Abstract
A novel gene encoding transglycosylating β-galactosidase (BGase) was cloned from Penicillium expansum F3. The sequence contained a 3,036-bp open reading frame encoding a 1,011-amino-acid protein. This gene was subsequently expressed on the cell surface of Saccharomyces cerevisiae EBY-100 by galactose induction. The BGase-anchored yeast could directly utilize lactose to produce galactooligosaccharide (GOS), as well as the by-products glucose and a small quantity of galactose. The glucose was consumed by the yeast, and the galactose was used for BGase expression, thus greatly facilitating GOS synthesis. The GOS yield reached 43.64% when the recombinant yeast was cultivated in yeast nitrogen base-Casamino Acids medium containing 100 g/liter initial lactose at 25°C for 5 days. The yeast cells were harvested and recycled for the next batch of GOS synthesis. During sequential operations, both oligosaccharide synthesis and BGase expression were maintained at high levels with GOS yields of over 40%, and approximately 8 U/ml of BGase was detected in each batch.
Galactooligosaccharides (GOS) are beneficial for human health as prebiotics that maintain the balance of normal flora in the intestine, enhance lactose tolerance and the digestibility of milk products, reduce serum cholesterol levels, increase Ca2+ absorption, synthesize B-complex vitamins, and reduce the risk of cancer (15, 18). Recently, a great deal of attention has been devoted to GOS synthesis, especially via enzymatic transglycosylation, since chemical synthesis of GOS is very tedious (16). GOS can be synthesized by β-galactosidase (BGase) from lactose by glycosyl transfer of one or more galactosyl units onto a galactose moiety of lactose or other structurally related galactosides (10). Both free and immobilized BGases from different microorganisms have been employed for GOS synthesis (12). Based on previous studies, using free enzymes has been associated with limitations, such as low stability and nonreusability of the enzymes. Using immobilized enzymes could overcome these problems, but there are still some drawbacks, including low recovery rates of enzyme activity, the gradual loss of enzyme during the reaction process, finite immobilized carriers, and large mass transfer resistance between some immobilized enzymes and substrates.
Recently, an alternative strategy to conventional enzyme immobilization was proposed in which the enzyme is anchored on the cell surfaces of engineered microorganisms, such as Escherichia coli and Saccharomyces cerevisiae (8, 13). S. cerevisiae is a highly advantageous host for cell surface display, as it may allow the accurate folding and glycosylation of recombinant proteins. It is generally regarded as safe in its applications in different fields. Yeast cell surface engineering has been demonstrated using the α-agglutinin receptor of S. cerevisiae to display foreign proteins on the cell surface. There were certain advantages to using cell surface-engineered yeast as an immobilized biocatalyst, e.g., the enzyme was anchored covalently on the cell surface without enzyme loss or additional treatments for immobilization, and the mass transfer resistance between the enzyme and the substrate was sharply reduced in contrast to conventional immobilization methods (17). Several studies have successfully used engineered yeast with immobilized target enzymes as a biocatalyst for a single use, such as the cell surface engineering of a β-glucosidase from Aspergillus oryzae for isoflavone aglycone production and a chitosanase from Paenibacillus fukuinensis for chitooligosaccharide production (7, 19). However, there have been no reports of the use of engineered yeast for consecutive batch production without loss of enzyme activity during the reaction process.
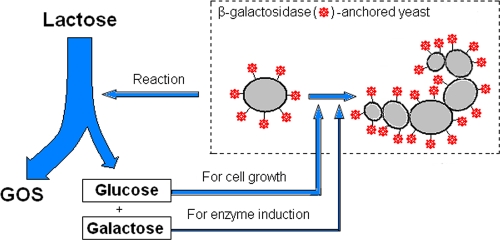
The objective of this work was to present a novel approach for GOS synthesis by anchoring BGase from Penicillium expansum F3 on the cell surface of S. cerevisiae as an immobilized enzyme. Figure 1 shows the main principle of this strategy. The BGase that was cell surface engineered and anchored to yeast (BGase-anchored yeast) could directly utilize lactose for GOS synthesis in batches without loss of enzyme activity. The carbon source (glucose) for cell growth and the inducer (galactose) for enzyme production were the by-products of lactose. The yield of GOS was greatly increased because of the removal of glucose and the continuous expression of BGase. The results showed that this method was especially suitable for GOS synthesis, and it has great promise for industrial oligosaccharide production in the future.
FIG. 1.
