Abstract
We used cultivation-independent methods to investigate the prokaryotic biogeography of the water column in six salt lakes in Inner Mongolia, China, and a salt lake in Argentina. These lakes had different salt compositions and pH values and were at variable geographic distances, on both local and intercontinental scales, which allowed us to explore the microbial community composition within the context of both contemporary environmental conditions and geographic distance. Fourteen 16S rRNA gene clone libraries were constructed, and over 200 16S rRNA gene sequences were obtained. These sequences were used to construct biotic similarity matrices, which were used in combination with environmental similarity matrices and a distance matrix in the Mantel test to discover which factors significantly influenced biotic similarity. We showed that archaeal biogeography was influenced by contemporary environmental factors alone (Na+, CO32−, and HCO3− ion concentrations; pH; and temperature). Bacterial biogeography was influenced both by contemporary environmental factors (Na+, Mg2+, and HCO3− ion concentrations and pH) and by geographic distance.
Biogeography aims to explain spatial patterns of diversity in the context of evolutionary events such as speciation, dispersal, extinction, and species interactions (42). Macroecologists have long studied the biogeography of higher plants and animals in various habitats (9, 13). In contrast, there is very little information available on the biogeography of prokaryotes. This stemmed from the difficulty of assessing microbial communities by cultivation methods, which only sampled 0.1 to 10% of the microbial community (30). However, with the advent of cultivation-independent sequencing techniques, microbial communities of many environments have been characterized, including soil (43), the Arctic and Antarctic Oceans (5), and the Sargasso Sea (61). This in turn facilitated prokaryotic biogeography studies in a number of environments on scales ranging from 20,000 km to 0.002 km (42).
A study of the biogeography of soil bacteria across the Americas showed that differences were largely attributed to soil pH, with higher diversity observed in neutral soils (20). Bacterial communities in an estuary in Massachusetts were found to vary with the salinity gradient (14). Such studies demonstrated that environmental parameters influenced biogeographical patterns in microbial diversity. Further studies demonstrated that biogeography of hot spring cyanobacteria, hyperthermophilic archaea, and Pseudomonas strains was influenced by geographic distance, which led to isolation of disparate populations and subsequent genetic divergence (12, 51, 63). The apparent allopatric speciation demonstrated in these studies therefore contested the idea that prokaryotes were not affected by limits to dispersal due to their small size, abundance, and metabolic plasticity (i.e., “everything is everywhere” [see below]) (21).
A simple framework was suggested to distinguish between the effects of evolutionary events and contemporary environmental conditions on the spatial variation of microbial diversity (42). At the center of this framework were four hypotheses. The null hypothesis stated that microorganisms were distributed randomly over space. Upon rejection of the null hypothesis, the second hypothesis stated that spatial variation reflected the influence of contemporary environmental variation. It assumed that geographic distance did not affect diversity due to the wide dispersal of microorganisms. This hypothesis represents the famously quoted “everything is everywhere; the milieu selects” by Baas-Becking (4, 6). The third hypothesis stated that variation was shaped by evolutionary events (geographic distance) that limited dispersal and that past environmental conditions led to genetic divergence between different microbial assemblages. The fourth hypothesis stated that the biogeography of microorganisms was determined by both contemporary environmental conditions and past evolutionary events (geographic distance). It is important to note here the possibility that evolutionary events can be represented by geographic distances. (For more details on this framework, see reference 42.)
Many studies have been carried out on salt lakes and salterns around the world (28), but few have tried to explain variations in microbial community composition. Those that did identified salinity, altitude, redox and ionic concentration, pO2, and seasonal events as relevant factors (7, 11, 16, 17, 34, 35, 38, 65). To our knowledge, only two studies have looked at the effect of intercontinental geographic distances on microbial community composition in salt lakes. Foti and colleagues looked specifically at the biogeography of Thioalkalivibrio in soda lakes across Mongolia, Kenya, California, Egypt, and Siberia and found that these bacteria showed a tendency for endemism; hence, geographic distance was a significant factor in influencing community composition (22). A further study looked at the biogeography of Salinibacter ruber strains from salterns in the Mediterranean, Atlantic, and Peruvian regions using a metabolomic approach. Geographically distinct strains were distinguished by characteristic metabolites (58).
We examined the prokaryotic community composition in several salt lakes using ribosomal DNA methods. Six of the salt lakes in this study were situated on the Inner Mongolian steppe, northwest of Beijing, which had an average elevation of 1,000 to 2,000 m above sea level. The lakes were mostly several hundred kilometers apart (0.147 to 395.2 km) and were in different climate and vegetation zones: from typical grassland steppe in the north and east to desert steppe bordering the Gobi desert in the south and west (70). The lakes were Bagaejinnor, Chagannor, Ejinnor, Erliannor, Shangmatala, and an unnamed lake near Xilinhot. Lakes Ejinnor and Erliannor were extensively developed into salterns. Salar Guayatayoc Lake was situated in the same basin as the Salinas Grandes in the Argentine Altiplano at an elevation of 3,432 m, north-west of the city Salta, ∼18,000 km from the other lakes. All salt lakes were athalassohaline, located in arid climates, and subjected to high solar radiation and wide ranges of temperature. The lakes had different salt compositions and allowed us to explore the microbial community composition within the context of both contemporary environmental conditions and geographic distance.
Here we describe the microbial diversity of six salt lakes in Inner Mongolia and one salt lake in Argentina. Using the framework previously described, we present evidence that biogeography of Archaea in these salt lakes was significantly influenced (P < 0.05) by environmental factors (Na+, CO32−, and HCO3− ion concentrations, pH, and temperature), but not geographic distance, consistent with the previously stated hypothesis 2. We also show that the biogeography of Bacteria was significantly influenced (P < 0.05) by both environmental factors (Na+, Mg2+, and HCO3− ion concentrations and pH) and geographic distance, consistent with the previously stated hypothesis 4.
MATERIALS AND METHODS
Descriptions of sampling sites.
All sites were remote from centers of population and usually involved long drives over unmade roads. Our ability to transport equipment was limited. Lakes themselves were often fringed by deep mud, making sampling hazardous. The first four lakes were in areas of grassland steppe, the last two lakes in desert steppe, and the Argentine lake in an arid high-altitude plateau. Temperature, pH, and chemical analyses of the brines are shown in Table 1.
TABLE 1.
Chemical composition of the salt lake brines
| Sample | Environment | Temp (°C) | pH | Conductivity (mS/cm)/total salinity (g/liter) | Concn of:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cl (M) | K (mM) | Mg (M) | Na (M) | S (M) | HCO3 (mM) | CO3 (mM) | |||||
| Lake Bagaejinnor | Salt lake | 20.5 | 8.5 | 474/333 | 4.61 | 33.2 | 0.35 | 5.32 | 1.07 | 7.4 | 3.3 |
| Lake Chagannor | Salt lake | 17.1 | 10.5 | 202/118 | 1.08 | 14 | 0.001 | 2.89 | 0.43 | 360 | 410 |
| Lake Ejinnor | Saltern | 27.6 | 7.5 | 464/397 | 4.36 | 68.9 | 2.08 | 2.82 | 0.94 | 9.84 | 23.3 |
| Lake Erliannor | Saltern | 17.9 | 8 | 482/312 | 5.33 | 39 | 0.86 | 4.2 | 0.48 | 4.1 | 8.3 |
| Lake Shangmatala | Salt lake | 20.8 | 8.5 | 487/346 | 4.69 | 150 | 0.26 | 5.38 | 0.81 | 7.4 | 13 |
| Unknown lake | Salt lake | 23.6 | 8.5 | 463/356 | 5.4 | 53.1 | 0.085 | 5.06 | 0.33 | 13.9 | 1.7 |
| Pool | Small pool | 21 | 9.5 | 287/196 | 2.21 | 17.1 | 0.035 | 2.01 | 0.06 | 14.3 | 0 |
| Salar Guayatayoc | Salt lake | 10 | 7.5 | 5.5 | 173 | 0.061 | 4.9 | 0.031 | 9.84 | 0 | |
Lake Bagaejinnor was a hypersaline lake, whose coordinates were N45°08.527′E116°36.167′, north of the town Qog Ul. It has a surface area of 5 km2 during the wet season (66). It had evaporated over the summer, exposing salt-encrusted mud flats, and had been reduced to a number of small pools and lagoons. The brine was colorless, but the salt crystals had a pink coloration, indicating the presence of haloarchaea.
