey, M. I. Daneshvar, S. E. Roof, R. H. Orsi, E. D. Fortes, S. R. Millilo, H. C. den Bakker, M. Wiedmann, B. Swaminathan, and B. D. Sauders, submitted for publication).
MATERIALS AND METHODS
Description of data and study area.
To study the spatial distribution of Listeria spp., we used data collected as part of a larger study, which is described by Sauders (38). Briefly, over a 2-year period (2001 and 2002), a total of 907 samples of soil, water (including ponds, lakes, puddles, river streams, runoff water, and swamps), and vegetation (including pond algae, decaying logs, field grass, grass, leaf debris, and moss) were collected in the following four areas in New York State (NYS), representing the natural environment: Finger Lakes National Forest (FLNF), Adirondack Park, Catskill Park, and the Connecticut Hill Wildlife Management Area (CHWMA). Specifically, in 2001, samples were obtained through two to three visits per study area throughout the spring, summer, and autumn, while in 2002 each study area was visited once in the spring, summer, and autumn. Geospatial location data for each sample were collected with a Garmin Emap handheld global positioning system (GPS). Also, the date of sample collection was recorded. Samples were collected into sterile Whirl-Pak bags (Nasco, Fort Atkinson, WI) using sterile gloves and/or presterilized disposable plastic spatulas or scoops. Samples were held on wet ice for up to 24 h before being cultured. All samples were cultured for the presence of Listeria with selective enrichment in Listeria enrichment broth (Difco, Becton Dickinson, Sparks, MD). Isolated Listeria species were as follows: L. seeligeri (67%), L. welshimeri (24%), L. monocytogenes (6%), “L. marthii” sp. nov. (a new species; 2%), and L. innocua (<1%). All Listeria isolates were characterized by PCR amplification and sequencing of the partial open reading frame of the stress response gene sigB and, for isolates collected in 2001, the housekeeping gene gap. However, sample size limitations precluded statistical analyses at the species and genotype levels. There were 567 unique sampling locations in the Sauders study (38). Soil, vegetation, and water samples were collected at 303, 302, and 294 of these unique locations, respectively. Because all of the samples collected at the same location share an identical set of observations, we applied a simple rule to characterize the presence of Listeria in a location: one positive sample was sufficient to consider a location positive. The same rule was applied for isolation of Listeria from the soil, vegetation, and water layers of a location.
Spatial data modeling.
For each sampled location in the Sauders study (38), we obtained potentially relevant spatially referenced covariates (details and sources described below) describing the local ecology from readily available spatial data models. All the spatial data models and GPS data for sampling locations were imported into ArcGIS 9 (ESRI, Redland, CA), reprojected into the universal transverse mercator coordinate system, North American Datum of 1983, clipped to the study area, and overlaid with sampling locations to obtain information on environmental factors associated with each sampling location. In total, we obtained 77 variables (defined in Table 1) that could be grouped under the following categories: soil properties, precipitation, ambient temperature, alternating freezing and thawing temperatures (freeze-thaw cycles), geographic position, and calendar time. Soil property variables (shown in Table 1) were obtained for 13 NYS counties where sampling was performed, namely, Schuyler, Seneca, Hamilton, Tompkins, Greene, Delaware, Sullivan, Ulster, Essex, Fulton, Franklin, Herkimer, and St. Lawrence, from the compiled tabular and spatial Soil Survey Geographic (SSURGO) data (47). Information on precipitation prior to sample collection was obtained from the U.S. Historical Climatology Network Daily Temperature, Precipitation, and Snow Data (54). Specifically, for each sampling site, the closest weather station with available information on the precipitation on the day of and days before sample collection was identified. It is not known whether precipitation occurring closer to or farther before sample collection has more effect on isolation of Listeria from a location. Therefore, we created four variables describing the amount of rain on the day of sample collection, as well as on 1 day, 2 days, and 3 days before (Table 1). As the effect of precipitation probably lasts several days, the average amount of rain over a period of time may be a more important factor influencing isolation of Listeria spp. from a location. Therefore, we created 10 additional precipitation variables describing the average precipitation during an ever-increasing time window prior to sample collection (Table 1). For our analysis, all the rainfall measurements were converted from inches to millimeters (1 in. = 25.4 mm). In addition to precipitation, information was obtained on the ambient temperature on the day of and days prior to sample collection (54). However, again, it is unclear whether minimum, average, or maximum daily temperature most influences the recovery (isolation) of Listeria spp. from a location. Also, it is unclear whether the temperature on days closer to sample collection or on days farther before is more influential. Furthermore, the effect of ambient temperatures may be cumulative, and so an average of the measurements over several days may be a better predictor for Listeria recovery. Therefore, following the same approach used to derive and name precipitation variables, we created 14 variables for each of the minimum, average, and maximum daily ambient temperatures (Table 1). For our analysis, all temperature measurements were converted from degrees Fahrenheit to degrees Celsius {°C = [(F − 32)/9] × 5}. We were also interested in the effect of freeze-thaw cycles on the status of isolation of Listeria spp. from a location. The occurrence of freeze-thaw cycles was estimated from Williams et al. (54): if on any day during the 10-day period prior to sample collection or on any two consecutive days during that period the minimum daily temperature was below 0°C while the maximum was above 0°C, this was recorded as the occurrence of a freeze-thaw cycle. From this information, we created six “freeze-thaw cycle” variables (Table 1). We were also interested in whether the geographic position of a sampling location with respect to cardinal directions, described by easting and northing geographic coordinates (expressed in meters), had any effect on isolation of Listeria spp. from a location. Easting refers to the eastward measured distance from the “false easting,” which is uniquely defined in each universal transverse mercator zone, while northing refers to the northward measured distance from the equator. Finally, we were interested in whether the calendar time when samples were collected and the proximity to roads and water bodies (lakes, rivers, and streams) had any effect on isolation of Listeria spp. from a location (Table 1). To determine the proximity of a sampling location to the closest road, we used road data for the NYS counties where samples were collected as well as for two nearby counties, Oneida and Clinton (45). Spatial data on hydrography (31) were used to estimate the proximity of the sampling location to the nearest body of water. While potentially relevant, there was no information about the exposure of the location to sunlight, e.g., north versus south slope. As L. monocytogenes was the only pathogenic Listeria species represented in the data set used and as it represented only 6% of all isolated Listeria spp., the proximity and density of livestock and wildlife populations in the study area were not considered.
TABLE 1.
Names, descriptions, defining attributes, and units of measurement (where applicable) of the considered variables grouped into soil properties, precipitation, ambient temperature, freeze-thaw cycles, geographic position, and calendar timea
| Category and variable name | Description | Defining attribute | Unit of measurement |
|---|---|---|---|
| Soil properties | |||
| Loam.soil | Obtained by grouping all soil types in the SSURGO database as loam soil and not loam soil | Loam soil type | |
| Slope.gradient | Difference in elevation between two points, expressed as a percentage between two points and shown as the weighted avg slope gradient of all components in the unit of the SSURGO map | % | |
| Water.depth | The shallowest depth to a wet soil layer (water table) during the yr, expressed as cm from the soil surface for components whose composition in the SSURGO map unit is equal to or exceeds 15% | cm | |
| Water.Storage.25 | The vol of water that the soil to the specified depth can store that is available to plants and expressed as the weighted avg of all components in the SSURGO map unit | 25 cm | cm |
| Water.Storage.50 | As for Water.Storage.25 | 50 cm | cm |
| Water.Storage.100 | As for Water.Storage.25 | 100 cm | cm |
| Water.Storage.150 | As for Water.Storage.25 | 150 cm | cm |
| Drainage | The natural drainage condition of the soil (referring to the frequency and duration of wet periods) of the dominant drainage class for the SSURGO map unitb | ||
| Precipitation | |||
| Precipitation.0 | Amt of rain on the specified day | Day t0 | mm |
| Precipitation.1 | As for Precipitation.0 | Day t1 | mm |
| Precipitation.2 | As for Precipitation.0 | Day t2 | mm |
| Precipitation.3 | As for Precipitation.0 | Day t3 | mm |
| Precipitation.0_1 | Avg precipitation for the specified period | Period t0-t1 | mm |
| Precipitation.0_2 | As for Precipitation.0_1 | Period t0-t2 | mm |
| Precipitation.0_3 | As for Precipitation.0_1 | Period t0-t3 | mm |
| Precipitation.0_4 | As for Precipitation.0_1 | Period t0-t4 | mm |
| Precipitation.0_5 | As for Precipitation.0_1 | Period t0-t5 | mm |
| Precipitation.0_6 | As for Precipitation.0_1 | Period t0-t6 | mm |
| Precipitation.0_7 | As for Precipitation.0_1 | Period t0-t7 | mm |
| Precipitation.0_8 | As for Precipitation.0_1 | Period t0-t8 | mm |
| Precipitation.0_9 | As for Precipitation.0_1 | Period t0-t9 | mm |
| Precipitation.0_10 | As for Precipitation.0_1 | Period t0-t10 | mm |
| Ambient temp | |||
| Temperature.L.0 | Minimum daily temp on the specified day | Day t0 | °C |
| Temperature.L.1 | As for Temperature.L.0 | Day t1 | °C |
| Temperature.L.2 | As for Temperature.L.0 | Day t2 | °C |
| Temperature.L.3 | As for Temperature.L.0 | Day t3 | °C |
| Temperature.L.0_1 | Avg of the minimum daily temp in the specified period | Period t0-t1 | °C |
| Temperature.L.0_2 | As for Temperature.L.0_1 | Period t0-t2 | °C |
| Temperature.L.0_3 | As for Temperature.L.0_1 | Period t0-t3 | °C |
| Temperature.L.0_4 | As for Temperature.L.0_1 | Period t0-t4 | °C |
| Temperature.L.0_5 | As for Temperature.L.0_1 | Period t0-t5 | °C |
| Temperature.L.0_6 | As for Temperature.L.0_1 | Period t0-t6 | °C |
| Temperature.L.0_7 | As for Temperature.L.0_1 | Period t0-t7 | °C |
| Temperature.L.0_8 | As for Temperature.L.0_1 | Period t0-t8 | °C |
| Temperature.L.0_9 | As for Temperature.L.0_1 | Period t0-t9 | °C |
| Temperature.L.0_10 | As for Temperature.L.0_1 | Period t0-t10 | °C |
| Temperature.a.0 | Avg daily temp on the specified day | Day t0 | °C |
| Temperature.a.1 | As for Temperature.a.0 | Day t1 | °C |
| Temperature.a.2 | As for Temperature.a.0 | Day t2 | °C |
