Abstract
DNA sequence information has increasingly been used in ecological research on microbial eukaryotes. Sequence-based approaches have included studies of the total diversity of selected ecosystems, studies of the autecology of ecologically relevant species, and identification and enumeration of species of interest for human health. It is still uncommon, however, to delineate protistan species based on their genetic signatures. The reluctance to assign species-level designations based on DNA sequences is in part a consequence of the limited amount of sequence information presently available for many free-living microbial eukaryotes and in part a consequence of the problematic nature of and debate surrounding the microbial species concept. Despite the difficulties inherent in assigning species names to DNA sequences, there is a growing need to attach meaning to the burgeoning amount of sequence information entering the literature, and there is a growing desire to apply this information in ecological studies. We describe a computer-based tool that assigns DNA sequences from environmental databases to operational taxonomic units at approximately species-level distinctions. This approach provides a practical method for ecological studies of microbial eukaryotes (primarily protists) by enabling semiautomated analysis of large numbers of samples spanning great taxonomic breadth. Derivation of the algorithm was based on an analysis of complete small-subunit (18S) rRNA gene sequences and partial gene sequences obtained from the GenBank database for morphologically described protistan species. The program was tested using environmental 18S rRNA data sets for two oceanic ecosystems. A total of 388 operational taxonomic units were observed for 2,207 sequences obtained from samples collected in the western North Atlantic and eastern North Pacific oceans.
Ecological studies of aquatic microbial eukaryotes require identification and enumeration of organisms with extremely wide taxonomic diversity. The assemblages are typically dominated by phototrophic and heterotrophic protists (microalgae and protozoans), but microscopic metazoans belonging to a variety of animal phyla can also contribute significantly. Identification of protists in environmental samples is particularly difficult because most species have been defined morphologically (41, 84). Protistan identification involves a wide variety of procedures for collection, preservation, specimen preparation, and examination (34, 84), as well as many different types of taxonomic expertise. Very few studies have attempted to identify and enumerate all protistan taxa because of these complexities, which makes it difficult to evaluate ecological studies of protistan diversity, community structure, and biogeochemical function.
The growing database of DNA sequence information for a wide spectrum of microbial eukaryotes offers the possibility for greatly improving the existing tools for studying the phylogeny and ecology of these organisms. Much of the initial impetus for the acquisition of rRNA gene sequence information for microbial eukaryotes in the 1980s and 1990s arose from a desire to improve our understanding of the evolutionary relationships among the taxa, especially among the many protistan lineages (66, 73-75). Such research provided significant insights into the evolution of eukaryotic organisms and continues to facilitate the generation, testing, and modification of numerous hypotheses related to this topic (1, 7, 15, 39, 72).
Molecular taxonomy has several real or potential advantages for ecologists compared to traditional taxonomies, including (i) the fact that it can be applied to a wide range of taxa, including those possessing few distinctive morphological features; (ii) applicability to all life stages of a species; (iii) a reduced requirement for formal (i.e., morphological) taxonomic training; (iv) a standardized approach for sample processing, interpretation, and comparison across different studies; (v) the potential for automation of much of the processing of sample characterization; and (vi) the ability to taxonomically characterize the large numbers of samples that are typical of most ecological studies.
DNA sequence information has been used to establish distinctions among protistan species with few or variable morphological features (11, 14, 23, 50, 86), as an aid to characterize lineages of minute protists which largely lack morphological characteristics (3-5), and to identify and study specific protistan taxa in complex natural assemblages using fluorescence in situ hybridization and real-time quantitative PCR (37, 43, 71). This work has helped establish the spatiotemporal distributions of a number of ecologically important species, such as harmful algal species, and/or species that have significance for human health (6, 12, 19, 32, 55, 63, 88).
Genetic approaches have also been extensively used to assess the composition of natural assemblages of protists from a variety of ecosystems. Such studies have reported lists of gene sequences representing a wide array of protistan lineages from freshwater environments, various oceanic ecosystems ranging from polar to tropical, anoxic ecosystems, and deep-sea environments (20-22, 25, 29-31, 36, 44-47, 51, 54, 78, 80). Interpretation of the results of these investigations of protistan community composition and structure could be improved by a clearer understanding of how sequence information translates into taxonomic composition. Moreover, the effectiveness of statistical approaches for comparison of the structures of microbial communities is dependent on an accurate account of the number of taxa in the assemblages (67).
Methods utilizing DNA sequences for deriving microbial operational taxonomic units (OTUs) and subsequently determining species richness from sequence data are now appearing (68, 69). These approaches hold great promise for ecologists because they provide potentially powerful tools for examining community composition. Molecular taxonomy has been received enthusiastically by many workers in the ecological community, but with skepticism by some workers. Proponents have openly campaigned for the development of a DNA taxonomy to augment existing taxonomic schemes for microbes that rely primarily on morphology or physiology (9, 65, 81). Skeptics have noted technical and conceptual problems with the approach and have expressed concern that molecular taxonomies do not necessarily facilitate an understanding of the morphological, physiological, and behavioral characteristics of organisms (27, 64). For ecologists, the optimal situation might involve the use of genetic signatures (to facilitate sample analysis) combined with an understanding of how this information is related to morphology, physiology, and behavior in order to understand the biogeography of functional traits, not just taxonomic entities (35).
There has been very little effort to derive a practical, sequence-based protistan taxonomy for ecological research. The diversity studies to date have used a range of approaches and/or a range of sequence similarity values to create OTUs from eukaryotic sequence libraries, and there has been little consistency or justification of the choice of the values (see Discussion). This inconsistency has caused confusion in interpreting data and comparing data sets in different studies. Resistance by many researchers to infer protistan species identities from sequence information exists in part because the species concept for protists is problematic. Morphological features have traditionally been used for species descriptions, but reproductive and physiological criteria, and more recently DNA sequences, have also been incorporated (53, 56). This combination of disparate characters for defining protistan species has complicated the process of extrapolating the descriptions directly to species definitions based solely on DNA sequences. Regrettably, the complicated taxonomic schemes presently in use for protists are particularly difficult to apply in ecological studies.
The application of DNA sequence to ecological studies cannot await a resolution to the debate over the protistan species concept, if that ever happens (16). A practical method, recognizing the present limitations of this approach, could significantly improve our ability to interpret the large sequence data sets now appearing. The goal of this study was to establish a practical, reproducible approach for the use of DNA sequence information for defining molecular OTUs for ecological studies of microeukaryotic organisms, with a focus primarily on protistan taxa.
We designed and tested a computer program (Microbial Eukaryote Species Assignment [MESA]) to establish species-level OTUs from 18S rRNA sequence information. Sequence data obtained from the GenBank database for a wide variety of taxa were used to design and test this program. The program was then applied to sequence data obtained from environmental samples collected from the western North Atlantic and eastern North Pacific. A total of 388 taxonomic OTUs were derived from the combined sequence libraries containing a total of 2,207 partial 18S rRNA sequences. In this large database, only 54 of 388 of the OTUs were present in both the Atlantic and Pacific sample sets. Rare taxa (OTUs with ≤2 clones) comprised the majority of OTUs.
MATERIALS AND METHODS
The overall logic behind the design and application of the automated program for calling OTUs for protists was as follows. Full-length, 18S rRNA sequences of “well-defined” (i.e., morphologically defined) protistan species were selected from the GenBank database. The strains included multiple strains belonging to a variety of species and strains of multiple species in a number of genera across a wide phylogenetic range. Automated, pairwise alignment of all sequences was performed using ClustalW (83). Intraspecific sequence variability (for multiple strains in a species) and interspecific similarity (for different species in a genus) were analyzed based on the ClustalW alignments. A logical, overall demarcation value (percentage of similarity) for differentiating among the sequences at approximately the species level was determined based on an analysis of the alignments and the GenBank species identifications. The MESA program (available for download at http://www.usc.edu/dept/LAS/biosci/Caron_lab/index.html) was then developed for calling OTUs from 18S rRNA sequences using the percentage of similarity derived as described above. Finally, the MESA program was applied to an environmental sequence database to assess microbial-eukaryote diversity.