Lake Ejinnor was a hypersaline lake, at coordinates N45°14.452′E116°32.477′ north of the town of Qog Ul, 40 km from Lake Bagaejinnor. It was a large shallow lake, 0.05 to 0.3 m deep, with evaporating lagoons on the eastern side of the main body of water. The lake water sample was taken from a large saltern containing red brine and orange-pigmented salt crystals about 0.3 m deep.
An unnamed lake, located northwest of Xilin Hot, at coordinates of N47°55.355′E115°36.757′, was also sampled. It was a hypersaline lake situated near an abandoned soda works. The lake was divided by several causeways. A shallow lagoon was found cut off from the rest of the lake, which was where one of the sampling sites was located (XH). The lake had a thick white salt crust, while the brine was clear and colorless and contained brine shrimp (Artemia sp.). Leading from the lake was a drainage channel that connected to a 15-cm-deep pool of green brine (147 m from the lake), where a second sample was taken (X).
Lake Shangmatala, a hypersaline lake, was located in a shallow basin surrounded by hills at an elevation of 987 m, at coordinates N43°22.751′E114°01.361′. The lake had a surface area of 2.5 km2 and a depth of 0.1 to 0.15 m. The lake was surrounded by lush grassland and vegetation, which grew almost up to the water's edge. The soil nearest the lake appeared to be soda soil, which had a layer of lichen growing on the surface. It was noted that an unpleasant-smelling gas was emitted. A causeway led directly into the lake.
Lake Chagannor was a large hypersaline soda lake, situated near a soda works, 120 km south of Mandulatu, at coordinates of N43°16.131′E112°55.636′. Sampling took place on the south side of the lake. The brine appeared green, and the mud was gray and viscous, with a layer of fine salt.
Lake Erliannor was a hypersaline lake, located north of Erenhot on the Mongolian border and the trans-Siberian railway, at coordinates N43°44.426′E112°02.081′. It was reported to have a surface area of 8.75 km2 and a depth of 0.1 to 0.3 m. The natural lake was unrecognizable due to extensive development of salterns. The lake water sample was taken from a saltern (0.1-m depth) that contained colorless brine and a white salt crust.
Salar Guayatayoc Lake was a hypersaline lake on the north edge of the Salinas Grandes, Argentina. Its coordinates were S23°36.604′W65°51.998′. It was locally reported to have a depth of 30 m and was covered by an ∼1-m-thick salt crust. Samples were taken through a hole in this crust.
Sample collection.
Biomass from the water column from the Argentinean salt lake was sampled in July 2003 and from the Inner Mongolian salt lakes in September 2003. In Inner Mongolia, brine was sampled at a distance in 250-ml stainless steel beakers suspended on the end of a 1-m pole. In Argentina, brine was collected through a hole in the ∼1-m-thick salt crust. Water was filtered through sterile 0.45-μm-pore membrane filters (Millipore) in a 250-ml-capacity polycarbonate filter unit (Sartorius) using a Nalgene hand pump, which produced a vacuum of 40 to 50 cm Hg under field conditions. Water was processed in this way until flow stopped, which suggested that sufficient biomass was captured on the filter. Membrane filters were removed from the apparatus using sterile tweezers and placed immediately in cold sterile stabilization buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 2 M NaCl) and agitated to resuspend the cells. This was immediately placed on ice until further processing.
Measuring geographic distances.
GPS coordinates recorded at each sampling point were imported into MapSource according to the manufacturer's instructions to measure the geographic distances between the sites.
Measuring pH, temperature, and salinity of the salt lakes.
The pH of the salt lake water was measured using pH strips (Merck).
The temperature was measured using a Solomat 520C temperature monitor or a Hanna KType thermocouple with an SP weighted tanker probe (Jencons, Leighton Buzzard, United Kingdom) according to the manufacturer's instructions. The temperature was measured at a distance by attaching the probe to the end of a 1-m pole. Other physical and chemical analyses were performed on samples filtered through a 0.22-μm-pore membrane and stored in sterile screw-cap vials.
The salinity was measured using a Hanna HI 8633 or HI9033 multirange conductivity meter (Jencons, Leighton Buzzard, United Kingdom), which was calibrated to 20°C with a temperature coefficient of 2% according to the manufacturer's instructions. All salt lake water readings were off the scale: hence, they were serially diluted with distilled water and readings were made at the 199.9-mS/cm range. Water conductivity gave an indication of total salt concentration in grams per liter (64).
Determining the chemical composition of salt lake water.
Chemical analysis of the salt lake water was carried out by inductively coupled plasma optical emission spectrometry. Samples were sent for analysis to the Geology Department at the University of Leicester.
Titrations of carbonate and bicarbonate.
Concentrations of carbonate (CO32−) and bicarbonate (HCO3−) ions were found by titration of lake water with H2SO4 using a Digital Titrator model 16900 (Hach Systems for Analysis) according to the manufacturer's instructions.
Community DNA extraction and PCR Amplification of 16S rRNA genes.
Community DNA from the Inner Mongolian environmental samples was extracted using the GenomicPrep cell and tissue DNA isolation kit (Amersham Biosciences). The initial stages of protein precipitation were carried out on site, and the sample was stored at −20°C until DNA purification could be carried out in the laboratory in Leicester. Community DNA from the Argentinean sample was extracted by freezing the filter in a small amount of liquid nitrogen, which was then homogenized in a precooled pestle and mortar (at −80°C). This material was transferred to a clean tube, and 960 μl of NET buffer (150 mM NaCl, 100 mM EDTA [pH 8.0], 50 mM Tris-HCl [pH 8.0]) containing 15 mg/ml of lysozyme was added, which was incubated at 37°C for 10 min. One hundred ninety-two microliters of 11 mg/ml proteinase K and 128 μl of 10% (wt/vol) sodium dodecyl sulfate were added, and this mixture was incubated for a further 30 min at 65°C. DNA was extracted by phenol chloroform and ethanol precipitation. The DNA pellet was resuspended in 150 μl of Tris-EDTA. Archaeal and bacterial 16S rRNA genes were PCR amplified as previously described (25).
Construction of 16S rRNA gene libraries and screening inserts.
PCR products were ligated into pGEM-T Easy cloning vector and transfected into Escherichia coli JM109 cells according to the manufacturer's instructions to make 16S rRNA gene libraries. Forty-eight white colonies containing recombinant plasmids were picked from each library and grown in Luria-Bertani broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl, adjusted to pH 7.0 with 1 M NaOH) containing 100 μg/ml of ampicillin at 37°C overnight. Inserts were amplified by colony PCR by the following method. Two to three microliters of cell culture was mixed with 20 μl of 0.25% (vol/vol) Tween 20 and boiled for 20 min to denature cellular proteins. Cell debris was pelleted by centrifugation at 10,000 × g for 10 min. Fifteen microliters of this cell lysate was used in a second round of PCR using flanking vector primers M13F (5′-GTTTTCCCAGTCACGAC-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) in a reaction previously described (see above), to a final volume of 25 μl. This gave an expected amplicon of 1.7 kb.