| Temperature.a.3 | As for Temperature.a.0 | Day t3 | °C |
| Temperature.a.0_1 | Avg of the avg daily temp in the specified period | Period t0-t1 | °C |
| Temperature.a.0_2 | As for Temperature.a.0_1 | Period t0-t2 | °C |
| Temperature.a.0_3 | As for Temperature.a.0_1 | Period t0-t3 | °C |
| Temperature.a.0_4 | As for Temperature.a.0_1 | Period t0-t4 | °C |
| Temperature.a.0_5 | As for Temperature.a.0_1 | Period t0-t5 | °C |
| Temperature.a.0_6 | As for Temperature.a.0_1 | Period t0-t6 | °C |
| Temperature.a.0_7 | As for Temperature.a.0_1 | Period t0-t7 | °C |
| Temperature.a.0_8 | As for Temperature.a.0_1 | Period t0-t8 | °C |
| Temperature.a.0_9 | As for Temperature.a.0_1 | Period t0-t9 | °C |
| Temperature.a.0_10 | As for Temperature.a.0_1 | Period t0-t10 | °C |
| Temperature.H.0 | Maximum daily temp on the specified day | Day t0 | °C |
| Temperature.H.1 | As for Temperature.H.0 | Day t1 | °C |
| Temperature.H.2 | As for Temperature.H.0 | Day t2 | °C |
| Temperature.H.3 | As for Temperature.H.0 | Day t3 | °C |
| Temperature.H.0_1 | Avg of the maximum daily temp in the specified period | Period t0-t1 | °C |
| Temperature.H.0_2 | As for Temperature.H.0_1 | Period t0-t2 | °C |
| Temperature.H.0_3 | As for Temperature.H.0_1 | Period t0-t3 | °C |
| Temperature.H.0_4 | As for Temperature.H.0_1 | Period t0-t4 | °C |
| Temperature.H.0_5 | As for Temperature.H.0_1 | Period t0-t5 | °C |
| Temperature.H.0_6 | As for Temperature.H.0_1 | Period t0-t6 | °C |
| Temperature.H.0_7 | As for Temperature.H.0_1 | Period t0-t7 | °C |
| Temperature.H.0_8 | As for Temperature.H.0_1 | Period t0-t8 | °C |
| Temperature.H.0_9 | As for Temperature.H.0_1 | Period t0-t9 | °C |
| Temperature.H.0_10 | As for Temperature.H.0_1 | Period t0-t10 | °C |
| Freeze-thaw cycles | |||
| Freeze.thaw.0 | Freeze-thaw cycle occurring on the specified day | Day t0 | NA |
| Freeze.thaw.1 | As for Freeze.thaw.0 | Day t1 | NA |
| Freeze.thaw.2 | As for Freeze.thaw.0 | Day t2 | NA |
| Freeze.thaw.3 | As for Freeze.thaw.0 | Day t3 | NA |
| Freeze.thaw.0_3 | Freeze-thaw cycle on any of the days in the specified period | Period t0-t3 | NA |
| Freeze.thaw.s.0_10 | Total no. of freeze-thaw cycles during the specified period | Period t0-t10 | NA |
| Geographic positions | |||
| Northing | Position with respect to the specified cardinal direction | To the North | m |
| Easting | Position with respect to the specified cardinal direction | To the East | m |
| Dtoroad | Proximity to the nearest road | m | |
| Dtowater | Proximity to the nearest body of water | m | |
| Park | Natural area where samples were collected | NA | |
| Calendar time | |||
| Season | Season when samples were collected | NA | |
| Month | Month when samples were collected | NA |
The day of sample collection is denoted as t0, the day before is t1, and so on until 10 days before sample collection (t10). NA, not applicable.
The original SSURGO categories were regrouped such that “excessively” and “somewhat excessively” drained classes formed the excessively drained class, “well-drained” and “moderately well-drained” classes became the moderately drained class, while “poorly,” “somewhat poorly,” and “very poorly” drained classes were recategorized into the poorly drained class.
Statistical analysis.
All statistical analyses were performed by using R (34). Independent variables were first evaluated for unconditional associations with the dependent variable (i.e., the overall, as well as soil-, vegetation-, and water-specific occurrence of Listeria) using a chi-square test for categorical data and t test for continuous data. Fisher's exact test was used when one or more of the expected cell frequencies in two-by-two tables were less than 5 and when more than 20% of the expected cell frequencies were less than 5 in larger tables. To assess whether continuous variables satisfied the normality assumption, required for application of the t test, the D'Agostino-Pearson omnibus test was performed. If a continuous variable did not satisfy the normality assumption, the Wilcoxon rank-sum test was used. Continuous variables with considerable numbers of ties (e.g., related to a common weather station) were assessed by the exact Wilcoxon rank-sum test that computes exact conditional (on the data) P values and quantiles using the shift algorithm (43). Associations between independent categorical variables were tested by use of the chi-square test. Correlations between independent continuous variables were assessed based on Pearson's correlation coefficient for the normally distributed variables. Correlations between variables that did not satisfy the normality condition were assessed based on Spearman's rho coefficient. Associations between independent categorical and continuous variables were assessed by the t test, the Wilcoxon rank-sum test, or the exact Wilcoxon rank-sum test as appropriate. In the univariate analysis, P values of ≤0.05 were considered statistically significant. Correction for multiple comparisons was not performed because of the exploratory character of the research to make sure that all important associations were identified. If more than one independent variable was associated with the dependent variable of interest at the 5% level, these variables were tested in a multivariable LR and CT. When two or more of the independent variables applicable for multivariable modeling were correlated, among the variables that had the most significant relationship with the dependent variable, the one that led to the greatest change in deviance in LR was retained in the LR and CT. Usually, this was the preferred discriminating factor in CT. Multivariable modeling was carried out on a subset of data, with complete observations on variables chosen to be included in the full LR model to assure a fair comparison between the LR and CT methods. The presence of spatial patterns in the Listeria spp. isolation data was analyzed by examination of the nearest-neighbor distance.
PCA.
Correlation analysis indicated that weather variables, including 14 variables grouped under the rainfall set, 42 variables describing ambient temperatures (14 variables for each of the minimum, average, and maximum daily temperature measurements), and 6 variables describing freeze-thaw cycles, were highly correlated. Therefore, the 62 weather-related variables were subjected to a principal component analysis (PCA) to examine whether the three weather variables (precipitation, ambient temperatures, and freeze-thaw cycles) do in fact emerge. Because variables were measured in different units, prior to PCA, the data were standardized by subtracting the mean and dividing by the standard deviation, i.e., we performed an eigenanalysis of the correlation matrix. The number of meaningful components to retain was determined based on the proportion of variance accounted and the interpretability criteria (16). According to the proportion of variance accounted criterion, only components accounting for >5% of the total variance were retained. According to the interpretability criteria, (i) there had to be at least three variables with major loadings on each retained component (a general rule of thumb for a value of a loading that designates a useful signal is ±0.2 to 0.35 [35]) and (ii) the rotated pattern had to demonstrate “simple structure.” Here, loading is a correlation coefficient between a variable and its principle component, while “simple structure” means that (a) most variables have relatively high factor loadings on only one component and near zero loadings on the other components and (b) most components have relatively high factor loadings for some variables and near zero loadings for the remaining variables.
The results of PCA were utilized in two scenarios. In the first scenario, weather variables loading on the same principal component were individually tested in univariate analysis and, among those statistically significantly associated with the dependent variable, one variable was chosen for the multivariable LR and CT modeling (as explained above). In the second scenario, weather variables in each retained principal component were substituted by a single continuous variable containing the predicted factor (component) scores. These variables were subject to univariate analysis and, where appropriate, used in the multivariable LR and CT modeling instead of all of the actual weather variables in the principal component.
LR.
The multivariable LR models were selected through an automatic stepwise regression (the “stepAIC” function in the MASS package) (49) based on the Akaike information criterion while obeying the principle of marginality. When applicable, the selected model was further simplified by extracting nonsignificant terms (P ≥ 0.05), starting with the most complex one. Each term deletion was followed by a likelihood ratio test. The assumption of a linear relationship between continuous explanatory variables and outcome was assessed by adding a quadratic term (the explanatory variable squared) to the model (9). When the quadratic term was found to be significant, the applicability of other polynomials was explored. An assessment of how the models fit the data was determined by using the Le Cessie-van Houwelingen-Copas-Hosmer test. Collinearity was investigated by calculating variance inflation factors (VIFs) for each of the explanatory variables in the multivariable model. To reduce high collinearity (VIF values of >10 and a mean of the VIFs considerably larger than 1) (4), one or more continuous variables in the model were centered (by subtracting the mean of the variable). Then the centered version of the variable was used in the model. An LR model can account for spatial autocorrelation in the data by inclusion of a spatial dependence variable and is then formally called an autologistic regression model (28). To assure a fair comparison with CT, which cannot directly consider spatial dependence in the data, we focused on analysis of the nonspatial LR models that did not consider spatial dependence. However, for the final nonspatial LR models that considered the actual weather variables (i.e., not the predicted component scores from PCA), variograms of the residuals were examined for evidence of spatial autocorrelation and the models were modified by inclusion of an autocovariate term. These autologistic regression models were analyzed to assess whether capturing spatial dependency in an LR model would change parameter estimates and their significance as well as the predictive performance of these models.
CT.
CT were built using the rpart package (44). The Gini index was used as a measure of node impurity. In pruning the tree to its optimal size, we used 10-fold cross-validation to choose the tree with the smallest misclassification error based on the “1 − standard error” (1 − SE) rule. Microbial cultures used to detect a pathogen in an environmental sample usually have a very high specificity (meaning that there are very few if any false positive [FP] isolation results) and low sensitivity (meaning that many negatives are actually false negative [FN] results). Consequently, all the positives in the data set are most likely to be truly positive, while many negatives may in fact be FN. To account for that, FN were penalized more than FP; costs of an FN and FP were set to 4 and 1, respectively. These costs produced trees with at least two nodes and were chosen through a trial-and-error method.
Testing the predictive performance of LR and CT.