Analysis of intra- and interspecies sequence similarity.
The design of the protistan OTU-calling program included use of publicly available sequence information (GenBank) for morphologically defined protistan taxa to establish an appropriate level of sequence similarity for use in the program. Morphologically defined species were employed because the overall purpose was to establish a link between DNA sequence similarity or dissimilarity and species identity based on traditional taxonomic schemes for protists. A wide range of taxa were specifically selected, including taxa with extensive morphological features, as well as taxa whose morphologies are variable or nondescript (e.g., amoebae and minute, nonflagellated algae) and whose ultrastructure, physiology, or behavior have been used to delineate species. Our logic was that the species chosen might represent classifications ranging from those of taxonomic “lumpers” to those of taxonomic “splitters.” Full-length 18S rRNA gene sequences were used because eukaryotic databases now contain sufficient numbers of these sequences to begin to allow meaningful comparisons.
Both intraspecific and interspecific comparisons were conducted to develop a program that would call OTUs with approximately species-level resolution. Seventeen species encompassing a total of 211 sequences were used to examine intraspecific sequence variability. The number of strains in the species varied from 4 to 56 (Table 1). Thirty-one genera were used to examine interspecific sequence variability. The number of species in the genera varied from 3 to 36 (Table 2). Sequence similarity among taxa above the genus level was not examined because the intent was to identify species-level distinctions, and it was assumed that the sequence-to-sequence variability among species belonging to different genera would be greater than the variability between congeners. The species employed in these analyses included amoebae, minute chlorophytes, euglenoids, kinetoplastids, dinoflagellates, ciliates, diplomonads, heterokonts (diatoms, chrysophytes), and prymnesiophytes. No attempt was made to equalize or normalize sample numbers across these diverse species.
TABLE 1.
Seventeen species whose complete 18S rRNA sequences were obtained from the GenBank database and were used to examine intraspecies sequence variability in full-length small-subunit (18S) rRNA genes
| Species | No. of strains | Avg intraspecies similarity (%) |
|---|---|---|
| Acanthamoeba castellanii | 12 | 96.5 |
| Acanthamoeba lenticulata | 12 | 84.9 |
| Alexandrium catenella | 16 | 99.9 |
| Alexandrium tamarense | 37 | 98.2 |
| Chlamydomonas noctigama | 6 | 99.5 |
| Entamoeba histolytica | 4 | 99.0 |
| Euglena gracilis | 6 | 93.1 |
| Euglena mutabilis | 6 | 95.1 |
| Euplotes aediculatus | 4 | 99.9 |
| Euplotes vannus | 4 | 98.7 |
| Giardia intestinalis | 9 | 97.5 |
| Gymnodinium beii | 5 | 99.7 |
| Nannochloropsis gaditana | 10 | 99.9 |
| Phaeocystis globosa | 8 | 99.6 |
| Plasmodium knowlesi | 12 | 97.0 |
| Thalassiosira rotula | 4 | 99.4 |
| Trypanosoma cruzi | 56 | 98.4 |
| Total | 211 | 98.0 |
TABLE 2.
Thrity-one genera employed to examine interspecies sequence variability in full-length small-subunit (18S) rRNA genes
| Genus | No. of species | Avg interspecies similarity (%) |
|---|---|---|
| Acanthamoebaa | 20 | 83.0 |
| Alexandriuma | 13 | 94.6 |
| Amphidinium | 8 | 88.6 |
| Bodo | 7 | 85.2 |
| Chaetoceros | 5 | 93.2 |
| Chlamydomonasa | 26 | 94.2 |
| Chrysochromulina | 8 | 96.0 |
| Cryptomonas | 8 | 82.1 |
| Dinophysis | 5 | 98.5 |
| Entamoebaa | 12 | 76.0 |
| Euglenaa | 29 | 70.4 |
| Euplotesa | 13 | 91.3 |
| Giardiaa | 4 | 89.5 |
| Gymnodiniuma | 6 | 95.3 |
| Gyrodinium | 10 | 95.4 |
| Leishmania | 5 | 99.6 |
| Mallomonas | 9 | 96.3 |
| Nannochloropsisa | 6 | 98.8 |
| Oxytricha | 4 | 94.7 |
| Paramecium | 13 | 93.6 |
| Paraphysomonas | 6 | 91.4 |
| Phaeocystisa | 5 | 97.3 |
| Plasmodiuma | 12 | 86.5 |
| Prorocentrum | 9 | 93.1 |
| Pyramimonas | 6 | 97.4 |
| Scrippsiella | 3 | 98.3 |
| Synura | 6 | 95.9 |
| Tetrahymena | 17 | 98.8 |
| Thalassiosiraa | 8 | 95.0 |
| Tintinnopsis | 4 | 94.4 |
| Trypanosomaa | 36 | 87.8 |
| Total | 323 | 87.0 |
Species in this genus were employed in the intraspecies comparison whose results are shown in Table 1.
Primary-read, full-length 18S rRNA sequences (in FASTA file format) were prepared for pairwise alignment by trimming each sequence (if necessary) at the 5′ and 3′ ends using an automated method that read from the end of the sequence toward the center and removed base sequences that contained more than five questionable (N) residues per 25 bp. This process did not affect the full-length sequences obtained from the GenBank database to test the MESA program, but it was necessary for the environmental sequence databases. Intraspecies sequence variability (strain-to-strain variability within a species) was determined using pairwise alignments of full-length 18S rRNA sequences for the 17 species examined; a total of 2,712 pairwise comparisons were examined, and for n strains in a species, the number of pairwise alignments in each species was n!/[(n − 2)! × 2!)] using ClustalW without additional manual alignment. Aligned sequences were truncated to remove any nonoverlapping sequences at the ends of each gene pair. Gaps were assigned one mismatch for each base pair difference. Pairwise alignments of full-length 18S rRNA sequences for 323 congeneric species distributed in 31 protistan genera were also examined to establish the level of sequence similarity appropriate for distinguishing different species in a genus. A total of 2,439 pairwise alignments of congeners were obtained using ClustalW, as indicated above.
The ClustalW alignments were not manually adjusted because our goal was to develop an approach that would allow comparison of large sequence databases with minimal human assistance rather than to obtain truly phylogenetically informative alignments. Similarity values were calculated from the total number of base pair mismatches for the overlapping fragments of two sequences for every pairwise intra- and interspecies comparison, and similarity matrices were constructed for the two data sets. Average similarity values were determined for each species for all strain-strain comparisons, and then a weighted average for intraspecific sequence similarity across all species was calculated. A similar analysis was performed for the pairwise comparisons for congeneric species to obtain an average interspecies sequence similarity.
The distributions of intra- and interspecific sequence variability were examined visually, and a similarity value was chosen that minimized discrimination among strains within each species but maximized discrimination among species within each genus. This similarity value was used in the design and application of the MESA program (95% sequence similarity) (see Results).
Derivation of the MESA program.
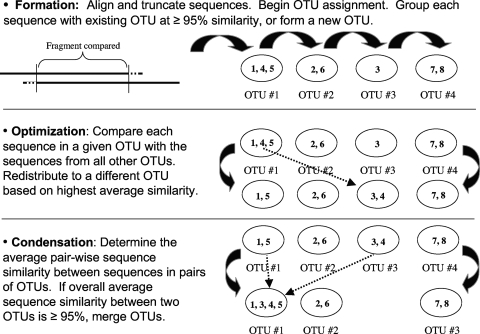
The algorithm for the MESA program is shown in Fig. 1. An initial round of sequence comparisons was conducted to place all sequences into provisional OTUs (formation) (Fig. 1). The first OTU was established by selecting the first sequence in a sequence file. The second sequence was compared to the first OTU sequence using the ClustalW alignment to determine sequence similarity. If the similarity value was ≥95%, then the sequences were placed together in OTU #1. If the similarity was <95%, then the second sequence formed a separate OTU (OTU #2). Each subsequent sequence was then compared to OTU #1. If the sequence similarity of the new sequence with any of the sequences in OTU #1 was <95%, then the new sequence was compared to the sequences in OTU #2 and so forth until each sequence was either placed in an existing OTU or formed a separate OTU.
