Abstract
A two-color fluorescence in situ hybridization assay that allows for the simultaneous identification of Cryptosporidium parvum and C. hominis was developed. The assay is a simple, rapid, and cost-effective tool for the detection of the major Cryptosporidium species of concern to public health.
Cryptosporidium (Apicomplexa) is a genus of protozoan parasites with species and genotypes that infect humans, domesticated livestock, companion animals, and wildlife worldwide (5, 6, 14, 15, 20, 23). The majority of cases of cryptosporidiosis in humans are caused by Cryptosporidium parvum or C. hominis (8, 10, 19, 24), although rare cases due to species such as C. meleagridis, C. felis, or C. canis have been reported (8, 9, 11-13, 17, 18, 22). The specific identification and characterization of Cryptosporidium species are central to the control of this disease in humans and a wide range of animals.
One of the most widely adopted techniques for the identification of microorganisms in complex microbial communities is fluorescence in situ hybridization (FISH) using rRNA-targeted oligonucleotide probes (2-4). This method relies on the hybridization of synthetic oligonucleotide probes to specific regions within the rRNA of the organism. While FISH has been applied for the detection of Cryptosporidium oocysts in water samples (21), no FISH probes that successfully differentiate C. hominis from C. parvum have been reported.
We have reported previously on the design of a species-specific probe, Cpar677, that detects C. parvum (1). In this study, we report on the design and validation of a C. hominis species-specific probe, Chom253. Together, the two probes were used here for the development of a two-color, microscopy-based FISH assay for the simultaneous detection of C. parvum and C. hominis.
C. hominis probe design and validation.
An alignment of 18S rDNA sequences of Cryptosporidium species obtained from the GenBank database was used to design a probe specific for C. hominis (Chom253, TCA CAT TAA TTG TGA TCC). The resulting Chom253 probe differs from the C. parvum 18S sequence by 2 nucleotides and from the Cryptosporidium horse genotype 18S sequence by 1 nucleotide. The Chom253 probe was synthesized with Cy3 attached (Proligo, Australia), and the probe specificity was tested against a panel of eight Cryptosporidium species (C. parvum, C. hominis, C. andersoni, C. muris, C. meleagridis, C. felis, Cryptosporidium cervine genotype, and Cryptosporidium rabbit genotype) by use of the FISH protocol developed for C. parvum (1). Specificity studies revealed that Ch253-Cy3 hybridized with C. hominis and Cryptosporidium rabbit genotype oocysts. Fluorescent signals were not observed with any of the other species examined. Hybridization of the C. hominis-specific probe to the Cryptosporidium rabbit genotype was due to the probe target region being 100% homologous between the rabbit genotype and C. hominis.
Two-color FISH for differentiation of C. parvum and C. hominis.
A two-color FISH assay for the simultaneous identification of C. parvum and C. hominis was developed using Cpar677-Cy3 and Chom253-fluorescein isothiocyanate (FITC) and the FISH protocol reported for C. parvum (1), except that the hybridization buffer contained both of the oligonucleotide probes at a final concentration of 1.5 mmol liter−1 each. The assay was validated using 50 human fecal samples positive for Cryptosporidium, and oocysts were isolated from feces by use of a purification procedure described previously (1). FISH results were compared to PCR-restriction fragment length polymorphism (RFLP) results by use of a DNA extraction method, an 18S rDNA nested PCR, and an RFLP analysis described previously (1). In the dual assay, of the 50 isolates examined, 23 tested positive with Chom253, with a bright-green fluorescence signal observed (Fig. 1A), indicating the presence of C. hominis. The remaining 27 isolates tested positive with Cpar677, resulting in a bright-red fluorescence signal (Fig. 1B), indicating the presence of C. parvum. PCR-RFLP analysis, using VspI, confirmed our FISH results, as demonstrated by specific banding profiles. The results confirmed the strong correlation between FISH and PCR-RFLP observed previously (1).
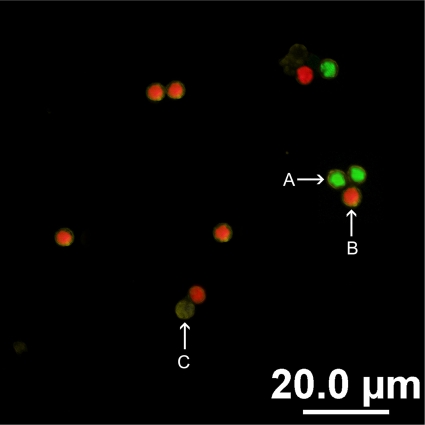
FIG. 1.