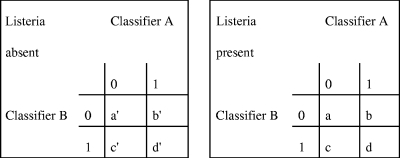
To compare models produced by two different statistical methods, one would need a common criterion. This criterion must be unbiased and independent of the method used to develop a particular model (48). Comparing CT with LR is difficult because none of the error rates and goodness-of-fit statistics computed by the methods satisfy these requirements. The solution is to compare the models on the basis of their predictive accuracy, that is, the ability to correctly classify new cases in an independent test data set (48). However, we did not have the luxury of a large data set that could be divided into learning and independent test data sets. Therefore, predictive performance of the LR and CT was assessed on the same set of 10 subsamples of data through a 10-fold cross-validation and compared on two statistics, sensitivity and specificity. The outcome values predicted from a CT are dichotomous and could be easily summarized in a confusion matrix, from which sensitivity and specificity could be easily computed. However, the values predicted from an LR model vary continuously between 0 and 1. To dichotomize results of an LR model, a cutoff value is required. Choosing 0.5 as the cutoff is reasonable only if the prior probabilities of class 1 and class 0 are the same in the population of interest and if the costs of an FP and FN are the same (48). However, in our data sets, neither of the conditions was met. To account for the low sensitivity and high specificity of microbial cultures (and the corresponding high number of FN and low number of FP) and to assure comparability with the CT method, costs of an FN and FP were set to 4 and 1, respectively. Using these class-conditional misclassification costs, costs were estimated over the full range of possible cutoffs obtained from 10-fold cross-validation. The cutoff with the lowest associated misclassification cost was considered optimal and was used to dichotomize the LR output so that the confusion matrix could be constructed and sensitivity and specificity could be calculated. Because both LR and CT models were evaluated on the same data sets, Linnet and Brandt's test, an adaptation of McNemar's test for comparison of correlated proportions, was applied to test whether an observed difference in the test performances was statistically significant (25). Positive and negative isolates were divided into four groups according to the combined predictions of the two classifiers, LR and CT (Fig. 1). Then, the test statistic was
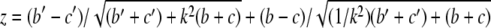 |
where 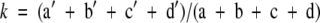 . The null hypothesis of no difference between LR and CT was rejected if the absolute value of z exceeded 1.96 (two-sided test; 2α = 0.05). If the Linnet and Brandt's test showed a statistically significant difference between LR and CT, two separate McNemar's tests were conducted to test the null hypotheses of equal sensitivities (from the “Listeria present” cross-tabulation in Fig. 1) and specificities (from the “Listeria absent” cross-tabulation in Fig. 1) obtained for the two classifiers (LR and CT).
. The null hypothesis of no difference between LR and CT was rejected if the absolute value of z exceeded 1.96 (two-sided test; 2α = 0.05). If the Linnet and Brandt's test showed a statistically significant difference between LR and CT, two separate McNemar's tests were conducted to test the null hypotheses of equal sensitivities (from the “Listeria present” cross-tabulation in Fig. 1) and specificities (from the “Listeria absent” cross-tabulation in Fig. 1) obtained for the two classifiers (LR and CT).
FIG. 1.
Cross-tabulations of isolation results (Listeria absent and Listeria present) for Linnet and Brandt's test to compare the predictive performance of any two classifiers A and B (e.g., LR and CT models). “0” indicates a negative isolation result, and “1” indicates a positive one. b′ is the number of negative samples classified correctly by classifier B and classified falsely by classifier A and conversely for c′ samples. Analogous reasoning applies to the positive samples.
RESULTS
Of the 567 unique sampling locations (Fig. 2), 159 (28%) tested positive for Listeria spp. Listeria bacteria were isolated from samples collected at 19% (57/303), 34% (103/302), and 16% (48/294) of the unique sampling locations of soil, vegetation, and water, respectively. Among 303 unique soil sampling locations, one location had a duplicate sample collected (both positive). Among 302 locations where vegetation was sampled, five locations had a duplicate sample collected (four with one positive and one negative sample and one with both samples positive) and one had three samples collected (all three positive). No duplicate samples of water were collected from the same location. The median of the nearest-neighbor distance was 52 m (range, 1 to 11,750 m) for all sampling locations, 95 m (range, 2 to 12,660 m) for soil, 87 m (range, 2 to 12,660 m) for vegetation, and 60 m (range, 1 to 19,040 m) for water. In all four data sets, the median of the nearest-neighbor distance was greater among positive locations than among negative locations, but the ranges of these measurements overlapped considerably. Weather data were obtained from the weather station closest to the sampling site. The average distance to the closest weather station was 38 km (range, 4 to 71 km).
FIG. 2.
Study area.
Univariate analyses.
Table 2 shows a cross-tabulation of data between each of the response variables (i.e., occurrence of Listeria in [i] a spatial location as well as in [ii] the soil, [iii] vegetation, and [iv] water layers) and significantly associated categorical explanatory variables. The season and month of sample collection were associated with all four response variables. However, because these independent variables are only proxies for seasonal weather characteristics, they were not considered further in the multivariable LR and CT analyses and are not shown in Table 2. Table 3 shows medians and interquartile ranges of continuous variables for locations with and without Listeria isolates significantly associated with the response variables. For variables that were subjected to PCA (Tables 2 and 3), we show only variables that loaded on only one of the principal components (Table 4). These variables did not show a complex structure and were therefore considered for testing in univariate analysis and, where applicable, in multivariable LR and CT.
TABLE 2.
Categorical independent variables for isolation of Listeria spp. from the environmenta
| Variable group, variable, and categoryb |
Listeria absent
|
Listeria present
|
OR | OR lowerc | OR upperc | P value | ||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | |||||
| Spatial location | ||||||||
| Position | ||||||||
| Park | NA | NA | NA | NA | NA | NA | NA | NA |
| Adirondacks | 95 | 0.23 | 23 | 0.14 | 1 | NA | NA | NA |
| Catskill | 112 | 0.27 | 49 | 0.31 | 1.81 | 1.03 | 3.18 | 0.039 |
| FLNF | 120 | 0.29 | 42 | 0.26 | 1.45 | 0.81 | 2.57 | 0.208 |
| CHWMA | 81 | 0.2 | 45 | 0.28 | 2.29 | 1.28 | 4.11 | 0.005 |
| Soil properties | ||||||||
| Loam.soil | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 154 | 0.42 | 34 | 0.23 | 1 | NA | NA | NA |
| 1 | 214 | 0.58 | 115 | 0.77 | 2.43 | 1.58 | 3.76 | 0.000 |
| Drainage | NA | NA | NA | NA | NA | NA | NA | NA |
| Excessive | 30 | 0.09 | 5 | 0.04 | 1 | NA | NA | NA |
| Poor | 149 | 0.46 | 42 | 0.3 | 1.69 | 0.62 | 4.63 | 0.302 |
| Well | 145 | 0.45 | 94 | 0.67 | 3.89 | 1.46 | 10.38 | 0.004 |
| Freeze-thaw cycle | ||||||||
| Freeze.thaw.2 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 247 | 0.61 | 128 | 0.81 | 1 | NA | NA | NA |
| 1 | 161 | 0.39 | 31 | 0.19 | 0.37 | 0.24 | 0.58 | 0.000 |
| Freeze.thaw.3 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 269 | 0.66 | 140 | 0.88 | 1 | NA | NA | NA |
| 1 | 139 | 0.34 | 19 | 0.12 | 0.26 | 0.16 | 0.44 | 0.000 |
| Soil layer | ||||||||
| Freeze-thaw cycle | ||||||||
| Freeze.thaw.0 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 170 | 0.69 | 48 | 0.84 | 1 | NA | NA | NA |
| 1 | 76 | 0.31 | 9 | 0.16 | 0.42 | 0.2 | 0.9 | 0.022 |
| Freeze.thaw.3 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 175 | 0.71 | 49 | 0.86 | 1 | NA | NA | NA |
| 1 | 71 | 0.29 | 8 | 0.14 | 0.4 | 0.18 | 0.89 | 0.022 |
| Vegetation layer | ||||||||
| Soil properties | ||||||||
| Loam.soil | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 63 | 0.35 | 19 | 0.19 | 1 | NA | NA | NA |
| 1 | 118 | 0.65 | 79 | 0.81 | 2.22 | 1.23 | 3.99 | 0.007 |
| Drainage | NA | NA | NA | NA | NA | NA | NA | NA |
| Excessive | 22 | 0.13 | 4 | 0.04 | 1 | NA | NA | NA |
| Poor | 58 | 0.33 | 28 | 0.3 | 2.66 | 0.83 | 8.44 | 0.089 |
| Well | 94 | 0.54 | 61 | 0.66 | 3.57 | 1.17 | 10.86 | 0.018 |
| Freeze-thaw cycle | ||||||||
| Freeze.thaw.0 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 133 | 0.67 | 84 | 0.82 | 1 | NA | NA | NA |
| 1 | 66 | 0.33 | 19 | 0.18 | 0.46 | 0.26 | 0.81 | 0.007 |
| Freeze.thaw.2 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 121 | 0.61 | 85 | 0.83 | 1 | NA | NA | NA |
| 1 | 78 | 0.39 | 18 | 0.17 | 0.33 | 0.18 | 0.59 | 0.000 |
| Freeze.thaw.3 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 131 | 0.66 | 92 | 0.89 | 1 | NA | NA | NA |
| 1 | 68 | 0.34 | 11 | 0.11 | 0.23 | 0.12 | 0.46 | 0.000 |
| Water layer | ||||||||
| Position | ||||||||
| Park | ||||||||
| Adirondacks | 57 | 0.23 | 6 | 0.13 | 1.00 | NA | NA | NA |
| Catskill | 68 | 0.28 | 17 | 0.35 | 2.38 | 0.88 | 6.42 | 0.082 |
| FLNF | 78 | 0.32 | 4 | 0.08 | 0.49 | 0.13 | 1.81 | 0.274 |
| CHWMA | 43 | 0.17 | 21 | 0.44 | 4.64 | 1.72 | 12.48 | 0.001 |
| Soil properties | ||||||||
| Loam.soil | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 104 | 0.47 | 10 | 0.23 | 1.00 | NA | NA | NA |
| 1 | 117 | 0.53 | 34 | 0.77 | 3.02 | 1.42 | 6.42 | 0.003 |
| Freeze-thaw cycle | ||||||||
| Freeze.thaw.3 | NA | NA | NA | NA | NA | NA | NA | NA |
| 0 | 173 | 0.70 | 42 | 0.88 | 1.00 | NA | NA | NA |
| 1 | 73 | 0.30 | 6 | 0.13 | 0.34 | 0.14 | 0.83 | 0.014 |
NA, not applicable; variables are defined in Table 1.
“0” indicates a negative isolation result, and “1” indicates a positive one.
OR lower and OR upper indicate lower and upper bounds of a 95% confidence interval, respectively.
TABLE 3.