FIG. 1.
Flow diagram of the MESA algorithm for calling protistan OTUs using full-length 18S rRNA gene sequences.
An optimization step was performed once all sequences had been placed into provisional OTUs in order to determine the best possible placement of each sequence in the OTUs (Fig. 1). The average level of sequence similarity of each sequence to all other sequences in an OTU was determined and compared to the average level of similarity of the same sequence to sequences in all other OTUs. Any sequence that exhibited greater average similarity to the sequences in another OTU was moved to the OTU with which it had greater average similarity.
Finally, a condensation step was conducted to determine whether any two OTUs possessed overall an average similarity that warranted condensation of the two OTUs into a single OTU (Fig. 1). The average sequence similarities for the sequences in two OTUs were compared for every pair of OTUs. If the average similarities were ≥95%, the two OTUs were condensed into a single OTU.
Testing the reliability of the MESA program.
An initial test of the OTU-calling program was conducted using two replicate 18S rRNA clone libraries constructed from a single water sample obtained in the western North Atlantic. The purpose of this exercise was to test how closely OTUs were called from two clone libraries constructed independently from the same water sample. Replication of the cloning and sequencing approach employed for environmental samples was an inherent component of the evaluation.
The water collection, sample processing, cloning, and sequencing protocols used have been described previously (21). Briefly, water was collected using Niskin bottles from the subsurface euphotic zone at a station along the United States continental shelf (36°21′N, 75°14′W), and samples were pooled to create a single sample. The sample was prefiltered through a 200-μm Nitex screen to remove most metazoans and filtered onto a 47-mm glass fiber GF/F filter (Whatman International, Ltd., Florham Park, NJ). DNA was released from cells using 1 ml lysis buffer (100 mM Tris [pH 8], 40 mM EDTA [pH 8], 100 mM NaCl, 1% sodium dodecyl sulfate) at 70°C with bead beating (0.5-mm zircon beads), followed by 1% hexadecyltrimethylammonium bromide (Sigma), and then extracted in phenol-chloroform and precipitated with isopropanol (33).
The resulting DNA was divided into two aliquots, and each aliquot was used in independent PCRs. Full-length 18S rRNA genes were amplified from the genomic DNA extracts using universal eukaryotic primers Euk-A (5′-AACCTGGTTGATCCTGCCAGT-3′) and Euk-B (5′-GATCCTTCTGCAGGTTCACCTAC-3′) (52). Amplicons that were the appropriate length were excised, gel purified, ligated into plasmids using a pGEM-T Easy vector kit (Promega), and used to transform Electro10Blue electrocompetent cells (Stratagene) using procedures described previously (21). DNA sequencing was carried out with a Beckman-Coulter CEQ8000 automated DNA sequencer (Fullerton, CA) used according to the manufacturer's specifications. A single sequencing read was performed using Euk-570F (5′-GTAATTCCAGCTCCAATAGC-3′) (87). The sequences obtained ranged from 400 to 700 bp long. The resulting partial sequences were checked for chimeric sequences using Ribosomal Database Project Chimera Check (http://rdp8.cme.msu.edu/html/), possible chimeric sequences were eliminated, and the remaining sequences were analyzed using pairwise alignments and placed into OTUs using the procedures described above. The lengths of the aligned sequences used for estimation of the similarity values varied because of the variable read lengths.
Application of the MESA program to a large environmental data set.
The ability of the OTU-calling program to handle a large environmental data set was examined by applying it to a database containing 2,207 partial sequences derived from previously published data for samples collected in the North Atlantic (970 sequences) (21) and from a study site in the coastal eastern North Pacific (1,237 sequences). The latter data set was comprised of clone libraries constructed from water samples collected on a single date at depths of 1, 20, 42, 150, 500, and 880 m at the San Pedro Ocean Time Series station located midway between Santa Catalina Island and the United States mainland in the San Pedro Channel (33°33′N, 118°24′W). This location is the site of an ongoing microbial observatory, and a complete analysis of the data set will be presented elsewhere (GenBank accession numbers GQ382277 to GQ383513 [North Pacific] and DQ917930 to DQ919024 [North Atlantic]). Sample collection and processing and DNA extraction, amplification, cloning, and sequencing were conducted as described above previously (18). Seawater used in the study was prefiltered through 200-μm screens, and particulate material was collected on GF/F glass fiber filters (Whatman International Ltd., Florham Park, NJ). Sequencing of the libraries was conducted using Euk-570F (5′-GTAATTCCAGCTCCAATAGC-3′) to provide compatibility with the North Atlantic data. Libraries from individual depths contained 137 to 257 sequences, but sequence information from all depths for the Pacific samples was combined for the present analysis.
The resulting partial sequences from the combined North Atlantic and North Pacific samples were processed as a single data set, placed into OTUs using the MESA program, and then separated according to sampling site. Taxonomic information pertaining to the 50 most abundant OTUs was obtained using BLAST (2) with the NCBI (8) and ARB (48) databases. Searches were conducted using all of the members of each OTU.
RESULTS
Construction and evaluation of the MESA program.
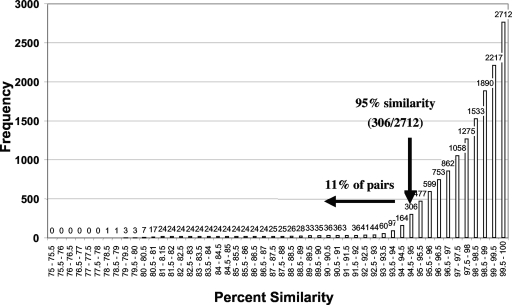
The analysis of intraspecies sequence variability indicated that there was a high level of sequence similarity across the 211 strains in the 17 species examined (Fig. 2). Overall, the sequence similarity between strains of the same species was high, and the average value was 98% similarity for all 2,712 pairwise comparisons (Table 1), although a small percentage of the comparisons yielded relatively low values. A total of 89% of the strain-strain comparisons resulted in placement in the same OTU by the MESA program using a level of sequence similarity of 95%. Most of the 11% of the comparisons that had similarity values less than 95% involved a single amoeba species, Acanthamoeba lenticulata. For this species the overall average value in pairwise comparisons was particularly low (85%) compared with all other species.
FIG. 2.
Cumulative intraspecific sequence similarity for 211 full-length small-subunit rRNA gene sequences distributed among 17 species (see Table 1). The total number of pairwise comparisons was 2,712.
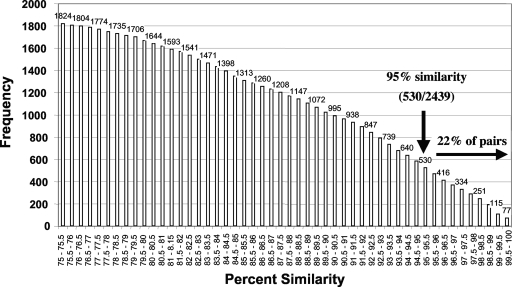
The results of the intrageneric, interspecific comparisons were less decisive than the results of the intraspecific comparisons with respect to a similarity value that clearly demarcated species (Fig. 3). For 78% of the interspecies pairwise alignments the sequence similarity was ≤95%, while 22% of the pairs showed >95% similarity (that is, 22% of the time different species were placed in the same OTU). The overall sequence similarity among species in the same genus was 87% for all 2,439 pairwise comparisons (Table 2). The congeneric species for which the sequence similarity values were highest were species in the genera Tetrahymena, Leishmania, and Nannochloropsis.
FIG. 3.
Cumulative intrageneric, interspecific sequence similarity for 323 full-length small-subunit rRNA gene sequences distributed among 31 genera (see Table 2). The total number of pairwise comparisons was 2,439.