Schematic of GOS synthesis by BGase-anchored yeast. The BGase-anchored yeast is represented by modified ovals. The surface BGase converted lactose into GOS, glucose, and a small quantity of galactose. The undesirable glucose was consumed by the yeast for cell growth, and the galactose induced the continuous expression of BGase. Thus, BGase-anchored yeast cells were grown and successively utilized lactose to produce GOS. After a batch reaction, BGase-anchored yeast with higher BGase activity could be harvested and recycled for another batch of GOS synthesis under the same cultivation conditions as the first batch.
MATERIALS AND METHODS
Strains and media.
P. expansum F3 was isolated from soil and was cultivated at 28°C in medium containing 10 g/liter glucose, 10 g/liter peptone, 10 g/liter yeast extract, and 3 g/liter NaCl. E. coli DH5α and S. cerevisiae EBY-100 were cultivated according to the protocols in the pYD1 Yeast Display Vector Kit (Invitrogen; 2002).
Plasmids, enzymes, and reagents.
Plasmid pYD1, the enzymes, and the reagents for RNA manipulation were purchased from Invitrogen. Enzymes for DNA manipulation and the pMD18-TA cloning kit were obtained from TaKaRa (Japan). o-Nitrophenyl-β-d-galactoside was purchased from Sigma.
cDNA cloning.
Total RNA was isolated by the Trizol method (Invitrogen). The first strand of cDNA was synthesized using the First-Strand cDNA Synthesis kit and an oligo(dT) primer (Invitrogen). All the PCR primers are listed in Table 1. A conserved cDNA fragment (bgaF3c) from P. expansum F3 BGase was amplified by PCR using the degenerate primers D1 and D2, which were designed on the basis of nucleotide sequences encoding the BGases. PCR was performed using TaKaRa LA Taq polymerase, and successive reactions were repeated for 30 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s. The upstream fragment of bgaF3c was amplified by PCR using the gene-specific reverse primer G1 and a degenerate forward primer, D3, from the N-terminal sequence of the wide-type enzyme (ELLQKYVTWD). The full-length cDNA fragment was obtained using a 3′/5′-Full RACE Core Set following the manufacturer's protocol (Invitrogen). The primers used in the 3′ rapid amplification of cDNA ends (RACE) reaction were an oligo(dT)-3′ site adaptor primer (3AP), a 3′ site adaptor primer (AUAP), and a bgaF3 gene-specific primer (3F). The primers used for the 5′ RACE reaction were a reverse transcription primer (5R1), the first PCR primers (5R2 and 5AP), and nested PCR primers (5R3 and AUAP). The PCR products were cloned into the pMD18-T vector and sequenced. Sequence analysis and multiple alignments were performed using the BLAST Tool (http://www.ncbi.nlm.nih.gov/BLAST/) and the ClustalW program (http://www.ebi.ac.uk/clustalw/).
TABLE 1.
Primers used in gene cloning
| Primer | DNA sequence (5′→3′) |
|---|---|
| Oligo(dT) | TTTTTTTTTTTTTTTTTT |
| D1 | AACGARGGTGGWCTGTACGC |
| D2 | GGARGTAGGWCCKATGTTG |
| 5R1 | CGACGTAGAAAGACACGCAGTTGAATCCCAATG |
| 5R2 | TAGCAAGTAGATTCCAGCCTCTTTTGCCGC |
| 5R3 | ATCCAGGGAAACCACCGCCGGAAACTTCGG |
| 5AP | GGCCACGCGTCGACTAGTACCCCCCCCCCCCCCCCC |
| AUAP | GGCCACGCGTCGACTAGTAC |
| D3 | GGAAMCTGYTGCAGAAATAYGTCACTTGGGA |
| G1 | GATAGAAGATCAAGGTCAAAGCTGGCACTGTAGAAG |
| 3AP | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT |
| 3F | CACCAGCCCAAGCCATCCACCAAG |
| Pf | CATGAATTCGGAGTTGCTTCAAAAATATGTGACTTGGGa |
| Pr | GCATGCGGCCGCGTACGCCCCCTTTCGAGACb |
The EcoRI site is underlined.
The NotI site is underlined.
Expression of BGase.
The cDNA encoding the mature BGase was amplified by PCR using the primers Pf and Pr (Table 1). The PCR products were digested with the restriction enzymes EcoRI and NotI and purified with a Gel Extract Kit (BioFlux, Japan). The purified fragment was inserted into plasmid pYD1, which had been digested with the same restriction enzymes. The identity of the recombinant plasmid was confirmed by restriction enzyme digestion and gel electrophoresis. The recombinant plasmid (pYD1/bgaF3) was then transformed into S. cerevisiae EBY-100 competent cells by electroporation.