Continuous independent variables for isolation of Listeria spp. from the environmenta
| Variable group and variable |
Listeria absent
|
Listeria present
|
P value | ||||
|---|---|---|---|---|---|---|---|
| Median | 25th % | 75th % | Median | 25th % | 75th % | ||
| Spatial location | |||||||
| Position | |||||||
| Northing (103) | 4,702 | 4,670 | 4,711 | 4,693 | 4,670 | 4,707 | 0.036 |
| Soil properties | |||||||
| Slope.gradient | 6.00 | 2.80 | 10.00 | 12.00 | 5.00 | 20.00 | 0.000 |
| Water.depth | 31.00 | 0.00 | 38.00 | 31.00 | 0.00 | 54.00 | 0.007 |
| Water.storage.50 | 5.42 | 4.78 | 6.65 | 6.60 | 5.42 | 8.05 | 0.000 |
| Water.storage.100 | 7.76 | 5.78 | 10.10 | 8.10 | 6.78 | 11.35 | 0.003 |
| Water.storage.150 | 7.78 | 6.78 | 11.35 | 8.96 | 7.22 | 11.83 | 0.005 |
| Precipitation | |||||||
| Precipitation.2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.41 | 0.006 |
| Precipitation.0_3 | 0.38 | 0.00 | 1.84 | 0.95 | 0.19 | 2.03 | 0.048 |
| Precipitation.0_6 | 1.09 | 0.18 | 3.63 | 1.16 | 0.22 | 4.35 | 0.002 |
| Temp | |||||||
| Temperature.a.0_2 | 9.82 | 6.30 | 17.41 | 14.63 | 7.22 | 21.30 | 0.003 |
| Temperature.a.0_3 | 8.33 | 5.97 | 17.78 | 15.14 | 8.06 | 20.28 | 0.000 |
| Temperature.a.0_4 | 9.44 | 6.11 | 18.78 | 14.33 | 7.11 | 20.56 | 0.000 |
| Temperature.a.0_5 | 9.63 | 5.56 | 19.44 | 13.70 | 6.57 | 20.56 | 0.000 |
| Temperature.a.0_6 | 8.41 | 6.19 | 19.52 | 13.10 | 6.51 | 21.43 | 0.001 |
| Temperature.a.0_7 | 7.43 | 6.67 | 19.44 | 13.06 | 6.46 | 22.01 | 0.008 |
| Temperature.a.0_8 | 8.15 | 6.70 | 19.14 | 13.02 | 6.54 | 22.28 | 0.011 |
| Temperature.a.0_9 | 8.28 | 6.50 | 19.22 | 13.22 | 7.28 | 22.33 | 0.006 |
| Temperature.L.0_6 | 3.49 | 1.03 | 12.62 | 6.59 | 2.78 | 14.84 | 0.000 |
| Temperature.L.0_7 | 2.57 | 2.15 | 12.43 | 6.60 | 2.29 | 15.00 | 0.003 |
| Temperature.L.0_8 | 3.09 | 2.01 | 12.65 | 6.85 | 1.67 | 15.19 | 0.010 |
| Freeze-thaw cycle | |||||||
| Temperature.a.3 | 15.56 | 2.22 | 20.56 | 17.22 | 8.89 | 20.56 | 0.011 |
| Temperature.L.0 | 5.56 | −0.56 | 12.78 | 9.44 | 2.78 | 15.00 | 0.000 |
| Temperature.L.3 | 8.89 | −1.67 | 13.33 | 11.11 | 2.78 | 14.44 | 0.007 |
| Temperature.L.0_1 | 4.17 | 0.56 | 10.83 | 7.78 | 3.19 | 14.86 | 0.000 |
| PCA scores | |||||||
| PC.temperature | 1.90 | −3.04 | 3.75 | −0.81 | −3.97 | 2.85 | 0.000 |
| PC.rain | 0.56 | −0.36 | 1.19 | 0.33 | −0.36 | 1.15 | 0.004 |
| Soil layer | |||||||
| Position | |||||||
| Easting (103) | 495.5 | 353.1 | 550.4 | 363.1 | 352.7 | 511.6 | 0.012 |
| Soil property | |||||||
| Water.depth | 31.00 | 0.00 | 48.00 | 41.00 | 20.00 | 54.00 | 0.045 |
| Precipitation | |||||||
| Precipitation.0 | 0.00 | 0.00 | 0.25 | 0.00 | 0.00 | 4.06 | 0.011 |
| Precipitation.2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 8.13 | 0.004 |
| Freeze-thaw cycle | |||||||
| Temperature.L.0_1 | 5.00 | 0.56 | 14.72 | 7.78 | 5.00 | 11.67 | 0.041 |
| PCA score | |||||||
| PC.freeze.thaw | −0.27 | −0.84 | 0.26 | −0.03 | −0.31 | 0.15 | 0.045 |
| Vegetation layer | |||||||
| Position | |||||||
| Northing (103) | 4,694 | 4,680 | 4,711 | 4,693 | 4,669 | 4,707 | 0.047 |
| Precipitation | |||||||
| Precipitation.2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.41 | 0.024 |
| Temperature | |||||||
| Temperature.a.0_2 | 9.82 | 6.30 | 17.41 | 16.67 | 9.82 | 21.48 | 0.000 |
| Temperature.a.0_3 | 9.10 | 5.97 | 18.19 | 17.36 | 8.33 | 21.25 | 0.000 |
| Temperature.a.0_4 | 9.44 | 6.11 | 18.44 | 18.11 | 7.69 | 21.64 | 0.000 |
| Temperature.a.0_5 | 9.63 | 5.56 | 18.89 | 18.06 | 7.34 | 21.67 | 0.000 |
| Temperature.a.0_6 | 8.41 | 6.19 | 19.52 | 17.78 | 7.22 | 22.08 | 0.000 |
| Temperature.a.0_7 | 7.43 | 6.46 | 19.44 | 17.78 | 7.29 | 22.17 | 0.000 |
| Temperature.a.0_8 | 8.15 | 6.54 | 19.14 | 17.84 | 7.07 | 22.38 | 0.001 |
| Temperature.a.0_9 | 8.28 | 6.50 | 19.22 | 17.44 | 7.51 | 22.92 | 0.001 |
| Temperature.L.0_6 | 3.49 | 1.03 | 12.06 | 10.79 | 2.96 | 15.97 | 0.000 |
| Temperature.L.0_7 | 2.57 | 2.15 | 12.43 | 10.83 | 2.41 | 16.25 | 0.000 |
| Temperature.L.0_8 | 3.09 | 1.67 | 12.65 | 10.93 | 2.49 | 16.48 | 0.000 |
| Freeze-thaw cycle | |||||||
| Temperature.a.3 | 15.56 | 2.22 | 20.56 | 18.33 | 10.00 | 21.11 | 0.000 |
| Temperature.L.0 | 5.56 | −0.56 | 11.67 | 10.00 | 3.33 | 15.00 | 0.000 |
| Temperature.L.3 | 8.89 | −1.67 | 13.33 | 12.22 | 3.33 | 15.56 | 0.000 |
| Temperature.L.0_1 | 4.17 | 0.35 | 10.83 | 8.89 | 5.00 | 15.00 | 0.000 |
| Temperature.H.3 | 17.78 | 6.11 | 26.67 | 23.89 | 17.22 | 27.22 | 0.005 |
| PCA score | |||||||
| PC.temperature | 2.10 | −2.81 | 3.97 | −2.36 | −4.37 | 2.82 | 0.000 |
| Water layer | |||||||
| Position | |||||||
| Northing (103) | 4,706 | 4,668 | 4,711 | 4,691 | 4,670 | 4,693 | 0.022 |
| Soil properties | |||||||
| Slope.gradient | 6 | 1.9 | 9 | 12 | 5.75 | 20 | 0.000 |
| Water.storage.50 | 5.2 | 4.78 | 6.6 | 6.6 | 5.795 | 8.05 | 0.000 |
| Water.storage.100 | 6.42 | 5.57 | 10.09 | 9.98 | 7.91 | 12.54 | 0.000 |
| Water.storage.150 | 7.22 | 6.5 | 11.35 | 9.98 | 7.943 | 15.57 | 0.001 |
| Precipitation | |||||||
| Precipitation.0 | 0 | 0 | 0.254 | 0.127 | 0 | 0.762 | 0.011 |
| Precipitation.0_4 | 0.76 | 0.20 | 1.89 | 0.81 | 0.46 | 5.08 | 0.022 |
| Precipitation.0_5 | 0.85 | 0.17 | 1.64 | 1.21 | 0.85 | 4.23 | 0.008 |
| Precipitation.0_6 | 1.07 | 0.18 | 3.63 | 3.63 | 1.09 | 4.35 | 0.001 |
| PCA score | |||||||
| PC.rain | 0.69 | −0.08 | 1.46 | 0.31 | −0.35 | 0.99 | 0.045 |
PC.rain, PC.temperature, and PC.freeze.thaw denote component scores predicted from PCA.rain, PCA.temperature, and PCA.freeze.thaw, respectively; other variables are defined in Table 1. 25th % and 75th % indicate 25th percentile and 75th percentile, respectively.
TABLE 4.