The efficacy of the MESA program was also examined using partial sequences (approximately 600 bp of the 18S rRNA gene, beginning at 570F) of the species and strains employed in the full-length sequence analysis described above. This analysis was conducted to determine if the program could produce results with partial sequences that were similar to the results obtained with full-length sequences. Many of the sequences in the GenBank database that were generated from environmental 18S rRNA clone libraries are partial sequences obtained using 570F or a nearby primer for sequencing (20, 25, 42, 44, 47, 89). The results of the analysis using partial sequences and the results of the analysis using full-length sequences were virtually identical. The overall weighted average for intraspecies similarity for the partial 18S rRNA sequences was the same as that for the full-length sequences (98%). The interspecies comparison yielded a value of 90% for all 2,439 pairwise alignments when partial sequences were used, compared to a value of 87% when the full-length sequences were used.
Based on the analyses described above, a sequence similarity value of 95% was chosen for use in the MESA program to provide approximately species-level distinctions among 18S rRNA sequences of protists. This value represented a compromise between identifying multiple strains of a single species as a single OTU on the one hand and separating congeneric species into separate OTUs on the other hand.
Analysis of replicate clone libraries.
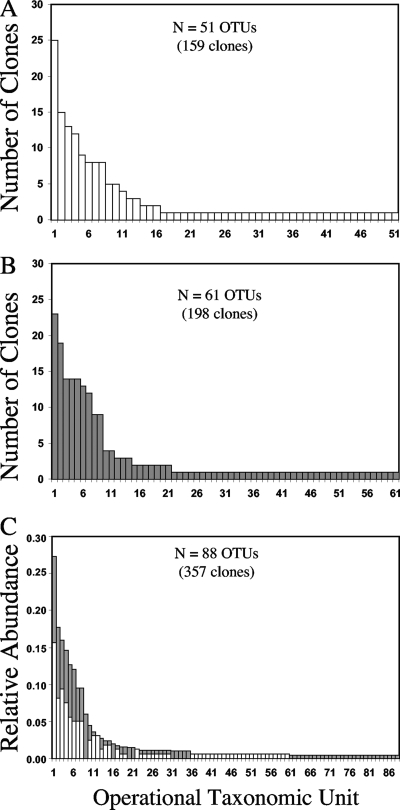
A total of 357 partial sequences were obtained for the two replicate clone libraries from the North Atlantic sample. The libraries were combined for OTU calling and then separated for comparison. Use of these sequences with the MESA program yielded 51 and 61 OTUs for the two libraries (Fig. 4). The general shapes of the rank abundance curves for each library were similar. Twenty-four of the OTUs were observed in both clone libraries, while 64 OTUs were unique to one of the libraries. The 24 OTUs observed in both libraries were among the most abundant OTUs in the combined data set. That is, the MESA program yielded “common” taxa that were observed in both libraries at quite similar relative abundances, with a few exceptions (Fig. 4C). The presence of many “rare” OTUs (OTUs represented by a single sequence) that were unique to one of the libraries was not surprising given the relatively low number of clones that were sequenced for each library and the potentially large numbers of the sequence types in natural samples.
FIG. 4.
OTU calling for replicate clone libraries. The clone libraries (A and B) were established using DNA subsamples taken from the same water sample collected from the North Atlantic. The libraries were constructed independently, but the sequences were combined into a single data set for OTU calling (C). Note the different axes for panels A and B and panel C. The OTU rank order differs from panel to panel, and the overlap of common OTUs is shown in panel C.
Analysis of North Atlantic and North Pacific environmental clone libraries.
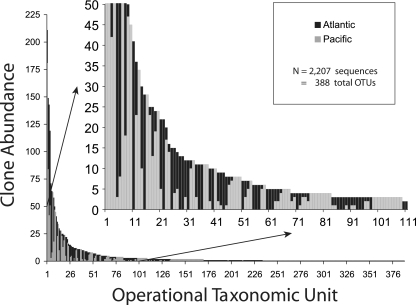
A total of 2,207 partial sequences from the combined North Atlantic and North Pacific clone libraries were analyzed using the MESA program (Fig. 5). These sequences yielded a total of 388 OTUs using a sequence similarity value of ≥95%. The rank abundance curve for these OTUs revealed that a relatively small number of OTUs (18% of the total) were composed of five or more sequences, while a large number of OTUs contained only one or two sequences.
FIG. 5.
OTUs established for sequences obtained from combined environmental clone libraries constructed for samples collected from the western North Atlantic and eastern North Pacific.
Most of the OTUs in the combined data set were present in libraries obtained at one of the sites but not at both sites (Table 3). Only 54 of the OTUs (14%) were present in clone libraries constructed using samples from both study sites. A great diversity of taxonomic groups was represented in the overall data set. Of the 50 most abundant OTUs in the combined data sets, 12 were metazoans (mostly copepods), while 38 returned best sequence matches that identified them as protistan taxa (Table 4). A substantial number of the latter sequences (17 of 38) had the closest phylogenetic affinity with unclassified alveolate taxa. Approximately one-half of the 50 most abundant OTUs were present in the data set for either the North Atlantic site or the North Pacific site but not in the data sets for both sites.
TABLE 3.
Microbial eukaryote OTU distribution in the Pacific and Atlantic clone libraries, including the numbers of OTUs that were unique to either the Pacific or Atlantic library and the numbers of OTUs that were present in both librariesa
| Supergroup | OTU category | No. of OTUs
|
|||
|---|---|---|---|---|---|
| Total | Unique to Pacific database | Unique to Atlantic database | In both databases | ||
| “Rhizaria” | Polycystinean | 11 | 11 | 0 | 0 |
| Acantharean | 9 | 9 | 0 | 0 | |
| Sticholonchid | 6 | 6 | 0 | 0 | |
| Cercozoan | 5 | 2 | 3 | 0 | |
| “Chromalveolata” | Stramenopile | 51 | 22 | 21 | 8 |
| Ciliate | 38 | 11 | 16 | 11 | |
| Dinoflagellate | 26 | 16 | 7 | 3 | |
| Apicomplexan | 1 | 0 | 1 | 0 | |
| Haptophyte | 4 | 2 | 1 | 1 | |
| Cryptophyte | 2 | 0 | 1 | 1 | |
| Group I alveolate | 21 | 19 | 1 | 1 | |
| Group II alveolate | 48 | 30 | 11 | 7 | |
| Unclassified alveolate | 76 | 51 | 15 | 10 | |
| Perkinsean | 1 | 1 | 0 | 0 | |
| “Plantae” | Chlorophyte | 11 | 4 | 5 | 2 |
| Rhodophyte | 3 | 1 | 2 | 0 | |
| Streptophyte | 2 | 2 | 0 | 0 | |
| “Excavata” | Euglenozoan | 15 | 12 | 3 | 0 |
| “Opisthokonta” | Arthropod | 27 | 10 | 12 | 5 |
| Cnidarian | 5 | 2 | 2 | 1 | |
| Ctenophore | 3 | 2 | 0 | 1 | |
| Echinodermate | 1 | 0 | 1 | 0 | |
| Urochordate | 4 | 2 | 1 | 1 | |
| Choanoflagellate | 5 | 4 | 1 | 0 | |
| Fungi | 5 | 3 | 0 | 2 | |
| Unclassified metazoan | 1 | 0 | 1 | 0 | |
| Unresolved lineages | Cryothecomonad | 1 | 0 | 1 | 0 |
| Ichthyosporean | 1 | 1 | 0 | 0 | |
| Unknown | Unclassified eukaryote | 5 | 5 | 0 | 0 |
| Total | 388 | 228 | 106 | 54 | |
The OTUs are organized as described by Teckle et al. (82).
TABLE 4.