The transformants were grown in yeast nitrogen base (YNB)-Casamino Acids (CAA)-glucose medium containing 6.7 g/liter YNB without amino acids, 5 g/liter CAA, and 20 g/liter glucose at 30°C. When the cell density reached 2.0 at 600 nm, the cells were harvested and cultivated at 25°C in YNB-CAA-galactose medium (the same composition as YNB-CAA-glucose medium except for the presence of galactose instead of glucose for the induction of BGase expression).
Immunofluorescence microscopy.
Immunofluorescence labeling was performed as reported previously, with slight modifications (11). Cells were incubated with the primary antibody against V5 (a small epitope presented on the P and V proteins of paramyxovirus simian virus 5) at a dilution of 1:3,000 at room temperature for 30 min. After the cells were washed three times with phosphate-buffered saline (10 mM; pH 7.5), the second antibody, fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G at a dilution of 1:500, was added to the cells at room temperature for 1 h. After being washed, the cells were observed under a fluorescence microscope.
BGase assay.
BGase activity was measured by adding 450 μl of 2 mM o-nitrophenyl-β-d-galactoside to yeast cells that were harvested from a 200-μl culture. The reaction was performed at 50°C for 10 min and then stopped by adding 1 ml of 500 mM Na2CO3. The amount of o-nitrophenol released was measured with a spectrophotometer at 400 nm. One unit of enzyme activity was defined as the amount of enzyme required to liberate 1 μM of o-nitrophenol per minute under the assay conditions.
GOS synthesis by BGase-anchored yeast.
GOS was produced by cultivating the BGase-anchored yeast in YNB-CAA-lactose medium, which has the same composition as YNB-CAA-glucose medium except for the presence of lactose instead of glucose. The cell density of the BGase-anchored yeast was adjusted to 8.00 at 600 nm, which contained 2.10 U/ml BGase. The effects of initial lactose concentrations (50, 100, 150, 200, and 250 g/liter) on GOS synthesis were investigated at 25°C. Samples from the culture broth (1 ml) were periodically withdrawn to analyze the GOS yield. The yield of GOS was monitored by high-performance liquid chromatography as described in our previous study (9). After one batch synthesis of GOS, the culture broth remaining in the flask was centrifuged (3 min at 3,000 × g and 4°C) to separate the GOS solution from the BGase-anchored yeast cells. The biomass and BGase activity of the yeast were assayed. The yeast cells were used for the next batch of GOS synthesis under the same conditions as the first batch.
Nucleotide sequence accession number.
The nucleotide sequence containing the BGase open reading frame was submitted to GenBank with accession no. EU543998.
RESULTS
Cloning the BGase gene and sequence analysis.
A 285-bp conserved cDNA fragment (bgaF3c) from the BGase gene (bgaF3) was amplified with degenerate primers. Sequences downstream from bgaF3c (708 bp) were amplified by 3′ RACE. Sequences upstream from bgaF3c were amplified with primers G1 and D3 (2,587 bp), as well as 5′ RACE (388 bp). Assembly of the four fragments produced a 3,464-bp full-length cDNA containing a 3,036-bp open reading frame encoding a protein of 1,011 amino acids.
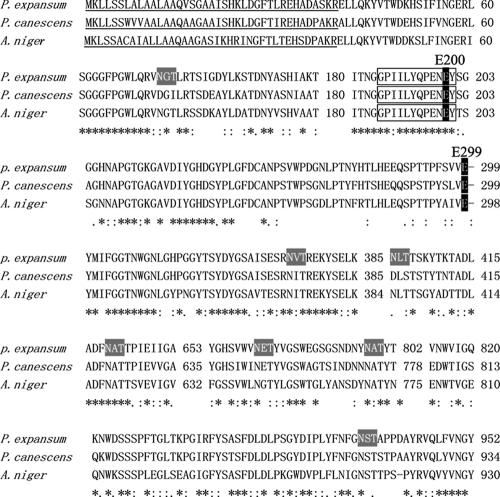
The putative protein was scanned with NCBI BLAST. The results showed that BGase belongs to glycoside hydrolase family 35 based on amino acid sequence similarity. BGase also belongs to glycoside hydrolase superfamily 42 based on hydrophobic cluster analysis (5). The P. expansum F3 protein was homologous to other fungal BGases. It had 84% identity with the BGase from Penicillium canescens, 71% identity with the BGase from Aspergillus niger CBS513, 70% with the BGase from Aspergillus phoenicis, 69% with the BGase from Aspergillus fumigatus Af293, 66% with the BGase from Talaromyces emersonii, and 36% identity with the BGase from Neosartorya fischeri NRRL181 (GenBank accession no. CAA49852, CAK44114, AAY21925, EAL90749, AAL32052, and EAW22192, respectively). As shown in Fig. 2, the aligned BGases possess catalytic residues corresponding to known Penicillium sp. amino acids essential for enzyme activity (E200 and E299) (14). Also, they contain a 39-amino-acid signal peptide and seven potential N-glycosylation sites (a conserved NXS/T motif, where X is not proline).