Principal component analysis of weather variables in data sets of (i) all unique spatial locations and of unique locations with sampled (ii) soil, (iii) vegetation, and (iv) watera
| Name of variable | Spatial location
|
Soil layer
|
Vegetation layer
|
Water layer
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | |
| Precipitation.0 | 0.04 | −0.33 | 0.19 | 0.03 | 0.29 | −0.12 | 0.03 | 0.28 | −0.15 | |||
| Precipitation.1 | −0.02 | 0.40 | −0.24 | −0.02 | 0.40 | −0.13 | 0.04 | −0.40 | 0.34 | |||
| Precipitation.2 | −0.01 | −0.42 | 0.00 | −0.01 | 0.37 | 0.14 | 0.00 | 0.37 | 0.08 | 0.02 | −0.38 | −0.12 |
| Precipitation.0_2 | −0.01 | −0.48 | 0.14 | −0.01 | 0.46 | −0.14 | −0.01 | 0.46 | −0.09 | 0.03 | −0.46 | 0.21 |
| Precipitation.0_3 | −0.01 | −0.49 | 0.11 | −0.01 | 0.47 | −0.09 | −0.01 | 0.46 | −0.06 | 0.03 | −0.47 | 0.13 |
| Precipitation.0_6 | 0.05 | −0.42 | −0.06 | 0.06 | 0.39 | 0.17 | 0.05 | 0.39 | 0.10 | −0.05 | −0.42 | −0.11 |
| Temperature.a.3 | −0.23 | −0.02 | −0.28 | −0.23 | 0.00 | 0.28 | ||||||
| Temperature.a.0_2 | −0.25 | 0.02 | 0.09 | −0.28 | −0.01 | −0.10 | −0.25 | −0.01 | −0.09 | |||
| Temperature.a.0_3 | −0.25 | 0.01 | −0.01 | −0.28 | −0.01 | 0.01 | −0.25 | −0.01 | 0.01 | 0.34 | 0.03 | 0.02 |
| Temperature.a.0_4 | −0.25 | 0.01 | −0.03 | −0.28 | −0.02 | 0.04 | −0.25 | −0.01 | 0.03 | 0.34 | 0.04 | 0.01 |
| Temperature.a.0_5 | −0.25 | 0.02 | −0.03 | −0.29 | −0.02 | 0.03 | −0.25 | −0.02 | 0.02 | 0.34 | 0.04 | 0.01 |
| Temperature.a.0_6 | −0.25 | 0.02 | 0.00 | −0.29 | −0.02 | 0.00 | −0.25 | −0.02 | −0.01 | 0.34 | 0.05 | 0.03 |
| Temperature.a.0_7 | −0.25 | 0.02 | 0.04 | −0.29 | −0.02 | −0.04 | −0.25 | −0.01 | −0.04 | 0.34 | 0.04 | 0.05 |
| Temperature.a.0_8 | −0.25 | 0.01 | 0.06 | −0.28 | −0.01 | −0.06 | −0.25 | −0.01 | −0.06 | |||
| Temperature.a.0_9 | −0.24 | 0.01 | 0.07 | −0.28 | −0.01 | −0.07 | −0.24 | 0.00 | −0.07 | |||
| Temperature.L.0 | −0.21 | 0.05 | 0.32 | −0.21 | −0.04 | −0.32 | ||||||
| Temperature.L.3 | −0.22 | 0.00 | −0.29 | −0.22 | −0.02 | 0.28 | ||||||
| Temperature.L.0_1 | −0.22 | 0.02 | 0.28 | −0.22 | −0.01 | −0.28 | ||||||
| Temperature.L.0_6 | −0.25 | 0.02 | 0.05 | −0.28 | −0.02 | −0.03 | −0.25 | −0.02 | −0.05 | 0.34 | 0.04 | 0.05 |
| Temperature.L.0_7 | −0.25 | 0.02 | 0.07 | −0.28 | −0.02 | −0.06 | −0.25 | −0.02 | −0.07 | 0.34 | 0.04 | 0.07 |
| Temperature.L.0_8 | −0.25 | 0.02 | 0.09 | −0.28 | −0.02 | −0.08 | −0.25 | −0.01 | −0.09 | |||
| Temperature.H.3 | −0.22 | −0.04 | −0.27 | −0.22 | 0.02 | 0.27 | ||||||
| Freeze.thaw.0 | 0.13 | 0.00 | −0.55 | 0.16 | −0.03 | 0.56 | 0.13 | −0.03 | 0.52 | −0.18 | 0.01 | −0.37 |
| Freeze.thaw.2 | 0.18 | 0.18 | 0.34 | 0.21 | −0.14 | −0.49 | 0.18 | −0.15 | −0.36 | −0.27 | 0.14 | 0.37 |
| Freeze.thaw.3 | 0.17 | 0.17 | 0.27 | 0.19 | −0.13 | −0.51 | 0.17 | −0.13 | −0.30 | −0.25 | 0.13 | 0.38 |
| SD (% of variance; cumulative %) | 4.01 (0.67; 0.67) | 1.95 (0.16; 0.83) | 1.35 (0.08; 0.90) | 3.49 (0.61; 0.61) | 2.12 (0.22; 0.83) | 1.10 (0.06; 0.89) | 4.00 (0.64; 0.64) | 2.12 (0.18; 0.82) | 1.37 (0.08; 0.90) | 2.88 (0.52; 0.52) | 2.06 (0.26; 0.78) | 1.22 (0.09; 0.87) |
Bold numbers indicate variables loading on the principle component (PC) represented by the column, i.e., the correlation coefficient between a variable and the corresponding PC. Variables are defined in Table 1. PC1, PC2, and PC3 indicate PC.temperature, PC.rain, and PC.freeze.thaw, respectively.
PCA.
Weather variables (precipitation; ambient temperature, including average, minimum, and maximum daily measurements; and freeze-thaw cycles) in the four analyzed data sets representing (i) all unique spatial locations and unique locations with sampled (ii) soil, (iii) vegetation, and (iv) water were subjected to a PCA. In all four data sets, only the first three components accounted for more than 5% of the total variance (Table 4). Combined, the three first components accounted for 87% or more of the total variability. Variables and corresponding factor loadings are presented in Table 4. In interpreting the rotated factor pattern, an item was said to load on a given component if an absolute value of the factor loading was 0.24, 0.25, 0.24, and 0.35 or greater for that component in all locations and soil, vegetation, and water layers, respectively, and was less than that for the other two components. Several variables describing average and minimum daily temperatures averaged over a period of time prior to sample collection were found to load on the first component, which was subsequently labeled as the temperature component (“PC.temperature”). Several precipitation variables loaded on the second component, subsequently labeled as the precipitation component (“PC.rain”). Finally, variables describing the occurrence of freeze-thaw cycles as well as minimum, maximum, or average daily temperature on days just prior to sample collection were loaded on the third component and were labeled as the “PC.freeze.thaw” component. These results demonstrate the simple structure of the weather data. For each retained component, a corresponding new variable was created with assigned predicted component scores.
LR.
The results of nonspatial multiple LR for the four data sets (i.e., [i] all unique spatial locations and unique locations with sampled [ii] soil, [iii] vegetation, and [iv] water) are presented in Table 5. These results allowed us to calculate the probability of isolation of Listeria spp. (and uncertainty around the estimated probability) as a function of the determinants in the model. For example, a location with loam soil, 10 cm of water stored in up to 50 cm of soil depth, and no history of a freeze-thaw cycle had a 67% (±1 SE = 37% and 87%) probability of harboring Listeria spp.: P(isolation) = 1/{1 + e−[−0.95 + 0.611 + (0.04)(10 − 6.7) + 0.281(10 − 6.7)]} = 0.67. The probability of isolation of Listeria spp. decreased to 30%, albeit with overlapping confidence intervals (±1 SE = 21% and 42%), if the same location did not have loam soil. Examining interaction effects identified for isolation of Listeria spp. from a spatial location indicated that the probability of isolation of Listeria spp. decreased with increasing water storage if a location was exposed to a freeze-thaw cycle, whereas it increased in the absence of a freeze-thaw cycle. However, confidence intervals around these predictions overlapped considerably.
TABLE 5.
Nonspatial multiple logistic regression models obtained for isolation of Listeria spp. from (i) all spatial locations and from the (ii) soil, (iii) vegetation, and (iv) water layersj
| Parameter description | Parameter estimate | SE | P value |
|---|---|---|---|
| Spatial location | |||
| Intercept | −0.95 | 0.26 | 0.000 |
| Northing.ca | −0.01 | 0.00 | 0.013 |
| Slope.gradient.cb | 0.03 | 0.01 | 0.025 |
| Water.storage.50.cc | 0.04 | 0.07 | 0.632 |
| Freeze.thaw.3=1d | −6.68 | 2.19 | 0.002 |
| Loam.soil=1 | 0.61 | 0.30 | 0.041 |
| (Freeze.thaw.3=1)*(Loam.soil=1)e | 5.20 | 2.08 | 0.013 |
| (Water.storage.50.c)*(Loam.soil=1) | 0.28 | 0.12 | 0.024 |
| (Slope.gradient.c)*(Freeze.thaw.3=1) | 0.09 | 0.05 | 0.066 |
| (Water.storage.50.c)*(Freeze.thaw.3=1) | −0.64 | 0.35 | 0.069 |
| Soil layer | |||
| Intercept | −1.79 | 0.19 | 7.3E−22 |
| Precipitation.2 | 0.13 | 0.03 | 7.7E−05 |
| Vegetation layer | |||
| Intercept | −2.99 | 0.96 | 0.002 |
| Northing.cf | −0.03 | 0.01 | 0.025 |
| Temperature.a.0_5.cg | 0.45 | 0.21 | 0.028 |
| Freeze.thaw.3=1 | 8.38 | 5.16 | 0.104 |
| Loam.soil=1 | 2.84 | 0.98 | 0.004 |
| (Temperature.a.0_5.c)*(Loam.soil=1) | −0.45 | 0.21 | 0.032 |
| (Temperature.a.0_5.c)*(Freeze.thaw.3=1) | 1.20 | 0.66 | 0.070 |
| (Northing.c)*(Loam.soil=1) | 0.04 | 0.02 | 0.022 |
| (Northing.c)*(Temperature.a.0_5.c) | 0.01 | 0.00 | 0.016 |
| (Northing.c)*(Temperature.a.0_5.c)*(Loam.soil=1) | −0.01 | 0.00 | 0.007 |
| Water layer | |||
| Intercept | −3.46 | 0.62 | 0.000 |
| Northing.ch | −0.01 | 0.01 | 0.085 |
| Freeze.thaw.3=1 | −1.68 | 0.59 | 0.004 |
| Slope.gradient | 0.04 | 0.02 | 0.022 |
| Water.storage.50 | 0.34 | 0.09 | 0.000 |
| (Northing.c2̂)i | 0.00 | 0.00 | 0.013 |
Prior to fitting a model for all spatial locations, the variable was centered by subtracting 4,706,297 and dividing by 1,000 to express in kilometers.
Prior to fitting a model for all spatial locations, the variable was centered by subtracting 11.6.
Prior to fitting a model for all spatial locations, the variable was centered by subtracting 6.7.
“=1” indicates presence of a risk factor.
“*” indicates interaction.
Prior to fitting a model for the vegetation layer, the variable was centered by subtracting 4,707,076 and dividing by 1,000 to express in kilometers.
Prior to fitting a model for the vegetation layer, the variable was centered by subtracting 13.61705.
Prior to fitting a model for the water layer, the variable was centered by subtracting 4,704,903 and dividing by 1,000 to express in kilometers.
“2̂” indicates a quadratic term.
Variables are defined in Table 1.
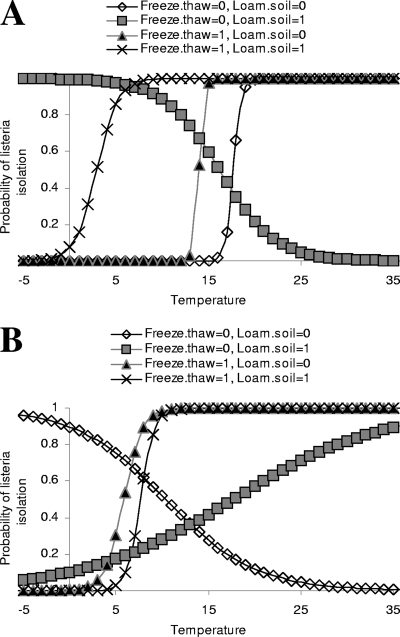
The best LR model for isolation of Listeria spp. from soil samples had only one variable (Table 5): precipitation on day 2 before sample collection with a positive but modest effect (odds ratio [OR] = 1.14 per mm of rain). Considering that several variables were associated with isolation of Listeria spp. from soil in univariate analyses, the presence of confounding was investigated but not detected. The best LR model for the occurrence of Listeria in the vegetation layer (Table 5) showed a complex structure. Calculating the probability of isolation of Listeria spp. as a function of the determinants in the model shows that in the northernmost part of the study area, the probability of isolation of Listeria spp. decreases with increasing ambient temperature for locations that are unexposed to a freeze-thaw cycle but have loam soil (Fig. 3A). The probability of isolation of Listeria spp. increases with increasing temperatures for any other type of soil. However, the signs of slopes changed to the opposite direction in the southernmost part of the study area; there, the probability of isolation of Listeria spp. increases as a function of temperature in the presence of loam soil, while it decreases in the absence of loam soil (Fig. 3B).