Taxonomic groups of the most abundant OTUs in the North Atlantic and North Pacific data set
| Rank | Taxonomic group |
|---|---|
| 1 | Arthropod (Opisthokonta) |
| 2 | Dinoflagellate (Chromalveolata) |
| 3 | Cnidarian (Opisthokonta) |
| 4 | Ciliate (Chromalveolata) |
| 5 | Ciliate (Chromalveolata) |
| 6 | Group II alveolate (Chromalveolata) |
| 7 | Group II alveolate (Chromalveolata) |
| 8 | Arthropod (Opisthokonta) |
| 9 | Ctenophore (Opisthokonta) |
| 10 | Acantharean (Rhizaria) |
| 11 | Group II alveolate (Chromalveolata) |
| 12 | Arthropod (Opisthokonta) |
| 13 | Polycystinean (Rhizaria) |
| 14 | Unclassified alveolate (Chromalveolata) |
| 15 | Ciliate (Chromalveolata) |
| 16 | Arthropod (Opisthokonta) |
| 17 | Unclassified alveolate (Chromalveolata) |
| 18 | Unclassified alveolate (Chromalveolata) |
| 19 | Group I alveolate (Chromalveolata) |
| 20 | Chlorophyte (Plantae) |
| 21 | Arthropod (Opisthokonta) |
| 22 | Unclassified alveolate (Chromalveolata) |
| 23 | Chlorophyte (Plantae) |
| 24 | Haptophyte (Chromalveolata) |
| 25 | Arthropod (Opisthokonta) |
| 26 | Ciliate (Chromalveolata) |
| 27 | Chlorophyte (Plantae) |
| 28 | Arthropod (Opisthokonta) |
| 29 | Ciliate (Chromalveolata) |
| 30 | Stramenopile (Chromalveolata) |
| 31 | Dinoflagellate (Chromalveolata) |
| 32 | Group II alveolate (Chromalveolata) |
| 33 | Group II alveolate (Chromalveolata) |
| 34 | Group II alveolate (Chromalveolata) |
| 35 | Stramenopile (Chromalveolata) |
| 36 | Ciliate (Chromalveolata) |
| 37 | Unclassified alveolate (Chromalveolata) |
| 38 | Group I alveolate (Chromalveolata) |
| 39 | Urochordate (Opisthokonta) |
| 40 | Ciliate (Chromalveolata) |
| 41 | Group I alveolate (Chromalveolata) |
| 42 | Arthropod (Opisthokonta) |
| 43 | Ciliate (Chromalveolata) |
| 44 | Arthropod (Opisthokonta) |
| 45 | Stramenopile (Chromalveolata) |
| 46 | Unclassified alveolate (Chromalveolata) |
| 47 | Ciliate (Chromalveolata) |
| 48 | Unclassified alveolate (Chromalveolata) |
| 49 | Sticholonchid (Rhizaria) |
| 50 | Group I alveolate (Chromalveolata) |
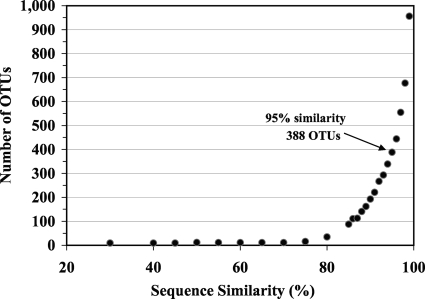
The effect of the similarity value employed in the MESA program on the number of OTUs estimated for the environmental data set was examined by processing the 2,207 sequences using a range of similarity values (Fig. 6). The value used dramatically affected the number of OTUs constructed by the program, particularly for the range of similarity values that have been generally employed in protistan molecular diversity studies. For example, increasing the similarity value from 95% to 99% resulted in a 2.5-fold increase in the number of OTUs for the sequences in the combined data set (from 388 to 956 OTUs).
FIG. 6.
Effect of the sequence similarity value used on the number of OTUs estimated from sequences obtained from combined environmental clone libraries collected in the western North Atlantic and eastern North Pacific.
DISCUSSION
Toward a DNA taxonomy.
The development of a DNA taxonomy for microbial eukaryotes would provide a much-needed tool for ecological studies of natural microbial communities, but the impediments to this goal include both technical and conceptual problems. The technical problems include potential artifacts related to DNA extraction and amplification, cloning, sequencing, and sequence manipulation. These problems will undoubtedly lessen at the present pace of biotechnological and computational progress. Conceptual issues are more problematic, as is the choice of the gene(s) used in the taxonomies. For example, some species possess multiple RNA gene copies with somewhat different base pair compositions (70). These different sequences could conceivably produce multiple OTUs for a single specimen if the differences are large enough, although such instances appear to be relatively rare. Similarly, the use of rapidly evolving genes or intergenic spacer regions might result in the creation of multiple OTUs for individuals that would be grouped into a single species using other criteria (60).
The algorithm and specific levels of similarity described here were developed using 18S rRNA gene sequences for a broad range of taxa. We chose the 18S rRNA gene because a substantial amount of information is available for this gene in public databases. However, the heterogeneous rates of evolution that have been noted for this gene (13) may make it less useful for some taxonomic groups. Another gene might prove to be more useful for those taxa, and this approach can easily be adapted as the databases for other genes expand. The use of ecologically relevant genes as the basis for molecular taxonomy might aid reconciliation of molecular and traditional taxonomic schemes. Indeed, we anticipate that future protistan molecular taxonomies may involve the use of specific genes for specific taxa or the use of multiple genes, much the way that multiple gene phylogenies are presently employed to obtain integrated perspectives on the evolutionary history of microbial taxa (24, 38, 49, 57). The MESA program described here has not been applied yet in this way, but it provides a conceptual template for such adaptations.
More significantly, the debate regarding what constitutes a protistan species makes reconciliation of traditional and molecular taxonomies difficult. The morphological species concept that dominates protistan taxonomy has been challenged by some investigators and considered inadequate because it sometimes fails to differentiate physiologically or sexually distinct entities with identical or nearly identical morphotypes (10, 17, 58, 65). Taxonomists continue to debate the integration of the morphological species concept, the biological species concept, and the ecological species concept for describing protistan species and their global distributions (16). In this debate, the decision by protistan ecologists to incorporate DNA analysis as one more observational and experimental tool is a pragmatic one, as is the conceptual approach presented here. It is impossible to formulate and test hypotheses concerning the environmental factors determining the distributions of ecologically and commercially important taxonomic entities without rapid, reliable means to determine the presence and abundances of these entities. Thus, new tools are required to allow large-scale studies of the ecology of these taxa that would not be possible using cumbersome, time-consuming, and often inaccurate morphology-based taxonomies.
Establishing a protistan OTU-calling program.
Current approaches for establishing OTUs for microbial taxa are typically based on evolutionary distances (68). These approaches have the potential to construct a truly phylogeny-based taxonomy, but they are difficult to apply in ecological research because the computations typically require manual adjustments to multiple-sequence alignments to improve automated alignments provided by programs such as ClustalW (83). Unfortunately, this is counterproductive when workers are dealing with the potentially thousands of sequences that are often required in ecological studies. Such a study would require an enormous amount of preparatory work and considerable training, slowing the processing time for a data set. This situation may improve in the future as algorithms for sequence alignment improve (28, 62). Given the present state of the programs, however, a specific objective of this study was to develop a program that could be applied in a “hands-off” fashion to facilitate rapid processing of the large data sets characteristic of ecological studies.
Our objective when we developed the MESA program was to establish practical guidelines for establishing protistan OTUs. The protistan MESA program does not provide a phylogeny-based taxonomy, nor did we attempt to resolve the controversial and difficult issue of the “species concept” for protistan taxa. We merely used protistan species whose identities were determined by traditional methods to provide information for setting demarcations between taxonomic units to obtain approximately species-level distinctions for use in ecological research. Therefore, consecutively called OTUs do not necessarily have a close phylogenetic relationship, because the manner in which the program handles gaps and variable regions is not necessarily appropriate for phylogenetic analysis. The information obtained in the analysis of intraspecies and interspecies variability (Fig. 2 and 3) assisted in selecting the level of sequence similarity used in the present analysis. The similarity value can be altered to permit more or less stringent formation of OTUs. Once an OTU is formed, the sequences in it can be analyzed by using BLAST to determine the closest phylogenetic affiliation.
The three-step process by which the program creates protistan OTUs includes initial assignment, optimization of the placement of sequences in OTUs following the initial establishment, and finally a test for condensation of some of the OTUs that exhibit strong similarity (Fig. 1). The final step is an attempt to prevent the creation of artificial “microdiversity” by generating OTUs that have few distinctions from one another. The latter process was particularly important for OTUs with large numbers of sequences but generally did not affect the existence of OTUs composed of only one or two sequences. The condensation step was also affected somewhat by the size of the database (i.e., the number of sequences compared); that is, as more sequences are added to a database, the potential for some level of condensation increases. For example, the North Atlantic data set yielded 165 OTUs when it was analyzed alone but 160 OTUs when it was analyzed in combination with the North Pacific data set. Based on these characteristics of the program, we believe that it provides a conservative estimate of the microbial-eukaryote taxa in a sample.