Two-color FISH assay for the identification of C. parvum and C. hominis oocysts, carried out using Cpar677-Cy3 and Chom253-FITC. (A) Positive hybridization of C. hominis oocysts was revealed by bright-green fluorescence. (B) Positive hybridization of C. parvum oocysts was revealed by bright-red fluorescence. (C) A lack of fluorescence confirms FISH-negative oocysts.
Oocyst recovery efficiency following FISH.
The recovery efficiency of the assay was evaluated with polycarbonate filter membranes by performing FISH using Cpar677-Cy3 on a precise number of Cryptosporidium oocysts. Precise numbers of C. parvum oocysts (105 ± 12) were prepared for recovery determination by use of fluorescence-activated cell sorting (7). Five groups of triplicate samples underwent hybridization on membranes according to the vacuum manifold technique previously described by Davies et al. (16). Each triplicate set of five samples was processed on the same day. Counterstaining of oocysts with CRY104-FITC enabled the quantification of oocysts to be assessed microscopically. The mean oocyst recovery efficiency for each of the three groups and the overall efficiency were 76.7% ± 14.9%, 88.5% ± 8.6%, 80.9% ± 2.1%, and 82.1% ± 5.9%, respectively. When the data were analyzed by using a two-sample t test (based on the assumption that data for both populations [before and after FISH] were normally distributed and the assumption that the population standard deviations for both were the same), no significant differences in oocyst recovery were detected for these groups (P = 0.985). The levels of recovery increased significantly by use of this method (P < 0.001) compared to recoveries obtained when FISH was not performed according to the vacuum manifold technique (16).
Mixed-species detection.
To evaluate the sensitivity of the two-color assay for the detection of mixed infections, precise numbers of C. parvum and C. hominis oocyst seeds were also prepared. Triplicate samples containing a total of 200 oocysts were used, and percentages of C. parvum to C. hominis oocysts prepared were 10:90, 25:75, 50:50, 75:25, and 90:10. The two-color assay was then performed as described above and quantified using epifluorescence microscopy. Simultaneous detection of 80.6% ± 3.1% of C. parvum and C. hominis oocysts occurred at all ratios tested. In each case, as few as 10 C. parvum and C. hominis oocysts were detected using the two-color assay (Table 1).
TABLE 1.
Recovery of mixed-species suspensions of C. parvum and C. hominis oocysts subjected to the two-color FISH assaya
| Ratio (%) of C. parvum to C. hominis oocysts | Mean % recovery following two-color FISH
|
|
|---|---|---|
| C. parvum | C. hominis | |
| 10:90 | 6.1 ± 2.2 | 80.7 ± 5.8 |
| 25:75 | 18.7 ± 5.1 | 68 ± 4.3 |
| 50:50 | 40.5 ± 5.0 | 41.1 ± 4.8 |
| 75:25 | 69.2 ± 4.1 | 18.8 ± 3.2 |
| 90:10 | 82.3 ± 4.9 | 6.8 ± 2.0 |
C. parvum- and C. hominis-specific probes were used for the detection of viable oocysts. Oocysts were quantified using epifluorescence microscopy.
Our two-color FISH assay, based on species-specific probes for C. parvum and C. hominis, can distinguish between the two major species involved in human infections. We have shown here that species-specific FISH is a reliable alternative to PCR and RFLP for rapid identification within a 3-h time frame. In summary, the potential to detect and identify pathogenic Cryptosporidium species in clinical, water, and environmental samples within a 3-h time frame demonstrates that FISH offers an alternative to traditional molecular diagnostic methods that utilize PCR.
Acknowledgments
We thank Ann Cassidy of Douglass Hanly Moir Pathology, Sydney, Australia, and Rogan Lee of ICPMR, Westmead Hospital, Sydney, for supplying Cryptosporidium-positive fecal samples; Wendi Smart from Lethbridge Research Centre, Canada, for supplying C. andersoni samples; Rachel Chalmers from the United Kingdom Cryptosporidium Reference Unit for supplying C. meleagridis, C. felis, Cryptosporidium cervine genotype, and Cryptosporidium rabbit genotype samples; and Michelle Power, Liette Waldron, and Nanda Altavilla for technical support.