FIG. 2.
Alignment of the amino acid sequences of BGases. The active sites of P. expansum F3 BGase are thought to be E200 and E299, which are shaded in black. The conserved GHF35 sequence is boxed, and the putative N-glycosylation sites are shaded in gray. Asterisks, colons, and periods indicate invariant residues, similar amino acids, and less-similar amino acids in the aligned sequences, respectively. The signal peptide is underlined. All of the aligned sequences are available in GenBank.
Expression of bgaF3 on the cell surface of yeast.
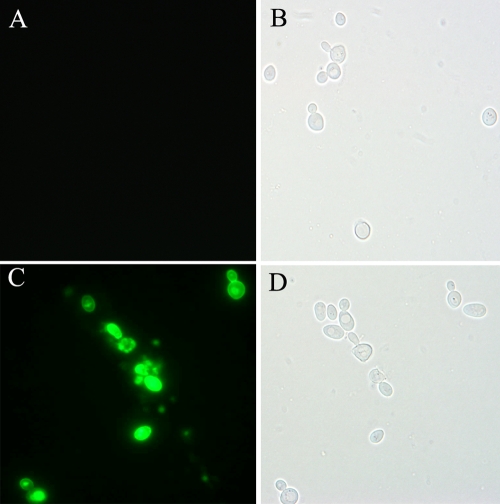
The bgaF3 gene was cloned into the pYD1 vector and subsequently expressed in S. cerevisiae EBY-100. As shown in Fig. 3, the cells containing the recombinant pYD1/bgaF3 vector were clearly labeled with green fluorescence. This confirmed that the BGase was successfully displayed on the cell surface of the yeast. The intensity of fluorescence varied from cell to cell, probably due to the different levels of expression in individual cells.
FIG. 3.
Confirmation of BGase expression on the yeast surface. (A and B) S. cerevisiae EBY-100 with pYD1 as a control cell. (C and D) S. cerevisiae EBY-100 with pYD1/bgaF3. (A and C) Fluorescence micrographs. (C and D) White-light micrographs.
The time course for BGase expression was determined with a spectrophotometer, with cell density measured at 600 nm and BGase activity at 400 nm. The cell density and BGase activity peaked at 40 h, with a maximum activity of 2.16 U/ml. The yeast cells with anchored BGase on the surface were harvested, and the effects of temperature on the enzyme activity were investigated. BGase activity increased with increasing temperature up to 70°C. Since the optimal temperature for protein expression on the cell surface was between 20 and 25°C, according to the protocol for yeast surface display, the BGase-anchored yeast was cultivated at 25°C in subsequent experiments.
Recycling BGase-anchored yeast for repeated batch synthesis of GOS.
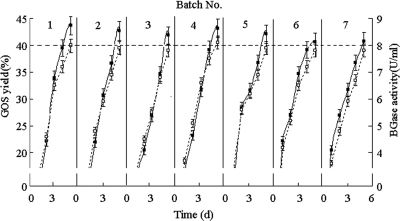
BGase-anchored yeast cells were resuspended and recultivated in YNB-CAA-lactose medium for repeated synthesis of GOS. The initial cell density was adjusted to an absorbance of 8.00 at 600 nm, and this had 2.10 U/ml of BGase activity. The effects of the initial lactose concentration were observed by following the time course of GOS synthesis. As shown in Table 2, the maximum yield of GOS (43.64%) was achieved with an initial lactose concentration of 100 g/liter for 5 days. The culture broth contained 54.99% lactose, 1.41% galactose, and no glucose. At that time, the cell density reached an absorbance of 40.52 at 600 nm and BGase activity increased to 8.08 U/ml. The BGase-anchored yeast was harvested and recycled for the next batch culture in YNB-CAA-lactose medium under the same conditions as the first batch. GOS synthesis and BGase expression were maintained at high levels during seven sequential batch cultures. Figure 4 shows GOS yields of more than 40%, and approximately 8 U/ml of BGase was present in each round of synthesis.