Choosing a similarity value for demarcating OTUs.
The absolute number of OTUs obtained from the large environmental clone libraries examined in this study was critically dependent on the sequence similarity value employed to distinguish among taxonomic units (Fig. 6). We chose a similarity value (95%) that was lower than the values that have been employed in most studies (97 to 99%). However, it is important to remember that our similarities result from automated, pairwise alignments without manual adjustment for hypervariable regions of the 18S rRNA gene. Manual alignment would have undoubtedly increased the levels of similarity between many pairs of sequences. As noted above, the omission of a manual alignment step was a conscious decision that allowed a “hands-off” procedure for processing very large data sets, such as the one shown in Fig. 5. This is a highly desirable approach for ecological investigations because of the large number of sequences that typically are processed in these studies, and it should facilitate direct comparisons between different investigations.
A similarity value of 95% was chosen purposefully as a conservative estimator of species richness in a natural sample. This decidedly conservative choice probably masked considerable physiological diversity in some OTUs. This conclusion is supported by the observation that congeneric species were placed in a single OTU in 22% of the 2,439 pairwise comparisons, and the congeners with the highest sequence similarity values were members of the genera Tetrahymena, Leishmania, and Nannochloropsis. Interestingly, these three genera include species whose taxonomic descriptions represent deviations from purely morphological descriptions. Mating type compatibility has been employed to separate morphologically indistinguishable species of Tetrahymena (17, 58), and DNA sequence information has been used to differentiate between Leishmania strains that vary in etiology (85, 86). The genus Nannochloropsis contains minute algae for which few morphological features are visible at the level of light microscopy and for which physiological and biochemical characteristics have been employed to supplement morphological features. It is not surprising, therefore, that a comparison of 18S rRNA sequences for species in these three genera might not be consistent with similarity values obtained for other protistan species. These genera exemplify the present state of confusion regarding the species concept for protists. Some researchers might consider the distinctions mentioned above strain-strain variability or “ecotypes” within morphospecies, while other researchers would confer species status (16). Nonetheless, refinement of the approach described here, specifically the use of taxon-specific similarity values, could bring the outcome of the MESA program more in line with the accepted taxonomic distinctions for various protistan lineages. We anticipate that future iterations of the MESA program might involve an initial basic grouping based on one level of sequence similarity and then apply a different level of similarity (or use a different gene) that is more informative for each group.
The overall average similarity value for intraspecies pairwise comparisons using the protistan sequences retrieved from the GenBank database in this study was 98% for all strains belonging to the 17 species examined (Fig. 2 and Table 1). Analysis of only A. lenticulata yielded a substantially lower average similarity value for strain-strain comparisons (85%), and the next lowest value observed was the value for Euglena gracilis (93%). Acanthamoebae are notoriously difficult to identify by morphological criteria, and A. lenticulata contains distinct clinical and genetic “types” that may represent cryptic species. Therefore, a similarity value of 98% might be more appropriate for species-level distinctions among most protists. For the data examined in the present analysis using the MESA program, the use of a similarity value of 98% for calling OTUs resulted in a 1.7-fold-greater number of OTUs, while the use of a value of 99% resulted in a 2.5-fold-greater number of OTUs (Fig. 6).
The use of a similarity value greater than 95% for the interspecies analysis carried out with full-length 18S rRNA sequences from the GenBank database resulted in better agreement between the number of OTUs called by the program and the number of congeneric species retrieved from the GenBank database (Fig. 3). However, the use of a similarity value greater than 95% in the MESA program also rapidly increased the frequency of placing strains of a single species into multiple OTUs in the intraspecies comparisons (Fig. 2). A value of 95% was chosen in the present study because it was a relatively conservative value for demarcating protistan OTUs in environmental sequence databases. Adjustment of the threshold value can be easily accommodated in the program, and the use of a range of similarity values may provide interesting insights into the microbial-eukaryote diversity present in a sample.
The molecular diversity studies of natural protistan assemblages conducted to date have employed a variety of methods and a variety of sequence similarity values for calculating the number of OTUs in 18S rRNA clone libraries. One approach has been to generate restriction fragment length polymorphism patterns from the full-length 18S rRNA genes in a clone library and then group the clones into taxonomic units based on these patterns (25, 42). This method has been employed to reduce the number of clones that need to be sequenced. A more common approach has been to partially sequence and align a large number of clones and use a specific sequence similarity value to group sequences into OTUs. Similarity values ranging from 95 to 98% have generally been employed (20, 21, 59, 77, 79). Worden (89) examined OTU calling with four different sequence similarity values (range, ≥96 to 99%), Doherty et al. (26) used a range of 88 to 99%, and Jeon et al. (40) examined the number of OTUs with a wide range of similarity values (50 to 99%). We are aware of no analysis of the type that we conducted here to provide a rationale for the specific value employed. The justifications for the values used in other studies have often been omitted, but the implication has been that they approximate species-level distinctions. The level of discrimination seems to be vaguely linked to empirical observations that small-subunit (16S) rRNA gene sequences of bacterial species differ by values on the order of 1 to 2% for well-aligned sequences. Our analysis is the first analysis of 18S rRNA sequences for species of protists that have been identified by traditional approaches.
Calling OTUs for the North Atlantic and North Pacific clone libraries.
The application of the MESA program to a large environmental clone library allowed automated processing of 2,207 sequences in the data set. The rank abundance curve generated from the data set is indicative of the results of the program (Fig. 5). The generation of the matrix of pairwise alignments consumed the majority of the processing time and was, of course, highly dependent on the total number of sequences and the processor speed. Calling OTUs required comparatively little time. The program yielded 388 OTUs for the combined Atlantic and Pacific sequence databases at approximately the level of morphospecies. A significant number of the OTUs most closely matched metazoan taxa in the BLAST analysis, particularly copepods (Arthropoda). Prescreening samples through 200-μm Nitex mesh did not remove these species. This result indicates that future iterations of the MESA program for 18S rRNA environmental libraries must take into account appropriate demarcations for metazoan taxa as well as protists if the approach is to have general applicability for ecological studies of microbial eukyarotes.
The shape of the rank abundance curve of OTUs generated by the program indicated the presence of a very large number of “rare” OTUs (OTUs comprised of one or two clones) in the combined data set (Fig. 5). A relatively low percentage of OTUs (14%; 54 of 388) were observed at both study sites. It is not uncommon in comparisons of environmental clone libraries from different locales that “rare” taxa constitute the majority of the OTUs and that the rare taxa tend to be different at different locales (61). This finding may indicate that there is endemism of protistan species, but it is important to note that approximately one-half of the 50 most common phylotypes were observed at both oceanic sites in the limited databases generated in the present study. The presence of different rare taxa in the North Atlantic and North Pacific samples may simply indicate that there is very high local species richness in microbial communities and that severe undersampling at a given site cannot accurately reveal the presence of rare taxa. In addition, differences in environmental conditions and sampling depths presumably resulted in differences in relative abundance among the taxa at the two sites. It has been reported that minor changes in environmental conditions during bottle incubations resulted in rapid changes in the protistan assemblage at the North Atlantic site (21). The inability of other molecular diversity studies to attain sampling saturation supports this conjecture (20, 21, 47, 76, 90).
Finally, it is noteworthy that the use of a sequence similarity value of 95% in the MESA program generated 388 unique OTUs for 2,207 sequences. The use of a higher value resulted in substantially more unique OTUs. The overall conclusion from this finding is that if protist taxonomists generally accept the incorporation of physiological and behavioral data into the present morphological species concept employed for protistan taxa, then the estimates of the species richness of natural protistan assemblages could be dramatically higher than those obtained when a similarity value of 95% is used. Molecular taxonomy holds the most promise for ecologists dealing with the staggering diversity of forms and functions of these organisms.