This work and Anitha Alagappan were supported by an Environmental Biotechnology CRC scholarship.
Footnotes
Published ahead of print on 24 July 2009.
REFERENCES
- 1.Alagappan, A., N. A. Tujulaa, M. Power, C. M. Ferguson, P. L. Bergquist, and B. C. Ferrari. 2008. Development of fluorescent in situ hybridisation for Cryptosporidium detection reveals zoonotic and anthroponotic transmission of sporadic cryptosporidiosis in Sydney. J. Microbiol. Methods 75:535-539. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., and B. Fuchs. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339-348. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angus, K. W. 1983. Cryptosporidiosis in man, domestic animals and birds: a review. J. R. Soc. Med. 76:62-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angus, K. W. 1990. Cryptosporidiosis in ruminants. CRC Press, Boston, MA.
- 7.Bennett, J. W., and M. R. Gauci. 1999. A comparison of enumeration techniques for Cryptosporidium parvum oocysts. J. Parasitol. 85:1165-1168. [PubMed] [Google Scholar]
- 8.Caccio, S. M. 2005. Molecular epidemiology of human cryptosporidiosis. Parassitologia 47:185-192. [PubMed] [Google Scholar]
- 9.Cacciò, S. M., E. Pinter, R. Fantini, I. Mezzaroma, and E. Pozio. 2002. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caccio, S. M., and E. Pozio. 2006. Advances in the epidemiology, diagnosis and treatment of cryptosporidiosis. Expert Rev. Anti Infect. Ther. 4:429-443. [DOI] [PubMed] [Google Scholar]
- 11.Cama, V. A., C. Bern, I. M. Sulaiman, R. H. Gilman, E. Ticona, A. Vivar, V. Kawai, D. Vargas, L. Zhou, and L. Xiao. 2003. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J. Eukaryot. Microbiol. 50S:531-533. [DOI] [PubMed] [Google Scholar]
- 12.Cama, V. A., R. H. Gilman, A. Vivar, E. Ticona, Y. Ortega, C. Bern, and L. Xiao. 2006. Mixed Cryptosporidium infections and HIV. Emerg. Infect. Dis. 12:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalmers, R., K. Elwin, A. Thomas, and D. Joynson. 2002. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J. Infect. Dis. 185:270-271. [DOI] [PubMed] [Google Scholar]
- 14.Current, W. L. 1985. Cryptosporidiosis. J. Am. Vet. Med. Assoc. 187:1334-1338. [PubMed] [Google Scholar]
- 15.Current, W. L., and L. S. Garcia. 1991. Cryptosporidiosis. Clin. Microbiol. Rev. 4:325-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, C. M., N. Altavilla, M. Krogh, C. M. Ferguson, D. A. Deere, and N. J. Ashbolt. 2005. Environmental inactivation of Cryptosporidium oocysts in catchment soils. J. Appl. Microbiol. 98:308-317. [DOI] [PubMed] [Google Scholar]
- 17.Gatei, W., Y. Suputtamongkol, D. Waywa, R. W. Ashford, J. W. Bailey, J. Greensill, N. J. Beeching, and C. A. Hart. 2002. Zoonotic species of Cryptosporidium are as prevalent as the anthroponotic in HIV-infected patients in Thailand. Ann. Trop. Med. Parasitol. 96:797-802. [DOI] [PubMed] [Google Scholar]
- 18.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 20.O'Donoghue, P. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 21.Vesey, G., N. Ashbolt, G. Wallner, M. Dorsch, K. Williams, and D. Veal. 1995. Assessing Cryptosporidium parvum oocyst viability with fluorescent in-situ hybridisation using ribosomal RNA probes and flow cytometry, p. 133-138. In W. W. Betts, B. Casemore, D. Fricker, C. Smith, and H. Watkins (ed.), Protozoan parasites and water. Royal Society of Chemistry, Cambridge, United Kingdom.
- 22.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. Gilman, and A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 23.Xiao, L., and R. Fayer. 2008. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int. J. Parasitol. 38:1239-1255. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, L., and U. M. Ryan. 2004. Cryptosporidiosis: an update in molecular epidemiology. Curr. Opin. Infect. Dis. 17:483-490. [DOI] [PubMed] [Google Scholar]