To look for the restriction fragment length polymorphism (RFLP) pattern of each insert, the 1.7-kb PCR products were digested with HaeIII (NEB) in the manufacturer's reaction buffer at 37°C for 2 h. Fragments were visualized by electrophoresis in Tris-acetate-EDTA (TAE) using 2% agarose. Identical RFLPs were then identified by eye, and unique sequences were sent for sequencing to Lark Technologies, Ltd. (Essex, United Kingdom).
Computer analysis.
The CHIMERA_CHECK program available from the Ribosomal Database Project (RDP) (http://rdp8.cme.msu.edu/cgis/news.cgi) and Pintail (3) were used to check for chimeric sequences. Rarefaction curves were calculated from RFLP data using Analytical Rarefaction version 1.3 available at UGA Stratigraphy Lab (http://www.uga.edu/strata/software/index.html). 16S rRNA sequences were searched using Blastn (1). Sequences were aligned using MEGA version 3.1 (39). The values for the Jaccard index were determined using EstimateS, version 7.5 (Department of Ecology and Evolutionary Biology, University of Connecticut; http://viceroy.eeb.uconn.edu/estimates). The simple and partial Mantel tests were carried out using the zt program (8).
Definition of OTU.
The 16S rRNA gene sequences were aligned using MEGA version 3.1 (39), and the output file was used to define operational taxonomic units (OTU) using DOTUR (59). This was done using the furthest neighbor clustering algorithm (default setting). In this study, three of the commonly used OTU definitions were used (95%, 97% and 99%), which is equivalent to comparing taxonomic resolutions at the genus, species, and subspecies levels (31). From 217 nonchimeric sequences, 184 unique sequences were detected at an OTU definition of 99%, 135 unique sequences at 97%, and 110 unique sequences at 95%.
Construction of phylogenetic trees.
Phylogenetic analysis was done using MEGA version 3.1 (39), using the Jukes and Cantor nucleotide substitution model for sequence alignment and the neighbor-joining method of tree inference. The support for each node was determined by assembling a consensus tree of 1,000 bootstrap replicates using the same phylogenetic settings.
SNDE.
Raw environmental data were standardized to make the different environmental factors comparable. This was done with the equation SNDE = (x − mean of the raw data)/standard deviation of the raw data, where SNDE represents the standard normal deviate equivalents and x represents the raw data for one sampling site.
Coverage.
Library coverage was calculated using the equation C = [1− (n1/N)] 100, where n1 represents the number of RFLPs represented by a clone and N represents the total number of clones in the library (24).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences retrieved from the clone libraries have been deposited into the EMBL Nucleotide Sequence Database under accession no. FM210811 to FM211027.
RESULTS
Screening for chimeras.
The sequencing data set (219 16S rRNA gene sequences) were screened for suspected chimeras using the CHIMERA_CHECK program available from the RDP. Subsequently, 56 suspected chimeric sequences were analyzed by Pintail (3). No chimeric sequences were detected in the archaeal 16S rRNA gene libraries. However, two chimeric sequences were detected in the bacterial 16S rRNA gene libraries and were removed from further analysis. This observed frequency is less than previously reported (62). However, since only partial sequences were used in the analysis, fewer chimeras were likely to be found. Chimeras are more likely to be detected in a data set containing longer 16S rRNA gene sequences: i.e., if both ends of the 16S rRNA gene were sequenced.
Library coverage.
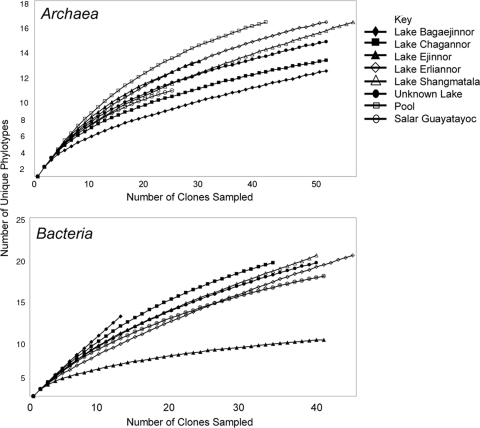
Rarefaction curves were used to identify when sampling was sufficient to determine species diversity with some level of confidence. The numbers of unique clones were plotted against the number of unique species (Fig. 1). Table 2 shows that library coverage was generally approaching plateau stages, with coverage calculated between 44 and 80%. The bacterial library from Lake Bagaejinnor was the clear exception, with coverage estimated at just 8%. Coverage in the archaeal libraries was generally higher than in the bacterial libraries, reflecting lower diversity in the former. Bacterial diversity in Lake Ejinnor was strikingly lower than in the other lakes, although the reason for this is unclear.
FIG. 1.
Rarefaction curves for sampling of the archaeal and bacterial 16S rRNA gene libraries.
TABLE 2.
Library coverage
| Environment | % of isolates in library
|
|
|---|---|---|
| Archaeal | Bacterial | |
| Lake Bagaejinnor | 75 | 8 |
| Lake Chagannor | 73 | 44 |
| Lake Ejinnor | 52 | 80 |
| Lake Erliannor | 70 | 56 |
| Lake Shangmatala | 67 | 50 |
| Unknown lake | 68 | 53 |
| Pool | 54 | 56 |
| Salar Guayatayoc | 63 | 46 |
Archaeal diversity.
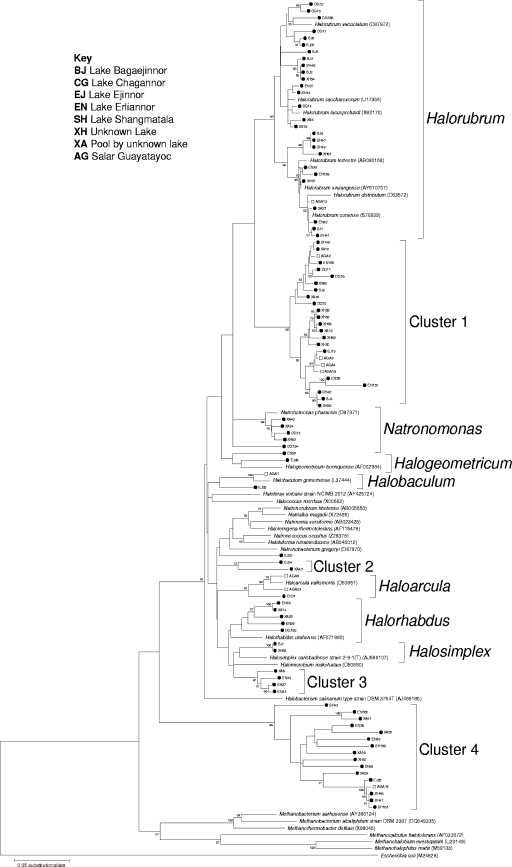
Phylogenetic analysis showed the assignment of clone sequences into seven monophyletic assemblages within the order Halobacteriales (Fig. 2). Clone sequences branched within the Halorubrum, Natronomonas, Halogeometricum, Halobaculum, Haloarcula, Halorhabdus, and Halosimplex lineages. The range of haloarchaea identified in this study was entirely consistent with the saline conditions of the environments sampled. There were additional, well-supported lineages that formed between these nodes, designated clusters 1 to 4 (bootstrap values between 74 and 99), which showed that these sequences were significantly different from any known species. Other known genera within the order Halobacteriales were represented in the tree; however, none of the sequences in the clusters affiliated closely with any of them. Cluster 1 contained sequences that were 99% identical to a clone found in crystallizer ponds in Australia (10). Clusters 2, 3, and 4 all showed low sequence similarity (<98%) to uncultured organisms (data not shown), so were therefore unique to the sites sampled. Moreover, sequence EJ22, found in Lake Ejinnor, did not affiliate with any lineage and was therefore unique to this saltern.