Acknowledgments
We gratefully acknowledge the assistance of the captains and crews of the R/V Endeavor and R/V Seawatch for technical assistance with sample collection. We thank D. Beaudoin, J. M. Rose, R. Schaffner, and M. Travao for assistance with collection and processing of the samples from the North Atlantic and North Pacific and K. B. Heidelberg for helpful comments on the manuscript.
Support for this study was provided by National Science Foundation grants MCB-0732066, MCB-0703159, and OCE-0550829 and by a grant from the Gordon and Betty Moore Foundation.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, R. A., and J. C. Bailey. 2002. Phylogenetic analysis of 32 strains of Vaucheria (Xanthophyceae) using the rbcL gene and its two flanking spacer regions. J. Phycol. 38:583-592. [Google Scholar]
- 4.Andersen, R. A., R. W. Brett, D. Potter, and J. P. Sexton. 1998. Phylogeny of the Eustigmatophyceae based upon 18S rDNA, with emphasis on Nannochloropsis. Protist 149:61-74. [DOI] [PubMed] [Google Scholar]
- 5.Andersen, R. A., Y. Van de Peer, D. Potter, J. P. Sexton, M. Kawachi, and T. LaJeunesse. 1999. Phylogenetic analysis of the SSU rRNA from members of the Chrysophyceae. Protist 150:71-84. [DOI] [PubMed] [Google Scholar]
- 6.Audemard, C., K. S. Reece, and E. M. Burreson. 2004. Real-time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl. Environ. Microbiol. 70:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 8.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank update. Nucleic Acids Res. 32:D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaxter, M. L. 2004. The promise of a DNA taxonomy. Philos. Trans. R. Soc. Lond. B 359:669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boenigk, J., S. Jost, T. Stoeck, and T. Garstecki. 2007. Differential thermal adaptation of clonal strains of a protist morphospecies originating from different climatic zones. Environ. Microbiol. 9:593-602. [DOI] [PubMed] [Google Scholar]
- 11.Boenigk, J., K. Pfandl, P. Stadler, and A. Chatzinotas. 2005. High diversity of the ‘Spumella-like’ flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 12.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinkmann, H., M. van der Giezen, and Y. Zhou. 2005. An empirical assessment of long-branch attraction artifacts in deep eukaryotic phylogenomics. Syst. Biol. 54:743-757. [DOI] [PubMed] [Google Scholar]
- 14.Brown, S., and J. F. De Jonckheere. 1999. A reevaluation of the amoeba genus Vahlkampfia based on SSU rDNA sequences. Eur. J. Protistol. 35:49-54. [Google Scholar]
- 15.Burki, F., K. Shalchian-Tabrizi, Å. Skjaeveland, S. I. Nikolaev, K. S. Jakobsen, and J. Pawlowski. 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 8:E790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caron, D. A. 2009. Protistan biogeography: why all the fuss? J. Eukaryot. Microbiol. 56:105-112. [DOI] [PubMed] [Google Scholar]
- 17.Coleman, A. W. 2002. Microbial eukaryote species. Science 297:337. [DOI] [PubMed] [Google Scholar]
- 18.Countway, P. D. 2005. Molecular ecology of marine protistan assemblages. University of Southern California, Los Angeles.
- 19.Countway, P. D., and D. A. Caron. 2006. Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl. Environ. Microbiol. 72:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Countway, P. D., R. J. Gast, M. R. Dennett, P. Savai, J. M. Rose, and D. A. Caron. 2007. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western N. Atlantic (Sargasso Sea and Gulf Stream). Environ. Microbiol. 9:1219-1232. [DOI] [PubMed] [Google Scholar]
- 21.Countway, P. D., R. J. Gast, P. Savai, and D. A. Caron. 2005. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukaryot. Microbiol. 52:95-106. [DOI] [PubMed] [Google Scholar]
- 22.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Jonckheere, J. F. 2004. Molecular definition and the ubiquity of species of the genus Naegleria. Protist 155:89-103. [DOI] [PubMed] [Google Scholar]
- 24.Dewhirst, F. E., Z. Shen, M. S. Scimeca, L. N. Stokes, T. Boumenna, T. C. Chen, B. J. Paster, and J. G. Fox. 2005. Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. Appl. Environ. Microbiol. 187:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diez, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty, M., B. A. Costas, G. B. McManus, and L. A. Katz. 2007. Culture independent assessment of planktonic ciliate diversity in coastal northwest Atlantic waters. Aquat. Microb. Ecol. 48:141-154. [Google Scholar]
- 27.Ebach, M. C., and C. Holdrege. 2005. More taxonomy, not DNA barcoding. BioScience 55:822-823. [Google Scholar]
- 28.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgcomb, V. P., D. T. Kysela, A. Teske, A. D. Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fawley, M. J., K. P. Fawley, and M. A. Buchheim. 2004. Molecular diversity among communities of freshwater microchlorophytes. Microb. Ecol. 48:489-499. [DOI] [PubMed] [Google Scholar]
- 31.Fawley, M. W., K. P. Fawley, and H. A. Owen. 2005. Diversity and ecology of small coccoid green algae from Lake Itasca, Minnesota, USA, including Meyerella planktonica, gen. et sp. nov. Phycologia 44:35-48. [Google Scholar]
- 32.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garces, and M. Magnani. 2004. Development of real-time PCR assay for rapid detection and quantification of Alexandrium minutum (dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gast, R. J., M. R. Dennett, and D. A. Caron. 2004. Characterization of protistan assemblages in the Ross Sea, Antarctica, by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2028-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gifford, D. J., and D. A. Caron. 1999. Sampling, preservation, enumeration and biomass of marine protozooplankton, p. 193-221. In ICES zooplankton methodology manual. Academic Press, London, United Kingdom.
- 35.Green, J. L., B. J. M. Bohannan, and R. J. Whitaker. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039-1043. [DOI] [PubMed] [Google Scholar]
- 36.Guillou, L., W. Eikrem, M. J. Chretiennot-Dinet, F. Le Gall, R. Massana, K. Romari, C. Pedros-Alio, and D. Vaulot. 2004. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 155:193-214. [DOI] [PubMed] [Google Scholar]
- 37.Handy, S. M., K. J. Coyne, K. J. Portune, E. Demir, M. A. Doblin, C. E. Hare, S. C. Cary, and D. A. Hutchins. 2005. Evaluating vertical migration behavior of harmful raphidophytes in the Delaware inland bays utilizing quantitative PCR. Aquat. Microb. Ecol. 40:121-132. [Google Scholar]
- 38.Harper, J. T., E. Waandersm, and P. J. Keeling. 2005. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 55:487-496. [DOI] [PubMed] [Google Scholar]
- 39.Hoppenrath, M., and B. S. Leander. 2006. Ebriid phylogeny and the expansion of the Cercozoa. Protist 157:279-290. [DOI] [PubMed] [Google Scholar]
- 40.Jeon, S.-O., J. Bunge, T. Stoeck, K. J.-A. Barger, S.-H. Hong, and S. S. Epstein. 2006. Synthetic statistical approach reveals a high degree of richness of microbial eukaryotes in an anoxic water column. Appl. Environ. Microbiol. 72:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, J. J., G. F. Leedale, and P. Bradbury. 2000. An illustrated guide to the protozoa. Allen Press, Inc., Lawrence, KS.