FIG. 3.
The probability of isolation of Listeria spp. from the vegetation layer in the northernmost (A) and southernmost (B) parts of the study area for different levels of average temperature over the 5 days before sample collection, for loam soil (denoted as “Loam.soil,” with “=0” and “=1” indicating its absence and presence, respectively), and for a freeze-thaw cycle occurring 3 days before sample collection (denoted as “Freeze.thaw,” with “=0” and “=1” indicating its absence and presence, respectively). The probability is calculated based on the corresponding LR model in Table 5.
Inclusion of an autocovariate term, estimated by an inverse distance-weighting scheme, into the final LR models resulted in deflation of parameter estimates and inflation of P values (data not shown). The results of LR using predicted component scores instead of the actual weather variables were very difficult to interpret and so are not shown here.
CT.
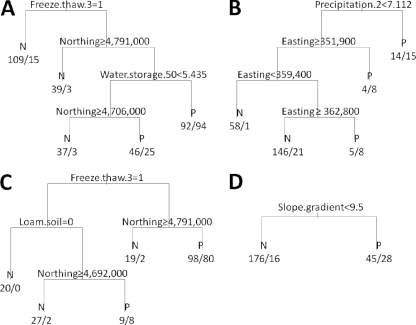
Figure 4A depicts a CT for the occurrence of Listeria in a spatial location. Occurrence of a freeze-thaw cycle prior to sample collection was the most important factor influencing the occurrence of Listeria spp. in a spatial location, indicated by its position closest to the root of the tree. The tree predicts that a location is more likely to show a positive result if it is located farther to the south (<4,791,000 northing) and has water storage of >5 cm to a soil depth of 50 cm. If water storage is <5 cm but the sampling site is located farther to the south, it is likely that isolation results will be positive. The best tree for the occurrence of Listeria in soil (Fig. 4B) showed precipitation occurring on 2 days before the sample collection and multiple nodes with the easting variable. If there was less than 7 mm of rain, the location was predicted to be negative. Multiple occurrences of easting in the tree as a splitting variable are difficult to interpret and may reflect differences among the four sampled areas. The optimal tree for the occurrence of Listeria in the vegetation layer of a location (Fig. 4C) predicted an interesting interplay among three variables (freeze-thaw cycle, loam soil, and northing). In a location, Listeria spp. will be isolated from the vegetation layer if there was no freeze-thaw cycle 3 days before sample collection and the site is located farther to the south from 4,791,000 northing. Even if there was a freeze-thaw cycle, a location will likely harbor Listeria if it has loam soil and is located farther to the south from 4,692,000 northing. The optimal tree for isolation of Listeria spp. from the water layer has only one predictor, slope gradient (Fig. 4D). In a location with a steep slope (>9.5%), it is likely that Listeria will be isolated from water. In models which considered component scores from PCA, CT were very similar to those that used the actual weather variables (and are therefore not shown here). Briefly, in the model for isolation of Listeria spp. from a spatial location and in the vegetation layer of a location, the freeze-thaw cycle at the root of the tree was replaced with the PC.temperature variable. In the soil model, precipitation was replaced by the PC.freeze.thaw variable. The optimal CT for the occurrence of Listeria in the water layer using component scores was identical to the one shown in Fig. 4D.
FIG. 4.
CT for isolation of Listeria spp. from a spatial location (A) as well as from the soil (B), vegetation (C), and water (D) layers of a spatial location. Freeze.thaw.3, freeze-thaw cycle occurring on day 3 before sample collection; Water.Storage.50, available water storage to a soil depth of 50 cm; Precipitation.2, the amount of rain on the second day before sample collection; Loam.soil, presence of loam soil type; Slope.gradient, slope gradient. On top of each node there is a rule used for data partitioning (for example, in panel A, the occurrence of a freeze-thaw cycle 3 days before sample collection is “Freeze.thaw.3=1”); the subset of data satisfying this rule will partition to the left daughter node, while the rest of the data will partition to the right daughter node. N in a terminal node denotes prediction of a negative result for Listeria, with, for example, “109/15” indicating the number of negative/positive observations, i.e., the number of true negatives/FN. P in a terminal node denotes the prediction of isolation of Listeria spp., with, for example, “92/94” indicating the number of negative/positive observations, i.e., the number of FP/true positives.
Predictive performance of LR and CT.
Table 6 summarizes the predictive performance of nonspatial LR and CT models. Overall, LR and CT models for the occurrence of Listeria in a spatial location and in the vegetation layer had relatively high sensitivities, while their specificities were quite low. The opposite was true for the models for isolation of Listeria spp. from the soil and water layers. The hypotheses of equal overall classification performances of LR and CT were rejected in three out of four analyses (Table 6). In these three analyses, CT had better sensitivity once and better specificity twice, i.e., it seemed to have a slightly better performance than LR. Inclusion of autocovariate terms into LR models significantly altered the predictive performance of LR models (quantitative results not shown). Compared with the nonspatial LR models, specificities of the corresponding autologistic regression models were significantly higher in predicting Listeria in all spatial locations and in the vegetation layer but significantly lower in soil and water layers. For the vegetation layer, the reduction in specificity was accompanied by a significantly better sensitivity. However, the predictive performances of all autologistic regression models were indistinguishable from the corresponding CT (quantitative results not shown). The predictive performance of models using component scores instead of the actual weather variables was evaluated as follows: CT had a very similar performance to those shown in Table 6, while either the performances of the nonspatial LR models were similar (all spatial locations model) or the models' sensitivity increased, albeit at the cost of reduced specificity (soil, vegetation, and water layer models) (quantitative results not shown).
TABLE 6.
Performance of nonspatial LR and CT modelsa
| Parameter | Optimal cutoff | Sensitivity | Specificity | Proportion correct | Probability of no difference between models | Probability of no difference between models' sensitivities | Probability of no difference between models' specificities |
|---|---|---|---|---|---|---|---|
| Spatial location | <0.001 | 0.01 | 0.001 | ||||
| LR | 0.18 | 0.91 | 0.41 | 0.56 | |||
| CT | NA | 0.86 | 0.49 | 0.6 | |||
| Soil layer | <0.001 | 0.001 | <0.001 | ||||
| LR | 0.3 | 0.28 | 0.94 | 0.81 | |||
| CT | NA | 0.51 | 0.77 | 0.72 | |||
| Vegetation layer | 0.002 | 0.30 | 0.01 | ||||
| LR | 0.16 | 0.91 | 0.34 | 0.54 | |||
| CT | NA | 0.86 | 0.43 | 0.58 | |||
| Water layer | 0.06 | 1.00 | <0.001 | ||||
| LR | 0.32 | 0.57 | 0.89 | 0.84 | |||
| CT | NA | 0.55 | 0.76 | 0.72 |
NA, not applicable.
DISCUSSION
In this study, we proposed a methodological framework for analysis of the spatially explicit factors affecting the local probability of pathogen isolation from the natural environment by using Listeria spp. as a model system. Specifically, LR and CT models were developed from common spatial data, and predictors for isolation of Listeria spp. were identified. Soil properties and weather characteristics were found to be the most important factors affecting isolation of Listeria spp. from a location, with combinations of factors differing for the soil, vegetation, and water layers of a location. In the following paragraphs, we discuss issues related to the findings, interpretations, limitations, and possible applications of the proposed methodological framework.
Appropriate inferential question and approach.
Utilization of GIS data starts with an understanding of the appropriate inferential question and inferential approach given the type of spatial data available. According to Waller (51), the three categories of spatial data are (i) spatial point process data, (ii) geostatistical data, and (iii) data from a set of regions partitioning the study area (referred to as lattice and regional data). In category i, spatial point process data, locations themselves are considered realizations of some random process, and we seek inference regarding the properties of the process. We may thus ask if observations are equally likely in all locations (as shown in reference 8). If not, where are observations more or less likely to occur? Category ii, geostatistical data, consists of a set of measurements taken at fixed locations, e.g., the temperature measured at each of a set of weather stations. In this case, the locations are set by design. Therefore, an inferential question of interest is the prediction of the same outcome at locations where no measurement was taken (as shown in reference 33). Category iii, regional data, generally involves summary measures for each region (such as the number of residents). Inferential questions often involve an accurate estimation of summaries from regions with small sample sizes or generalized linear modeling of outcomes and covariates measured on the same set of regions.
The occurrence of pathogens in the environment may be the realization of a random process. However, unless there are biological markers of pathogen presence, such as the occurrence of the associated disease, the presence of pathogens in the environment is unknown until a sample from a location chosen as part of a study design is collected and examined. For example, a common sampling strategy to study the occurrence of pathogens in the environment is to obtain a collection of convenience samples (such as in the Sauders data used here [38]) and/or samples from fixed entities, such as agricultural fields. Data collected in such a way fall into the geostatistical data category. The applicable inferential question for such data is the prediction of microbial presence in locations that were not or could not be sampled. However, if sampling locations are regularly distributed (such as in the study of Campylobacter spp. distribution in the environment [3]), the data could still be considered spatial point process data and analyzed for the presence of spatial patterns (e.g., clustering).
Factors affecting the probability of isolation of Listeria spp.
Isolation of Listeria is often considered indicative of conditions that pose an increased risk that food and the environment may be contaminated with the food-borne pathogen L. monocytogenes (for an example, see reference 46). The ecology of L. monocytogenes and Listeria spp. has been extensively studied in food processing environments (e.g., in the smoked fish processing plant environment [23]). However, an understanding of Listeria ecology in the natural environment is important as well, because a contaminated natural environment may be a source of L. monocytogenes contamination of feed for food-producing animals, raw food for human consumption, and for contamination of food processing environments and other environments (retail environments, home environments, etc.) that may lead to contamination of human foods (20). Our findings indicate that there is a strong association between weather and soil properties and the probability of isolation of Listeria spp. from locations in the natural environment. Furthermore, different factors and their combinations had distinct effects on the probability of isolation of Listeria spp. from soil, vegetation, and water layers.
The likelihood of isolation of Listeria spp. from any spatial location (Table 5), as well as from the vegetation (Table 5) (Fig. 4C) and water layers (Table 5) was generally higher if there was no freeze-thaw cycle prior to sample collection. This is consistent with the reported lethal or inhibitory effect of freeze-thaw cycles on microorganisms in natural environments and in foods (1, 12). The probability of isolation of Listeria spp. from the vegetation layer of a location was higher if the soil was of the loam type. Loam soil is generally considered ideal for growing crops. Thus, one could hypothesize that loam soil in a location provides for thriving vegetation, which then supports the survival of Listeria spp. in that location. However, in the absence of a freeze-thaw cycle, the effect of soil on the probability of isolation of Listeria spp. differed with geographic position (northing) (Fig. 3 and 4C) and temperature (Fig. 3): with increasing ambient temperatures, loam soil was negatively associated with the occurrence of Listeria bacteria in the northern part of the study area, while it was positively associated with their occurrence in the southern part. Because Listeria spp. are known to survive and multiply over a wide range of temperatures, from 1°C or 2°C to 45°C (21), this complex structure, if real, may be a consequence of adaptation of other microorganisms to different temperatures, as the microorganisms may compete with Listeria spp. in different soil types and geographic areas. Interestingly, precipitation increased the probability of isolation of Listeria spp. from soil, but it did not have any effect on isolation of Listeria spp. from vegetation. That is surprising, because it is generally considered that rain and water irrigation increase the probability of produce contamination with microorganisms (14).