FIG. 2.
Phylogenetic tree of the archaeal population. Closed circles indicate sequences from Inner Mongolia, and open squares indicate sequences from Argentina.
Twenty-eight clone sequences branched with the Halorubrum lineage. Sequences from both Inner Mongolia and Argentina were found in this group, which demonstrated its ubiquitous nature. Sequences from Lake Chagannor were most similar to haloalkaliphilic Halorubrum vacuolatum, which was consistent with the highly alkaline pH of this lake (pH 10.5). Similarly, sequences from Lake Chagannor and the small pool at the unnamed lake (pH 9.5) were affiliated with the haloalkaliphilic group Natronomonas. Twenty-five clone sequences branched with cluster 1, a well-supported lineage that is phylogenetically distinct from the Halorubrum branch (bootstrap value of 98). It was the second largest haloarchaeal group in this study and again was ubiquitous in the habitats studied. Cluster 4 contains 16 clone sequences from both Inner Mongolia and Argentina. It formed a lineage on the periphery of the Halobacteriales that does not show any resemblance to known Euryarchaeota, which suggested that adaptation to hypersaline environments may extend to Archaea outside the Halobacteriales.
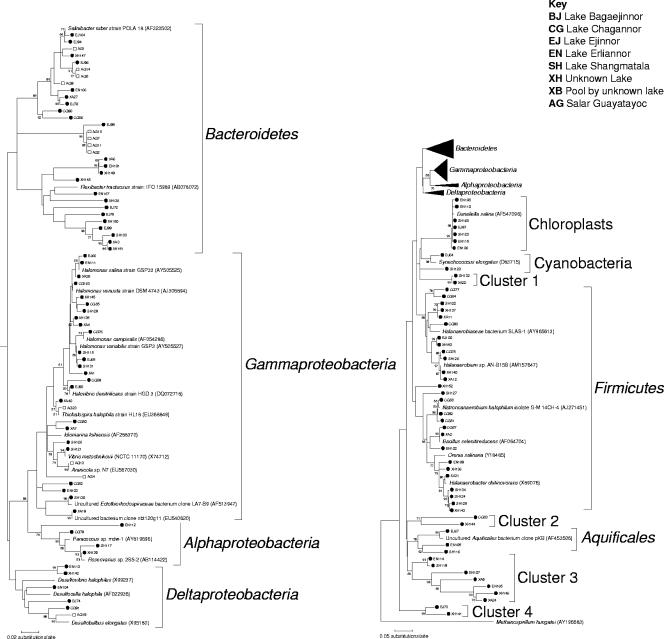
Bacterial diversity.
Phylogenetic analysis showed the distribution of clone sequences into seven monophyletic assemblages—Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, Bacteroidetes, Chlorophyceae, Cyanobacteria, and Aquificales—and two paraphyletic groups of the Firmicutes (Fig. 3). In addition, there were well-supported lineages that formed between these nodes, designated clusters 1 to 4 (bootstrap values between 50 and 99). Cluster 3 contained a sequence that was 99% identical to a clone found in Lake Chaka, an athalassohaline lake in China (35). Sequences in clusters 1, 2, and 4 were unique to these sites as they all had low sequence similarities to uncultured organisms (<98% identity; data not shown).
FIG. 3.
Phylogenetic tree of the bacterial population. Closed circles indicate sequences from Inner Mongolia, and open squares indicate sequences from Argentina. Black wedges on the right-hand side are shown in more detail on the left-hand side.
DNA from chloroplasts in Eukarya was clearly extracted in community DNA preparations, which resulted in several chloroplast 16S rRNA genes in the clone libraries (Chlorophyceae). These were related to chloroplasts found in Dunaliella salina, a typical salt lake inhabitant (49).
The Proteobacteria were the largest group, containing 38 clone sequences. This group was divided into the Gammaproteobacteria (28 sequences), Deltaproteobacteria (6 sequences), and Alphaproteobacteria (4 sequences). Sequences from both Inner Mongolia and Argentina were only affiliated with the former two divisions, while only Inner Mongolian sequences were affiliated with the Alphaproteobacteria. Many clone sequences were related to Halomonas, also typical inhabitants of salt lake environments (27). The second largest group was the Bacteroidetes, which consisted of 31 clone sequences from both Inner Mongolia and Argentina. Clone sequences from the saltern at Lake Ejinnor were affiliated with Salinibacter ruber, an extremely halophilic bacterium (2). The third largest group was the Firmicutes, which consisted of 27 clone sequences, all from Inner Mongolia. The majority of these clones were related to anaerobic species related to the Halanaerobiaceae, with three clones related to Bacillus sp.
Other clone sequences from Inner Mongolia, BJ67 (Lake Bagaejinnor), EN105 (Lake Erliannor), and SH116 (Lake Shangmatala), were also affiliated with or branched near to the deeply branching Aquificae (Fig. 3), which usually inhabit hot spring environments (55). This lineage appeared to be unique to the Inner Mongolian sites as no other Aquificales sequences have been found at other salt lakes. Owing to the low sequence similarity to existing 16S rRNA gene sequences, it may not be appropriate to construe the growth temperature ranges of these bacteria. Recently, mesophilic members of a deeply branching group have been discovered (46), so perhaps these clone sequences represent novel lineages distantly related to Aquificae that are adapted to lower temperatures.
Community composition and biotic similarity matrices.
Comparison of the overall archaeal community composition (Table 3) and bacterial community composition (Table 4) of the salt lakes demonstrates that they were unique. This was assessed using the Jaccard index, calculated using the program EstimateS for each definition of OTU. No pair of lakes scored a value of 1 (1 = identical biotic composition), as can be seen from the biotic similarity matrices.
TABLE 3.