- 42.Lefranc, M., A. Thénot, C. Lepere, and D. Debroas. 2005. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim, E. L. 1996. Molecular identification of nanoplanktonic protists based on small subunit ribosomal RNA gene sequences for ecological studies. J. Eukaryot. Microbiol. 43:101-106. [DOI] [PubMed] [Google Scholar]
- 44.López-García, P., A. Vereshchaka, and D. Moreira. 2007. Eukaryotic diversity associated with carbonates and fluid-seawater interface in Lost City hydrothermal field. Environ. Microbiol. 9:546-554. [DOI] [PubMed] [Google Scholar]
- 45.López-García, P., H. Philippe, F. Gail, and D. Moreira. 2003. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 100:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López-García, P., F. Rodríguez-Valera, C. Pedrós-Alió, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 47.Lovejoy, C., R. Massana, and C. Pedros-Alio. 2006. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Appl. Environ. Microbiol. 72:3085-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez-Murcia, A. J., L. Soler, M. J. Saavedra, M. R. Chacón, J. Guarro, E. Stackebrandt, and M. J. Figueras. 2005. Phenotypic, genotypic, and phylogenetic discrepancies to differentiate Aeromonas salmonicida from Aeromonas bestiarum. Int. Microbiol. 8:259-269. [PubMed] [Google Scholar]
- 50.Maslov, D. A., S. J. Westenberger, X. Xu, D. A. Campbell, and N. R. Sturm. 2007. Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J. Eukaryot. Microbiol. 54:57-65. [DOI] [PubMed] [Google Scholar]
- 51.Massana, R., L. Guillou, B. Diez, and C. Pedros-Alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 53.Modeo, L., G. Petroni, G. Rosati, and D. J. S. Montagnes. 2003. A multidisciplinary approach to describe protists: redescriptions of Novistrombidium testaceum and Strombidium inclinatum Montagnes, Taylor and Lynn 1990 (Ciliophora, Oligotrichia). J. Eukaryot. Microbiol. 50:175-189. [DOI] [PubMed] [Google Scholar]
- 54.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 55.Moorthi, S. D., P. D. Countway, B. A. Stauffer, and D. A. Caron. 2006. Use of quantitative real-time PCR to investigate the dynamics of the red tide dinoflagellate Lingulodinium polyedrum. Microb. Ecol. 52:136-150. [DOI] [PubMed] [Google Scholar]
- 56.Moran, D. M., O. R. Anderson, M. R. Dennett, D. A. Caron, and R. J. Gast. 2007. A description of seven Antarctic marine Gymnamoebae including a new species and a new genus: Platyamoeba contorta n. sp. and Vermistella antarctica n. gen. n. sp. J. Eukaryot. Microbiol. 54:169-183. [DOI] [PubMed] [Google Scholar]
- 57.Moreira, D., S. von der Heyden, D. Bass, P. López-Garcia, E. Chao, and T. Cavalier-Smith. 2007. Global eukaryote phylogeny: combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol. Phylogenet. Evol. 44:255-266. [DOI] [PubMed] [Google Scholar]
- 58.Nanney, D. L. 1999. When is a rose?: the kinds of tetrahymenas, p. 93-118. In R. W. Wilson (ed.), Species: new interdisciplinary essays. MIT Press, Cambridge, MA.
- 59.O'Brien, H. E., J. L. Parrent, J. A. Jackson, J.-M. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Mahony, E. M., W. T. Tay, and R. J. Paxton. 2007. Multiple rRNA variants in a single spore of the microsporidian Nosema bombi. J. Eukaryot. Microbiol. 54:103-109. [DOI] [PubMed] [Google Scholar]
- 61.Pedrós-Alió, C. 2006. Microbial diversity: can it be determined? Trends Microbiol. 14:257-263. [DOI] [PubMed] [Google Scholar]
- 62.Poirot, O., E. O'Toole, and C. Notredame. 2003. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31:3503-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Popels, L. C., S. C. Cary, D. A. Hutchines, R. Forbes, F. Pustizzi, C. J. Gobler, and K. J. Coyne. 2003. The use of quantitative polymerase chain reaction for the detection and enumeration of the harmful alga Aureococcus anophagefferens in environmental samples along the United States East Coast. Limnol. Oceanogr. 48:92-102. [Google Scholar]
- 64.Rubinoff, D., S. Cameron, and K. Will. 2006. Genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J. Hered. 97:581-594. [DOI] [PubMed] [Google Scholar]
- 65.Scheckenbach, F., C. Wylezich, A. P. Mylnikov, M. Weitere, and H. Arndt. 2006. Molecular comparisons of freshwater and marine isolates of the same morphospecies of heterotrophic flagellates. Appl. Environ. Microbiol. 72:6638-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlegel, M. 1994. Molecular phylogeny of eukaryotes. Trends Ecol. Evol. 9:330-335. [DOI] [PubMed] [Google Scholar]
- 67.Schloss, P. D. 2008. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2:265-275. [DOI] [PubMed] [Google Scholar]
- 68.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scholin, C. A., D. M. Anderson, and M. L. Sogin. 1993. Two distinct small-subunit ribosomal RNA genes in the North American toxic dinoflagellate Alexandrium fundyense (Dinophyceae). J. Phycol. 29:209-216. [Google Scholar]
- 71.Scholin, C. A., K. R. Buck, T. Britschgi, G. Cangelosi, and F. P. Chavez. 1996. Identification of Pseudo-nitzschia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia 35:190-197. [Google Scholar]
- 72.Simpson, A. G. B., and A. J. Roger. 2004. The real ‘kingdoms’ of eukaryotes. Curr. Biol. 14:R693-R696. [DOI] [PubMed] [Google Scholar]
- 73.Sogin, M. L. 1991. Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1:457-463. [DOI] [PubMed] [Google Scholar]
- 74.Sogin, M. L. 1989. Evolution of eukaryotic microorganisms and their small subunit ribosomal RNAs. Am. Zool. 29:487-499. [Google Scholar]
- 75.Sogin, M. L., H. J. Elwood, and J. H. Gunderson. 1986. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc. Natl. Acad. Sci. USA 83:1383-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stoeck, T., B. Hayward, G. T. Taylor, R. Varela, and S. S. Epstein. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31-43. [DOI] [PubMed] [Google Scholar]
- 78.Stoeck, T., G. T. Taylor, and S. S. Epstein. 2003. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stoeck, T., A. Zuendorf, A. Behnke, and H.-W. Breiner. 2007. A molecular approach to identify active microbes in environmental eukaryote clone libraries. Microb. Ecol. 53:328-339. [DOI] [PubMed] [Google Scholar]
- 80.Takishita, K., H. Miyake, M. Kawato, and T. Maruyama. 2005. Genetic diversity of microbial eukaryotes in anoxic sediment around fumaroles on a submarine caldera floor based on the small-subunit rDNA phylogeny. Extremophiles 9:185-196. [DOI] [PubMed] [Google Scholar]
- 81.Tautz, D., P. Arctander, A. Minelli, R. H. Thomas, and A. P. Vogler. 2003. A plea for DNA taxonomy. Trends Ecol. Evol. 18:70-74. [Google Scholar]
- 82.Tekle, Y. I., L. Wegener Parfrey, and L. A. Katz. 2009. Molecular data are transforming hypotheses on the origin and diversification of eukaryotes. BioScience 59:471-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tomas, C. R. 1997. Identifying marine phytoplankton. Academic Press, San Diego, CA.
- 85.Uliana, S. R. B., M. H. T. Affonso, E. P. Camargo, and L. M. Floeter-Winter. 1991. Leishmania: genus identification based on a specific sequence of the 18S ribosomal RNA sequence. Exp. Parasitol. 72:157-163. [DOI] [PubMed] [Google Scholar]
- 86.Uliana, S. R. B., K. Nelson, S. M. Beverley, E. P. Camargo, and L. M. Floeter-Winter. 1994. Discrimination amongst Leishmania by polymerase chain reaction and hybridization with small subunit ribosomal DNA derived oligonucleotides. J. Eukaryot. Microbiol. 41:324-401. [DOI] [PubMed] [Google Scholar]
- 87.Weekers, P. H. H., R. J. Gast, P. A. Fuerst, and T. J. Byers. 1994. Sequence variations in small-subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol. 11:684-690. [DOI] [PubMed] [Google Scholar]
- 88.Whipps, C. M., and M. L. Kent. 2006. Phylogeography of the cosmopolitan marine parasite Kudoa thyrsites (Myxozoa: Myxosporea). J. Eukaryot. Microbiol. 53:364-373. [DOI] [PubMed] [Google Scholar]
- 89.Worden, A. Z. 2006. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat. Microb. Ecol. 43:165-175. [Google Scholar]
- 90.Zuendorf, A., J. Bunge, A. Behnke, K. J.-A. Barger, and T. Stoeck. 2006. Diversity estimates of microbial eukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol. Ecol. 58:476-491. [DOI] [PubMed] [Google Scholar]