The probability of isolating Listeria spp. from the soil layer was strongly associated with the amount of rain 2 days prior to sample collection, and the specificity of the LR model with precipitation as a single variable was very high (94%). This finding is consistent with data from food processing plants where Listeria bacteria are more frequent in plants with water and a high level of moisture (40). Also, it has been reported that higher soil moisture increases the survival of L. monocytogenes (53) and other food-borne pathogens, including Salmonella spp. (17).
Slope gradient was found to be the best predictor of the occurrence of Listeria spp. in water (based on the CT method) (Fig. 4D). This finding is consistent with a study by Smith et al. (41), which reported that watersheds with large proportions of urban land cover and agriculture on steep slopes had a very high probability of being contaminated with pathogens. The probability of isolation of Listeria spp. from a location as well as from the vegetation and water layers seemed to decrease with the geographic position of the sampling location farther to the north, while the probability of its isolation from soil decreased farther to the east. These trends may be real, but they may also reflect the difference in the isolation of Listeria spp. probability between sampled areas.
Because of the intimate contact between soil and vegetation in a location and their shared routes of L. monocytogenes contamination, Ivanek et al. (20) proposed that in a predictive model of L. monocytogenes dynamics in the natural environment, the soil and vegetation layers could be modeled jointly. Along the same line of reasoning, the same control measures would control L. monocytogenes contamination in the two layers. However, findings of this study indicate that different environmental factors and their combinations have distinct effects on the probability of isolation of Listeria spp. (and likely of L. monocytogenes isolation) from the soil, vegetation, and water layers of a location. This suggests that soil, vegetation, and water represent three distinct ecological niches for Listeria spp. and should be modeled separately. Correspondingly, the three layers will probably require distinct approaches to control L. monocytogenes contamination.
Predictive performance of LR and CT methods.
The classification performance of CT was slightly better than that of nonspatial LR and indistinguishable from that of autologistic regression models. However, it should be borne in mind that the predictive performance of LR and CT may have been affected by modeling strategies applied to assure their fair comparison. Specifically, we restricted CT analysis to the subset of data with complete observations on variables used in the LR model. Because the CT method can use data with missing observations, this strategy resulted in a loss of eligible information and may have impaired CT performance. In the LR modeling, we used automatic stepwise regression for model selection so that model building would be comparable to the “automatic” variable selection in the CT. An alternative approach, potentially better for LR, would be to apply causal concepts to data analysis, i.e., to list effects and interactions of interest prior to model fitting based on the knowledge and understanding of the system (26). Because of these reasons and a small number of models in which the performance of LR and CT was compared, there are not enough grounds in this study to claim better performance of the CT method. Rather, we conclude that the predictive performances of the two methods were comparable.
While having a classification performance comparable to that of LR, CT demonstrated excellent interpretability, which may be particularly useful in field applications. Other advantages of using a CT over an LR method are related to its inherent nonparametric character; it handles very well highly skewed, multimodal, categorical (ordinal and nonordinal) predictors. It is robust to outliers, and missing predictor variables are not dropped from the analysis. Unlike LR and other parametric models, which are intended to uncover a single dominant structure in data, CT is designed to work with data that might have multiple structures (interactions). A disadvantage of CT methods noticed here is related to the treatment of continuous predictor variables as discrete categories. CT can represent a continuous factor only by a series of distinct subranges. We experienced this in our CT through multiple occurrences of a continuous factor as a partitioning variable (e.g., the easting variable in the soil model [Fig. 4B]). Therefore, LR is often better at capturing an algebraic relationship between the response variable and a continuous factor. The ability of LR to capture spatial dependence (in autologistic regression models) could be seen as an advantage over CT. However, the validity of autologistic regression has recently been questioned: an assessment using artificial simulation data with known properties indicated that autologistic regression models consistently underestimate the effect of environmental variables and give biased estimates compared with a nonspatial LR (10). Consistent with this, the inclusion of contagion terms in the LR models developed here resulted in deflated effects of independent variables and higher P values. Both LR and CT could use misclassification penalties to account for an imperfect sensitivity of microbial isolation methods. Generally, a higher penalty for an FN would result in better sensitivity and consequently lower specificity. This has an important practical implication because both LR and CT could be manipulated if the goal was to achieve better sensitivity or specificity, whichever is desired. To conclude, based on our findings, LR and CT methods have comparable predictive performances and complementary strengths and weaknesses in identifying risk factors. Therefore, for analysis of microbial presence in the environment, our recommendation would be to apply both methods whenever feasible.
Spatial autocorrelation.
Spatial autocorrelation may be an important source of bias in spatial analyses (39), leading to poorly specified models and inflated significance estimates for predictor variables (24). It occurs as a consequence of the direct relationship between distance and likeness and the fact that elements of an ecosystem close to one another are more likely to be influenced by the same generating process and will therefore be similar (29). The two main procedures used to minimize the effect of spatial correlation are subsampling and inclusion of the contagion term (39), such as that used in autologistic regression models. In this study, the systematic subsampling could not be performed because of a relatively small sample size. Inclusion of a contagion term has been reported to be more effective than subsampling, but it prevents extrapolation beyond the geographic range of the calibration data (39). That is of concern in analysis of geostatistical data, such as that used in this study, because we were interested in prediction of the same outcome at locations where no samples were collected. Furthermore, as stated earlier, the validity of autologistic regression has been questioned (10). It has been reported that CT are less vulnerable to the effect of spatial correlation than generalized linear models (39), which may have contributed to a slightly better predictive performance of CT than the nonspatial LR in our study. Our findings about the indistinguishable predictive performance of autologistic regression models and CT support this.
Value of principle component analysis.
Large, complex data sets are characteristic for environmental microbiology whose exploration would often benefit from multivariate methods (32). PCA is one of the most popular multivariate exploratory analyses. We used PCA with two objectives. The first was to explore weather data, which resulted in the valuable confirmation that three distinct variables (temperature, precipitation, and freeze-thaw cycles) do indeed exist among the considered weather variables. Second, we predicted component scores (linear combinations of the original variables that account for most of the variance in the data) and used them in multivariate modeling instead of the actual weather variables. This approach allowed us to utilize information from several weather predictors simultaneously without the need to pick one from among several highly correlated variables. However, for the most part, the predictive performance of the models developed using predicted component scores was comparable to that achieved with the models using weather variables. Also, component scores were particularly difficult to interpret as they did not correspond to any real ecological entity.
Methodological implications for the control of infectious and food-borne pathogens.
The generalizability of findings of our study is limited to Listeria spp. in natural environments. However, the methodological framework proposed here is adaptable to study ecological determinants for the presence of free-living stages of many infectious and food-borne agents (e.g., E. coli, Salmonella spp., and Vibrio parahaemolyticus) in natural as well as in agricultural environments. The proposed framework may be particularly relevant to the control of the respective diseases as part of precision farming as well as to biosafety and biosecurity. Precision farming is an agricultural concept, based on recognition of the within-field variability, that uses technologies such as GPS, GIS, and remote sensing to find optimal agricultural management practices (52). Management practices are optimized from several points of view, including profitability, increased crop quality, improved sustainability, lower management risk, product traceability, and environmental protection (36). However, we propose that farmers' management decisions could also be optimized for the production of microbiologically safer foods by linking precision farming with models that can predict microbial presence in a location based on the site-specific environmental, meteorological, and management factors (similar to the one proposed here). Similarly, predictive models of microbial presence in the environment could be coupled with a predictive model of pathogen fecal shedding (19) and be used by farmers that use GIS-based livestock production (2, 50) to predict when fields may have an increased risk of animal exposure if used for grazing.
From the biosafety and biosecurity perspective, the presence of free-living stages of infectious and food-borne pathogens in the natural environment is of obvious concern to human and animal health. The methodological approach proposed here complements GIS-based spatial analyses that have been applied to elucidate pathogen epidemiology. For example, spatial analysis of Cryptococcus gattii distribution in the Pacific Northwest region of Canada indicated that human-mediated dispersal vehicles and footwear may be important mechanisms for dispersal of this pathogen (22). Similarly, analysis of temporal and spatial distribution of V. parahaemolyticus in mussels indicated that water salinity, modulated by seawater temperature in periods and areas of reduced salinity, is a primary factor governing the occurrence of this food-borne pathogen (27). Understanding how environmental factors affect pathogen occurrence in the natural environment could be translated into risk maps indicating areas and times with a higher exposure hazard to humans and animals, as has been done for disease vectors (e.g., ticks [55] and mosquitoes [18]). Furthermore, to fully account for environment-pathogen-host interaction, the predictive modeling of free-living pathogens in a location could be linked with mathematical models of within-host population disease transmission. This would be similar to the modeling of environment-vector-host interaction (such as modeling of the climate-driven transmission of plague in prairie dogs [42]). It is accepted that changes in weather patterns in the coming decades will likely cause important changes in the incidence and distribution of diseases with free-living stages, such as E. coli, Campylobacter spp., Salmonella spp., and C. gattii (15). Understanding how environmental factors affect pathogen occurrence in the environment could be used to predict microbial occurrence as a consequence of global climate change.
Acknowledgments
This research was supported by USDA Special Research Grants 2004-34459-14296, 2005-34459-15625, and 2006-34459-16952 and USDA Hatch Grant NYC-143951.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Allocca, V., F. Celico, E. Petrella, G. Marzullo, and G. Naclerio. 2008. The role of land use and environmental factors on microbial pollution of mountainous limestone aquifers. Environ. Geol. 55:277-283. [Google Scholar]
- 2.Barbari, M., L. Conti, B. K. Koostra, G. Masi, F. Sorbetti Guerri, and S. R. Workman. 2006. The use of global positioning and geographical information systems in the management of extensive cattle grazing. Biosystems Eng. 95:271-280. [Google Scholar]
- 3.Brown, P. E., O. F. Christensen, H. E. Clough, P. J. Diggle, C. A. Hart, S. Hazel, R. Kemp, A. J. H. Leatherbarrow, A. Moore, J. Sutherst, J. Turner, N. J. Williams, E. J. Wright, and N. P. French. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, S., and B. Price. 1991. Regression analysis by example, 2nd ed. Wiley, New York, NY.