Biotic similarity matrix for Archaeaa
| Sample site and % OTU | Biotic similarity of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| BJ | CG | EJ | EN | SH | XH | X | AG | |
| BJ | 1 | |||||||
| CG | ||||||||
| 99 | 0 | 1 | ||||||
| 97 | 0 | |||||||
| 95 | 0 | |||||||
| EJ | ||||||||
| 99 | 0.066 | 0 | 1 | |||||
| 97 | 0.071 | 0 | ||||||
| 95 | 0.166 | 0 | ||||||
| EN | ||||||||
| 99 | 0.041 | 0 | 0 | 1 | ||||
| 97 | 0.045 | 0.043 | 0 | |||||
| 95 | 0.117 | 0.052 | 0 | |||||
| SH | ||||||||
| 99 | 0.187 | 0 | 0.062 | 0 | 1 | |||
| 97 | 0.307 | 0.058 | 0.066 | 0.142 | ||||
| 95 | 0.454 | 0.062 | 0.230 | 0.166 | ||||
| XH | ||||||||
| 99 | 0.157 | 0 | 0 | 0.035 | 0.095 | 1 | ||
| 97 | 0.285 | 0.055 | 0 | 0.086 | 0.266 | |||
| 95 | 0.545 | 0.058 | 0.133 | 0.1 | 0.357 | |||
| X | ||||||||
| 99 | 0.041 | 0 | 0 | 0.066 | 0 | 0.035 | 1 | |
| 97 | 0.043 | 0.041 | 0 | 0.192 | 0.086 | 0.130 | ||
| 95 | 0.111 | 0.105 | 0 | 0.315 | 0.157 | 0.210 | ||
| AG | ||||||||
| 99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 97 | 0.076 | 0 | 0.181 | 0.105 | 0.25 | 0.066 | 0.1 | |
| 95 | 0.3 | 0 | 0.181 | 0.125 | 0.363 | 0.230 | 0.117 | |
These matrices show biotic similarity determined by the Jaccard index for Archaea at the three OTU definitions listed from top to bottom for each sampling site. A value of 1 indicates identical microbial communities. BJ, Lake Bagaejinnor; CG, Lake Chagannor; EJ, Lake Ejinnor; EN, Lake Erliannor; SH, Lake Shangmatala; XH, unknown lake; X, pool by unknown lake; AG, Salar Guayatayoc.
TABLE 4.
Biotic similarity matrix for Bacteriaa
| Sample site and % OTU | Biotic similarity of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| BJ | CG | EJ | EN | SH | XH | X | AG | |
| BJ | 1 | |||||||
| CG | ||||||||
| 99 | 0 | 1 | ||||||
| 97 | 0.04 | |||||||
| 95 | 0.043 | |||||||
| EJ | ||||||||
| 99 | 0 | 0 | 1 | |||||
| 97 | 0 | 0 | ||||||
| 95 | 0 | 0 | ||||||
| EN | ||||||||
| 99 | 0.04 | 0 | 0.041 | 1 | ||||
| 97 | 0.043 | 0.032 | 0.047 | |||||
| 95 | 0.09 | 0.071 | 0.05 | |||||
| SH | ||||||||
| 99 | 0.037 | 0 | 0.038 | 0.028 | 1 | |||
| 97 | 0.041 | 0.064 | 0.045 | 0.068 | ||||
| 95 | 0.086 | 0.107 | 0.047 | 0.107 | ||||
| XH | ||||||||
| 99 | 0 | 0 | 0.038 | 0.028 | 0 | 1 | ||
| 97 | 0.076 | 0.058 | 0.04 | 0.096 | 0.129 | |||
| 95 | 0.076 | 0.062 | 0.086 | 0.096 | 0.206 | |||
| X | ||||||||
| 99 | 0 | 0 | 0 | 0 | 0 | 0.090 | 1 | |
| 97 | 0.130 | 0.096 | 0 | 0.066 | 0.137 | 0.2 | ||
| 95 | 0.130 | 0.142 | 0 | 0.103 | 0.269 | 0.241 | ||
| AG | ||||||||
| 99 | 0 | 0 | 0.066 | 0 | 0 | 0 | 0 | 1 |
| 97 | 0 | 0 | 0.076 | 0 | 0 | 0 | 0 | |
| 95 | 0 | 0 | 0.2 | 0 | 0 | 0.041 | 0.045 | |
These matrices show biotic similarity determined by the Jaccard index for Bacteria at the three OTU definitions listed from top to bottom for each sampling site. A value of 1 indicates identical microbial communities. BJ, Lake Bagaejinnor; CG, Lake Chagannor; EJ, Lake Ejinnor; EN, Lake Erliannor; SH, Lake Shangmatala; XH, unknown lake; X, pool by unknown lake; AG, Salar Guayatayoc.
Geographic distance matrix.
The matrix for the distances between each of the environments in this study is found in Table S1 in the supplemental material. This was calculated using the GPS coordinates measured during the expedition (see Materials and Methods).
Environmental similarity matrices.
To construct environmental similarity matrices, all raw values had to be standardized to make them comparable since this accommodates different units of different variables. Therefore, SNDE values were calculated for temperature, pH, and concentrations of Na+, Mg2+, K+, Cl−, S, CO32−, and HCO3− ions (see Tables S2 to S10 in the supplemental material). In order to construct a similarity matrix with this new standardized data, values of 1 − the Euclidean distance between the SNDE values of two lakes were calculated. A value of 1 therefore indicated that a particular environmental factor was identical for the two lakes. The environmental similarity matrices are found in the supplementary material.
Biogeography of Archaea.
The simple Mantel test was carried out using the zt program (8). This program calculated whether biotic similarity correlated with environmental factors and whether this correlation was statistically significant. The value of r2 was the correlation value, and positive or negative values reflected the type of relationship between the two matrices, while P was the probability associated with r2. Values of P were significant if they were <0.05; values that were >0.05 indicated that the null hypothesis applied. (The null hypothesis stated that distances in matrix A were independent of the distances in matrix B.)
Table 5 shows the results of the simple Mantel test. The results showed that geographic distance was not a significant factor in influencing archaeal community composition, since P values across all OTU levels were >0.05. Therefore, historical events and geographic barriers to dispersal have not affected archaeal community composition.
TABLE 5.
Simple Mantel test for the archaeal population in this studya
| Factor | 99% OTU
|
97% OTU
|
95% OTU
|
|||
|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |
| Cl− | 0.360 | 0.056 | 0.371 | 0.076 | 0.493 | 0.052 |
| CO32− | 0.342 | 0.067 | 0.443 | 0.032 | 0.532 | 0.027 |
| Distance | −0.336 | 0.116 | 0.094 | 0.473 | 0.087 | 0.486 |
| HCO3− | 0.336 | 0.118 | 0.440 | 0.032 | 0.527 | 0.035 |
| K+ | 0.072 | 0.373 | −0.164 | 0.289 | −0.110 | 0.391 |
| Mg2+ | 0.139 | 0.393 | 0.306 | 0.166 | 0.279 | 0.176 |
| pH | 0.431 | 0.010 | 0.497 | 0.005 | 0.478 | 0.019 |
| S2− | 0.089 | 0.334 | −0.045 | 0.415 | −0.137 | 0.264 |
| Temp | 0.401 | 0.043 | 0.090 | 0.378 | 0.102 | 0.352 |
| Na+ | 0.245 | 0.113 | 0.322 | 0.051 | 0.343 | 0.038 |
r2 is the correlation value; positive or negative values reflect the type of relationship between the two matrices, while P is the probability associated with r2. P values are significant if P is <0.05 (boldface).
Only contemporary environmental factors appear to significantly influence the archaeal community composition. At the genus (95% OTU) and species (97% OTU) levels, CO32− and HCO3− ion concentrations and pH were significant factors in influencing archaeal community composition since P values were <0.05. The Na+ ion concentration was significant at the genus level only. All r2 values were positive; hence, biotic similarity of the salt lakes increased as the similarity of the environmental factors increased. At the subspecies level (99% OTU), pH and temperature were the only significant factors. Again, all r2 values were positive integers. Removal of the Argentine data set from the analysis did not significantly change these results (50).
Biogeography of Bacteria.
Table 6 shows the results of the simple Mantel test. The results showed that geographic distance was a significant factor in influencing the bacterial community composition at the genus level (95% OTU) and species level (97% OTU), since P values across these OTU levels were <0.05. Both r2 values were negative; hence, biotic similarity of the salt lakes increased as the geographic distance similarity between the lakes increased (i.e., the closer the lakes, the more similar the bacterial populations). The larger the distance between sites, the higher the likelihood of barriers to dispersal. We therefore concluded that historical events and geographic barriers affected bacterial community composition. At the subspecies level (99% OTU), geographic distance was no longer a significant factor affecting bacterial community composition. Only environmental factors were significant: i.e., HCO3− ion concentration and pH. Omitting data from Lake Bagaejinnor due to the low coverage values does not affect the results at the 99% or 97% OTU definitions, but at the 95% level, temperature and pH are no longer significant (data not shown).
TABLE 6.
Simple Mantel test for the bacterial population in this studya
| Factor | 99% OTU
|
97% OTU
|
95% OTU
|
|||
|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |
| Cl− | 0.226 | 0.147 | −0.069 | 0.464 | −0.043 | 0.479 |
| CO32− | 0.350 | 0.090 | 0.135 | 0.275 | 0.166 | 0.399 |
| Distance | −0.125 | 0.166 | −0.484 | 0.0016 | −0.328 | 0.002 |
| HCO3− | 0.362 | 0.002 | 0.131 | 0.319 | 0.168 | 0.370 |
| K+ | 0.118 | 0.290 | 0.210 | 0.180 | 0.000 | 0.417 |
| Mg2+ | −0.230 | 0.127 | 0.282 | 0.159 | 0.226 | 0.145 |
| pH | 0.358 | 0.033 | 0.306 | 0.119 | 0.325 | 0.045 |
| S2− | 0.068 | 0.359 | −0.056 | 0.404 | −0.083 | 0.367 |
| Temp | −0.068 | 0.396 | 0.503 | 0.022 | 0.372 | 0.048 |
| Na+ | −0.184 | 0.192 | −0.381 | 0.002 | −0.374 | 0.010 |
r2 is the correlation value, and positive or negative values reflect the type of relationship between the two matrices, while P is the probability associated with r2. P values are significant if P is <0.05 (boldface).
In order to disentangle the effects of the environment versus geographic distance, the partial Mantel test was used (42). The effects of the environment on biotic similarity were tested at the genus and species levels, while controlling the effects of geographic distance (Table 7). Na+ and HCO3− ion concentrations and pH had a significant influence at the genus level (95% OTU) and species level (97% OTU), with P values of <0.05. The Mg2+ ion concentration was significant at the species level only. Curiously, both HCO3− and Na+ ion concentrations had negative r2 values.
TABLE 7.
Partial Mantel test testing the effects of various factors while controlling for the effect of geographic distancea
| Factor | 97% OTU
|
95% OTU
|
||
|---|---|---|---|---|
| r | P | r | P | |
| CO32− | 0.235 | 0.190 | 0.228 | 0.262 |
| HCO3− | −0.346 | 0.022 | −0.342 | 0.024 |
| pH | 0.475 | 0.014 | 0.425 | 0.026 |
| Temp | 0.256 | 0.137 | 0.208 | 0.185 |
| Na+ | −0.346 | 0.024 | −0.342 | 0.025 |
| Cl− | −0.028 | 0.472 | −0.013 | 0.507 |
| K+ | −0.105 | 0.295 | −0.253 | 0.091 |
| Mg2+ | 0.426 | 0.032 | 0.305 | 0.059 |
| S2− | −0.164 | 0.214 | −0.151 | 0.237 |
r2 is the correlation value, and positive or negative values reflect the type of relationship between the two matrices, while P is the probability associated with r2. P values are significant if P is <0.05 (boldface).
When the Argentinean data were removed from the analysis, environmental factors were again found to be significant, but not geographic distance (50).
DISCUSSION
Archaeal biogeography.
We report the phylogeny and distribution of Archaea in seven salt lakes across two continents at almost antipodean positions to each other. Statistical analyses demonstrated that this distribution was significantly influenced by environmental factors (Na+, CO32−, and HCO3− ion concentrations; pH; and temperature) (Table 5). All r2 values were positive integers, which indicated that as environmental similarity increased, biotic similarity increased. Geographic distance was not a significant factor. This was supported by phylogenetic analysis, which showed that the Argentinean sequences were interspersed throughout the phylogenetic tree; therefore, no lineages were specific to either the Inner Mongolian or Argentinean salt lakes (Fig. 2). Our results are in contrast to a previous finding that the distribution of hyperthermophilic archaea showed a tendency for endemism (63), despite the fact that some archaeal species can be airborne (52). This implies that unlike hyperthermophilic archaea, haloarchaea are more robust over long-distance travel: one apparent explanation for this is that hyperthermophilic archaea are less likely to survive at ambient temperatures.
The finding that pH was a significant environmental factor in influencing haloarchaeal biogeography at all three definitions of OTU was not unexpected. pH would allow different species to be selected in either slightly alkaline (pH 7.5) or highly alkaline (pH 10.5) environments, which was the pH range of the salt lakes in this study. Since pH is dependent on CO32−and HCO3− ions, it was also not unexpected that these were also significant factors (although surprisingly, they were not significant at the 99% OTU level). Phylogenetic analysis supported this finding as haloalkaliphilic species were only found in the alkaline lakes, while haloarchaeal species in the other lakes were consistent with environments of lower pH values. For example, clones relating to Halorubrum vacuolatum and Natronomonas sp. were only detected in Lake Chagannor (pH 10.5) and in the small pool at the unnamed lake (pH 9.5). However, it was unusual that other alkaliphilic groups within the Halobacteriales were not detected, such as those found, for example, in soda lakes in Kenya and Egypt (26, 36, 44, 53).
It appeared that temperature was a significant factor in driving haloarchaeal biogeography at the subspecies level only (99% OTU). This implied that seasonal changes in temperature were important in influencing haloarchaeal biogeography. Experiments with samples from a saltern showed that at 35°C, dense growths of haloarchaea were observed at 35% and 40% (wt/vol) salt, but at 25°C, very little haloarchaeal growth was observed (15). Temperature may also play a role in competition: temperature was found to be the deciding factor in competition between moderately halophilic bacteria and haloarchaea, with bacterial growth being favored at lower temperatures (57).
Na+ ion concentration was a significant factor affecting haloarchaeal community composition at the genus level (95% OTU). Haloarchaea adapt to high NaCl concentrations in the environment by accumulation of KCl to exclude NaCl from the cells, thereby achieving osmotic equilibrium (40). This is an adaptation that does not extend to Archaea outside the Halobacteriales (23). In addition, some haloarchaeal enzymes have evolved a requirement for high Na+ concentrations (19, 45).
Bacterial biogeography.
Statistical analysis showed that the distribution of Bacteria in the six salt lakes across two continents was significantly influenced by geographic distance (Table 6). When this analysis was repeated for the Inner Mongolian samples alone, geographic distance was not a significant factor (50; data not shown), suggesting that geographic distance does not have a biogeographical effect on a local spatial scale. The strong winds observed on the steppe at the time of sampling could allow dispersal of microorganisms over long distances (18, 33, 37), and so the fact that geographic distance became a significant factor once the Argentinean data was added to the analysis suggested that there may be a tendency toward endemism in halophilic bacteria. The phylogenetic analysis implies certain lineages may be implicated in this endemism (Fig. 3). The finding that geographic distance affects bacterial biogeography is consistent with the other studies (e.g., references 12, 22, and 51) and has been explained by the fact that, at a long geographic distance, barriers to dispersal are more likely, and so evolutionary events such as speciation and extinction can give rise to differences in two populations separated by such barriers (32). Our finding is consistent with previous studies on Thioalkalivibrio and Salinibacter ruber, which showed that strains of both bacterial species were endemic to certain regions, despite having cosmopolitan distributions (22, 58).