- 5.Clarke, K. C., S. L. McLafferty, and B. J. Tempalski. 1996. On epidemiology and geographic information systems: a review and discussion of future directions. Emerg. Infect. Dis. 2:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Aoust, J. Y. 1997. Foodborne pathogenic bacteria: Salmonella species, p. 129-158. In M. P. Doyle, L. Beuchat, and T. J. Montville (ed.), Food microbiology fundamentals and frontiers. ASM Press, Washington, DC.
- 7.De'Ath, G., and K. E. Fabricius. 2000. Classification and regression trees: a powerful yet simple technique for ecological data analysis. Ecology 81:3178-3192. [Google Scholar]
- 8.Dohn, M. N., M. L. White, E. M. Vigdorth, C. R. Buncher, V. S. Hertzberg, R. P. Baughman, A. G. Smulian, and P. D. Walzer. 2000. Geographic clustering of Pneumocystis carinii pneumonia in patients with HIV infection. Am. J. Respir. Crit. Care Med. 162:1617-1621. [DOI] [PubMed] [Google Scholar]
- 9.Dohoo, I. W. Martin, and H. Stryhn. 2003. Veterinary epidemiological research. AVC Inc., Charlottetown, PEI, Canada.
- 10.Dormann, C. F. 2007. Assessing the validity of autologistic regression. Ecol. Model. 207:234-242. [Google Scholar]
- 11.Doyle, P. M., T. Zhao, J. Meng, and S. Zhao. 1997. Foodborne pathogenic bacteria: Escherichia coli O157:H7, p. 171-191. In M. P. Doyle, L. Beuchat, and T. J. Montville (ed.), Food microbiology fundamentals and frontiers. ASM Press, Washington, DC.
- 12.El-Kest, S. E., A. E. Yousef, and E. H. Marth. 1991. Fate of Listeria monocytogenes during freezing and frozen storage. J. Food Sci. 56:1068-1071. [DOI] [PubMed] [Google Scholar]
- 13.Frenkel, J. K. 1990. Toxoplasmosis in human beings. J. Am. Vet. Med. Assoc. 196:240-248. [PubMed] [Google Scholar]
- 14.Girardin, H., C. E. Morris, C. Albagnac, N. Dreux, C. Glaux, and C. Nguyen-The. 2005. Behaviour of the pathogen surrogates Listeria innocua and Clostridium sporogenes during production of parsley in fields fertilized with contaminated amendments. FEMS Microbiol. Ecol. 54:287-295. [DOI] [PubMed] [Google Scholar]
- 15.Greer, A., V. Ng, and D. Fisman. 2008. Climate change and infectious diseases in North America: the road ahead. CMAJ 178:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatcher, L. 1994. A step-by-step approach to using SAS system for factor analysis and structural equation modeling. SAS Institute, Cary, NC.
- 17.Holley, R. A., K. M. Arrus, K. H. Ominski, M. Tenuta, and G. Blank. 2006. Salmonella survival in manure-treated soils during simulated seasonal temperature exposure. J. Environ. Qual. 35:1170-1180. [DOI] [PubMed] [Google Scholar]
- 18.Hoshen, M. B., and A. P. Morse. 2004. A weather-driven model of malaria transmission. Malar. J. 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanek, R., Y. T. Gröhn, A. J. Ho, and M. Wiedmann. 2007. Markov chain approach to analyze the dynamics of pathogen fecal shedding—example of Listeria monocytogenes shedding in a herd of dairy cattle. J. Theor. Biol. 245:44-58. [DOI] [PubMed] [Google Scholar]
- 20.Ivanek, R., Y. T. Gröhn, and M. Wiedmann. 2006. Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog. Dis. 3:319-336. [DOI] [PubMed] [Google Scholar]
- 21.Junttila, J. R., S. I. Niemela, and J. Hirn. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 65:321-327. [DOI] [PubMed] [Google Scholar]
- 22.Kidd, S. E., P. J. Bach, A. O. Hingston, S. Mak, Y. Chow, and L. MacDougall. 2007. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg. Infect. Dis. 13:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lappi, V. R., J. Thimothe, K. K. Nightingale, K. Gall, V. N. Scott, and M. Wiedmann. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J. Food Prot. 67:2500-2514. [DOI] [PubMed] [Google Scholar]
- 24.Legendre, P. 1993. Spatial autocorrelation: problem or new paradigm? Ecology 74:1659-1673. [Google Scholar]
- 25.Linnet, K., and E. Brandt. 1986. Assessing diagnostic tests once an optimal cutoff point has been selected. Clin. Chem. 32:1341-1346. [PubMed] [Google Scholar]
- 26.Martin, W. 2008. Linking causal concepts, study design, analysis and inference in support of one epidemiology for population health. Prev. Vet. Med. 86:270-288. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Urtaza, J., A. Lozano-Leon, J. Varela-Pet, J. Trinanes, Y. Pazos, and O. Garcia-Martin. 2008. Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the Rias of Galicia, Spain. Appl. Environ. Microbiol. 74:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J., and J. Franklin. 2002. Modeling the distribution of four vegetation alliances using general linear models and classification trees with spatial dependence. Ecol. Model. 157:117-147. [Google Scholar]
- 29.Miller, J., J. Franklin, and R. Aspinall. 2007. Incorporating spatial dependence in predictive vegetation models. Ecol. Model. 202:225-242. [Google Scholar]
- 30.Mitscherlich, E., and E. H. Marth. 1984. Microbial survival in the environment: bacteria and rickettsiae important in human and animal health. Springer-Verlag, New York, NY.
- 31.National Atlas of the United States. 1999. Hydrography features of New York State. U.S. Department of the Interior, Washington, DC. http://cugir.mannlib.cornell.edu/bucketinfo.jsp?id=7315. Accessed 6 February 2008.
- 32.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raso, G., B. Matthys, E. K. N′Goran, M. Tanner, P. Vounatsou, and J. Utzinger. 2005. Spatial risk prediction and mapping of Schistosoma mansoni infections among schoolchildren living in western Cote d'Ivoire. Parasitology 131:97-108. [DOI] [PubMed] [Google Scholar]
- 34.R Development Core Team. 2004. R: a language and environment for statistical computing. 3-900051-07-0. The R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
- 35.Richman, M. B., and X. Gong. 1999. Relationships between the definition of the hyperplane width to the fidelity of principal component loading patterns. J. Climate 12:1557-1576. [Google Scholar]
- 36.Robert, P. C. 2002. Precision agriculture: a challenge for crop nutrition management. Plant Soil 247:143-149. [Google Scholar]
- 37.Rocourt, J., and P. Cossart. 1997. Foodborne pathogenic bacteria: Listeria monocytogenes, p. 337-352. In M. P. Doyle, L. Beuchat, and T. J. Montville (ed.), Food microbiology fundamentals and frontiers. ASM Press, Washington, DC.
- 38.Sauders, B. D. 2005. Molecular epidemiology, diversity, distribution and ecology of Listeria. Ph.D. thesis. Cornell University, Ithaca, NY.
- 39.Segurado, P., M. B. Araujo, and W. Kunin. 2006. Consequences of spatial autocorrelation on niche-based models. J. Appl. Ecol. 43:433-444. [Google Scholar]
- 40.Slade, P. J. 1992. Monitoring Listeria in the food production environment. I. Detection of Listeria in processing plants and isolation methodology. Food Res. Int. 25:45-56. [Google Scholar]
- 41.Smith, J. H., J. D. Wickham, D. Norton, T. G. Wade, and K. B. Jones. 2001. Utilization of landscape indicators to model potential pathogen impaired waters. J. Am. Water Res. Assoc. 37:805-814. [Google Scholar]
- 42.Snäll, T., R. B. O'Hara, C. Ray, and S. K. Collinge. 2008. Climate-driven spatial dynamics of plague among prairie dog colonies. Am. Nat. 171:238-248. [DOI] [PubMed] [Google Scholar]
- 43.Streitberg, B., and J. Röhmel. 1986. Exact distributions for permutations and rank tests: an introduction to some recently published algorithms. Stat. Software Newsl. 12:10-17. [Google Scholar]
- 44.Therneau, T. M., and B. Atkinson. 2008. The rpart package. http://cran.r-project.org/web/packages/rpart/rpart.pdf.
- 45.U.S. Census Bureau. 2001. Census roads, New York State: Schuyler, Seneca, Hamilton, Tompkins, Greene, Delaware, Sullivan, Ulster, Essex, Fulton, Franklin, Herkimer, St. Lawrence, Oneida, and Clinton. U.S. Census Bureau Geography Division, U.S. Department of Commerce, Washington, DC. http://cugir.mannlib.cornell.edu/datatheme.jsp?id=130. Accessed 7 February 2008.
- 46.U.S. Department of Agriculture. 2003. FSIS risk assessment for Listeria monocytogenes in deli meat. Food Safety and Inspection Service, U.S. Department of Agriculture, Washington, DC.
- 47.U.S. Department of Agriculture. 2006. SSURGO: soil survey geographic database for New York State: Schuyler, Seneca, Hamilton, Tompkins, Greene, Delaware, Sullivan, Ulster, Essex, Fulton, Franklin, Herkimer, and St. Lawrence. Soil Survey Staff, Natural Resources Conservation Service, U.S. Department of Agriculture, Washington, DC. http://soildatamart.nrcs.usda.gov. Accessed 1 February 2008.
- 48.Vayssieres, M. P., R. E. Plant, and B. H. Allen-Diaz. 2000. Classification trees: an alternative non-parametric approach for predicting species distributions. J. Vegetation Sci. 11:679-694. [Google Scholar]
- 49.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S, 4th ed. Springer, New York, NY.
- 50.Wade, T. G., B. W. Schultz, J. D. Wickham, and D. F. Bradford. 1998. Modeling the potential spatial distribution of beef cattle grazing using a geographic information system. J. Arid Environ. 38:325-334. [Google Scholar]
- 51.Waller, L. A. 2004. Bayesian thinking in spatial statistics. Department of Biostatistics, Rollins School of Public Health, Emory University, Atlanta, GA.
- 52.Weiss, M. D. 1996. Precision farming and spatial economic analysis: research challenges and opportunities. Proceedings of the Annual Meeting of the American Agricultural Economics Association, San Antonio, TX. Am. J. Agric. Econ. 78:1275-1280. [Google Scholar]
- 53.Welshimer, H. J. 1960. Survival of Listeria monocytogenes in soil. J. Bacteriol. 80:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, C. N., R. S. Vose, D. R. Easterling, and M. J. Menne. 2006. United States historical climatology network daily temperature, precipitation, and snow data. ORNL/CDIAC-118, NDP-070. Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, Oak Ridge, TN.
- 55.Wimberly, M. C., A. D. Baer, and M. J. Yabsley. 2008. Enhanced spatial models for predicting the geographic distributions of tick-borne pathogens. Int. J. Health Geogr. 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]