Geographic distance was not a significant factor at the subspecies level (99% OTU). Only pH and HCO3− ion concentration were significant at this level of OTU. Again, the correlation with pH was not unexpected. The biogeography of bacteria in freshwater lakes has often been correlated with pH (41, 67). It is not hard to imagine that the same effect would occur with halophilic bacteria, but to our knowledge, no other studies have shown this biogeographical effect. Phylogenetic analysis supported this finding, which showed that clones from Lake Chagannor (pH 10.5) and the pool at the unnamed lake (pH 9.5) were related to haloalkaliphilic species such as Halomonas campisalis and “Natronoanaerobium halophilum.”
Once the effects of geographic distance were controlled using the partial Mantel test, the contemporary environmental factors found to be significant in influencing bacterial biogeography at the genus level (95% OTU) and species level (97% OTU) were Na+, Mg2+, and HCO3− ion concentrations and pH. The r2 values for pH and Mg2+ ion concentrations were positive (the higher the environmental similarity, the higher the biotic similarity) (Table 7). Surprisingly, the r2 values for Na+ and HCO3− ion concentrations were negative (the higher environmental similarity, the lower the biotic similarity), which does not fit what is currently known about microbial biogeography (42), where only positive correlations with environmental parameters have been observed.
Na+ ions were significant in influencing bacterial community composition at both the genus (95% OTU) and species (97% OTU) levels. However, unlike the haloarchaea, halophilic bacteria cope with high NaCl concentrations in the environment by accumulation of organic compatible solutes (56), with only a few exceptions (for example, the Halanaerobiales [19, 54] and Salinibacter ruber [48]). Other sodium salts may influence bacterial community composition since a previous study showed that the high salt requirement for a moderate halophilic bacterium was met by sodium salts other than NaCl (47). In addition, Na+ ions are important to some alkaliphilic bacteria as they replace protons as the coupling ion to cope with the high external pH, rather than increasing the electric potential difference across the cytoplasmic membrane (60).
The Mg2+ ion concentration was a significant factor in influencing the bacterial community composition at the species level only (97% OTU). Mg2+ favors the growth of haloarchaea (26), and so a possible explanation for this trend may be that only the halophilic bacteria that are tolerant to Mg2+ are able to proliferate and coexist with the haloarchaea. (MgCl2 is a chaotropic agent and is a limiting factor in the diversity of microbes in the environment [29].)
Conclusions.
Unlike other saline and soda lakes, such as those in the East African Rift Valley, these particular lakes are formed in depressions entirely by runoff from the surrounding topography. As such they are influenced by seasons and vary in salinity, depending on rainfall or spring melts. The lakes are shown as permanent sites on local maps of the areas (66, 68, 69). We sampled these lakes in summer months at or close to maximum salinity levels which last for several months: the particular prokaryote population we detected reflects the particular set of conditions we measured at that time. Clearly, in an ideal world, sequential sampling over an extended time period would be appropriate, but logistical and financial considerations preclude repeated visits to these remote sites. The expedition undertaken was designed to access the lakes during a relatively stable period of water chemistry but still must be a compromise in view of the seasonality of the sites.
Martiny et al. (42) suggested that the relative effect of the environment on microbial community composition relates to the geographical scale of sampling: at large geographic scales, distance seems to influence the community composition more, while environment seems not to have any effect; in contrast, at a small scale, it is the environment which has an effect but not distance (42). However, due to the small number of studies available thus far, this conclusion should be treated with caution. For example, studies at large-scale distances have been carried out on prokaryotes inhabiting extreme environments, in which the environmental parameters have a relatively small range, and focus on a particular prokaryotic species (22, 51, 63), while studies at small-scale distances target mixed bacterial populations (14). It is possible that this trend has arisen from the types of habitats sampled or the organism studied.
Similarly, we have presented a case in which microbial biogeography is not so clear-cut. It can be argued that extremophiles have larger limits to dispersal due to the lack of a suitable habitat. Certainly, this research has shown that bacterial biogeography is significantly affected by geographic distance and therefore barriers to dispersal. This implies that halophilic bacteria are less mobile or less robust across large distances. On the other hand, geographic distance was not a significant factor in affecting archaeal biogeography, but contemporary environmental factors were more significant. This implies that Archaea are dispersed more easily or are more robust over long-distance travel. The study of microbial biogeography is still in its infancy. Only further case studies will show any solid trends in this field.
Supplementary Material
Acknowledgments
This research was supported by the European Commission research program “Quality of Life and Management of Living Resources,” project Multigenome Access Technology for Industrial Catalysts (QLRT-2001-01972).
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Antón, J., R. Rosselló-Mora, F. Rodríguez-Valera, and R. Amann. 2000. Extremely halophilic Bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baas-Becking, L. G. M. 1934. Geobiologie of inleiding tot de milieukunde. W. P. van Stockum and Zoon, Gravenhange, The Netherlands.
- 5.Bano, N., S. Ruffin, B. Ransom, and J. T. Hollibaugh. 2004. Phylogenetic comparison of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beijerinck, M. W. 1913. De infusies en de ontdekking der backterien, p. 119-140. In F. Bruckmann et al. (ed.), Jaarboek van de Koninklijke Akademie voor Wetenschappen. Müller, Amsterdam, The Netherlands.
- 7.Benlloch, S., A. López-López, E. O. Casamayor, L. Øvreås, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Joint, F. Thingstad, C. Pedrós-Alió, and F. Rodríguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 8.Bonnet, E., and Y. Van der Peer. 2002. zt: a software tool for simple and partial Mantel tests. J. Stat. Software 7:1-12. [Google Scholar]
- 9.Brown, J. H., B. R. Riddle, and M. V. Lomolino. 2005. Biogeography. Sinauer Associates, Inc., Sunderland, MA.
- 10.Burns, D. G., H. M. Camakaris, P. H. Janssen, and M. L. Dyall-Smith. 2004. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 70:5258-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casamayor, E. O., R. Massana, S. Benlloch, L. Øvreås, B. Díez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodríguez-Valera, and C. Pedrós-Alió. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 12.Cho, J.-C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, C. B., and P. D. Moore. 2000. Biogeography: an ecological and evolutionary approach. Blackwell Science, Ltd., Oxford, United Kingdom.
- 14.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Moral, A., E. Quesada, V. Bejar, and A. Ramos-Cormenzana. 1987. Evolution of bacterial flora from a subterranean saline well by gradual salinity changes in enrichment media. J. Appl. Bacteriol. 62:465-471. [Google Scholar]
- 16.Demergasso, C. S., E. O. Casamayor, G. Chong, P. A. Galleguillos, L. V. Escudero, and C. Pedrós-Alió. 2004. Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol. Ecol. 48:57-69. [DOI] [PubMed] [Google Scholar]
- 17.Dimitriu, P. A., H. C. Pinkart, B. M. Peyton, and M. R. Mormile. 2008. Spatial and temporal patterns in the microbial diversity of a meromictic soda lake in Washington State. Appl. Environ. Microbiol. 74:4877-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echigo, A., M. Hino, T. Fukushima, T. Mizuki, M. Kamekura, and R. Usami. 2005. Endospores of halophilic bacteria of the family Bacillaceae isolated from non-saline Japanese soil may be transported by Kosa event (Asian dust storm). Saline Syst. 1:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Empadinhas, N., and M. S. da Costa. 2008. Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int. Microbiol. 11:151-161. [PubMed] [Google Scholar]
- 20.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 22.Foti, M., S. Ma, D. Y. Sorokin, J. L. W. Rademaker, J. G. Kuenen, and G. Muyzer. 2006. Genetic diversity and biogeography of haloalkaliphilic sulphur-oxidizing bacteria belonging to the genus Thioalkalivibrio. FEMS Microbiol. Ecol. 56:95-101. [DOI] [PubMed] [Google Scholar]
- 23.Galinski, E. A., and H. G. Trüper. 1994. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95-108. [Google Scholar]
- 24.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 25.Grant, S., W. D. Grant, B. E. Jones, C. Kato, and L. Li. 1999. Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles 3:139-145. [DOI] [PubMed] [Google Scholar]
- 26.Grant, W. D., M. Kamekura, T. J. McGenity, and A. Ventosa. 2001. Class III Halobacteria class nov., p. 294-334. In D. R. Boone, R. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 27.Grant, W. D. 2004. Life at low water activity. Philos. Trans. R. Soc. Lond. B 359:1249-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunde-Cimerman, N., A. Oren, and A. Plemenitaš. 2005. Adaptation to life at high salt concentrations in Archaea, Bacteria, and Eukarya. Springer, New York, NY.
- 29.Hallsworth, J. E., M. M. Yakimov, P. N. Golyshin, J. L. M. Gillion, G. D'Auria, F. de Lima Alves, V. La Cono, M. Genovese, B. A. McKew, S. L. Hayes, G. Harris, L. Giuliano, K. N. Timmis, and T. J. McGenity. 2007. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ. Microbiol. 9:801-813. [DOI] [PubMed] [Google Scholar]
- 30.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 31.Horner-Devine, M. C., M. Lage, J. B. Hughes, and B. J. M. Bohannan. 2004. A taxa-area relationship for bacteria. Nature 432:750-753. [DOI] [PubMed] [Google Scholar]
- 32.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua, N.-P., F. Kobayashi, Y. Iwasaka, G.-Y. Shi, and T. Naganuma. 2007. Detailed identification of desert-originated bacteria carried by Asian dust storms to Japan. Aerobiologia 23:291-298. [Google Scholar]
- 34.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, H., H. Dong, G. Zhang, B. Yu, L. R. Chapman, and M. W. Fields. 2006. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 72:3832-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, B. E., W. D. Grant, A. W. Duckworth, and G. G. Owenson. 1998. Microbial diversity of soda lakes. Extremophiles 2:191-200. [DOI] [PubMed] [Google Scholar]
- 37.Junfeng, L. 1997. Renewable energy development in China: resource assessment, technology status, and greenhouse gas mitigation potential. Appl. Energy 56:381-394. [Google Scholar]
- 38.Kulp, T. R., S. Han, C. W. Saltikov, B. D. Lanoil, K. Zargar, and R. S. Oremland. 2007. Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California soda lakes. Appl. Environ. Microbiol. 73:5130-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 40.Lanyi, J. K. 1974. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38:272-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindström, E. S., M. P. Kamst-van Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 71:8201-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Øvreås, A. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 43.McCraig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mesbah, N. M., S. H. Abou-El-Ela, and J. Wiegel. 2007. Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi An Natrun, Egypt. Microb. Ecol. 54:598-617. [DOI] [PubMed] [Google Scholar]
- 45.Mevarech, M., F. Frolow, and L. M. Gloss. 2000. Halophilic enzymes: proteins with a grain of salt. Biophys. Chem. 86:155-164. [DOI] [PubMed] [Google Scholar]
- 46.Nesbø, C. L., M. Dlutek, O. Zhaxybayeva, and W. F. Doolittle. 2006. Evidence for existence of “Mesotogas,” members of the order Thermotogales adapted to low-temperature environments. Appl. Environ. Microbiol. 72:5061-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Connor, K., and L. N. Csonka. 2003. The high salt requirement of the moderate halophile Chromohalobacter salexigens DSM3043 can be met not only by NaCl but by other ions. Appl. Environ. Microbiol. 69:6334-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oren, A., M. Heldal, S. Norland, and E. A. Galinski. 2002. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6:491-498. [DOI] [PubMed] [Google Scholar]
- 49.Oren, A. 2005. A hundred years of Dunaliella research: 1905-2005. Saline Syst. 1:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagaling, E. 2007. Ph.D. thesis. University of Leicester, Leicester, United Kingdom.
- 51.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographic isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 52.Radosevich, J. L., W. J. Wilson, J. H. Shinn, T. Z. DeSantis, and G. L. Andersen. 2002. Development of a high-volume aerosol collection system for the identification of air-bourne micro-organisms. Lett. Appl. Microbiol. 34:162-167. [DOI] [PubMed] [Google Scholar]
- 53.Rees, H. C., W. D. Grant, B. E. Jones, and S. Heaphy. 2004. Diversity of Kenyan soda lake alkaliphiles assessed by molecular methods. Extremophiles 8:63-71. [DOI] [PubMed] [Google Scholar]
- 54.Rengpipat, S., S. E. Lowe, and J. G. Zeikus. 1988. Effect of extreme salt concentrations on the physiology and biochemistry of Halobacteroides acetoethylicus. J. Bacteriol. 170:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reysenbach, A. 2001. Phylum BI. Aquificae phy. nov., p. 359-367. In D. R. Boone, R. Castenholz, and G. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 56.Roberts, M. F. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodríguez-Valera, F., F. Ruiz-Berraquero, and A. Ramos-Cormenzana. 1980. Behaviour of mixed populations of halophilic bacteria in continuous cultures. Can. J. Microbiol. 26:1259-1263. [DOI] [PubMed] [Google Scholar]
- 58.Rosselló-Mora, R., M. Lucio, A. Pena, J. Brito-Echeverría, A. López-López, M. Valens-Vadell, M. Frommberger, J. Antón, and P. Schmitt-Kopplin. 2008. Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME J. 2:242-253. [DOI] [PubMed] [Google Scholar]
- 59.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skulachev, V. P., H. Kobayashi, T. A. Krulwich, G. Schafer, R. H. Fillingame, R. K. Poole, G. M. Cook, M. J. Dimroth, W. N. Konings, and J. B. Stock. 1999. Bacterial energetics at high pH: what happens to the H+ cycle when the extracellular H+ concentration decreases? Bacterial response to pH. Novartis Found. Symp. 221:200-217. [DOI] [PubMed] [Google Scholar]
- 61.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 62.Wang, G. C., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 63.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 64.Williams, W. D., and J. E. Sherwood. 1994. Definition and measurement of salinity in salt lakes. Int. J. Salt Lake Res. 3:53-63. [Google Scholar]
- 65.Wu, Q. L., G. Zwart, M. Schauer, M. P. Kamst-van Agterveld, and M. W. Hahn. 2006. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan plateau, China. Appl. Environ. Microbiol. 72:5478-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiyu, Z., Z. Minggang, and D. Jihe. 1992. Salt lakes in Inner Mongolia. Science Press, Beijing, China.
- 67.Yannarell, A. C., and E. W. Triplett. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, G., S. P. Harrison, and B. Xue. 2001. Lake status records from China: data base documentation. MPI-BGC Technical Report 4.
- 69.Zheng, X., M. Zhang, J. Dong, Z. Gao, C. Xu, Z. Han, B. Zhang, D. Sun, and K. Wang. 1992. Salt lakes in Inner Mongolia of China. Science Press, Beijing, China.
- 70.Zhongling, L. 1963. An outline of stipa steppes in Inner Mongolia. Acta Phytoecol. Geobot. Sin. 1:156-158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.