TABLE 2.
GOS yields at different initial lactose concentrations
| Time (days) | GOS yield (%) at initial lactose concn (g/liter) ofa:
|
||||
|---|---|---|---|---|---|
| 50 | 100 | 150 | 200 | 250 | |
| 1 | 16.23 | 12.87 | 10.12 | 9.58 | 7.53 |
| 2 | 23.89 | 22.10 | 18.22 | 16.75 | 14.19 |
| 3 | 29.86 | 33.83 | 24.35 | 21.36 | 17.85 |
| 4 | 34.21 | 39.52 | 27.98 | 26.32 | 19.24 |
| 5 | 36.72 | 43.64 | 30.43 | 31.43 | 23.72 |
| 6 | 32.01 | 39.51 | 29.64 | 30.90 | 22.64 |
Standard deviations were ≤2%.
FIG. 4.
GOS yield (▪) and BGase activity (□) in each detected batch (from the first to the seventh batch).
DISCUSSION
Biocatalysts in the form of immobilized enzymes have several advantages over soluble enzyme preparations, including easier separation of reaction products from the incubation mixture, stabilization of the tertiary structure of the enzymes, and enhancement of the enzyme stability and operational lifetime (4, 12). During immobilization, the activity of an enzyme can be affected by many factors, such as the immobilization method and the size, shape, and composition of the carrier material. For practical applications, the success of enzyme immobilization depends on the properties of the carriers employed. The carrier material must be insoluble in water, must bind tightly to the enzyme, must have mechanical stability, and must not have deleterious effects on the activity of the enzyme. Various modified supports for covalent immobilization have been developed, including polymers, silica, and glass. However, there are limitations to each of these carriers, and the immobilization yield, which is defined as the ratio of the activity of the immobilized enzyme to the activity of the free enzyme, remains low at 22 to 83% (3). In our study, a novel BGase was anchored on the surfaces of yeast cells and used as an immobilized enzyme. The BGase-anchored yeast functioned as an immobilized biocatalyst and achieved one-step enzyme immobilization without any immobilized materials or extra treatments. It not only retained 100% of the initial enzyme activity, but the BGase activity of the yeast was also increased by lactose utilization during the process of GOS synthesis. The enzyme activity increased from 2.10 U/ml to 8.08 U/ml with yeast proliferation (from 8.00 to 40.52 at 600 nm). BGase activity was maintained on the surface of the yeast through several rounds of batch cultivation, and with each round there was a similar increase in enzyme activity as the cells proliferated.
The use of BGase-anchored yeast had other advantages for GOS synthesis. According to some studies, GOS synthesis by BGases is a dynamic equilibrium-controlled process (10). Thus, most attempts to improve GOS yields have focused on controlling the kinetics of the transglycosylation reaction using high concentrations of lactose, high temperature, or low water concentrations (6). To reduce the amount of water, oligosaccharide synthesis was performed in organic solvents, which shifted the reaction equilibrium to GOS synthesis (2). However, most enzymes have lower activity in organic solvents than in aqueous solution, and the solubility of lactose was also reduced when water was limited. High temperatures may increase GOS hydrolysis and reduce enzyme activity. The accumulation of large amounts of by-products (glucose and galactose) in the reaction mixture also inhibits transglycosylation and GOS synthesis. In one study, the yield of GOS improved with a yeast fermentation method in which glucose was consumed by cell growth (Sterigmatomyces elviae cells producing BGase), but most of these yeast cells were wasted because there was not sufficient production of enzyme for batch synthesis. In this study, the engineered yeast functioned as an immobilized biocatalyst that directly utilized lactose for GOS synthesis, as well as fermenting glucose to remove by-products. It also continued expressing BGase through galactose induction and thus greatly improved the yield of GOS.
The maximum yield of GOS from the BGase-anchored yeast reached 43.64%, which was much higher than the yield from wild-type immobilized enzyme in calcium alginate (28.7%) (9). Our GOS yield was also higher than yields produced by other immobilization methods, such as the 40% yield obtained with Bacillus circulans BGase immobilized on ion-exchange resins and silica gel or the 34.8% yield produced by Thermus aquaticus BGase entrapped in agarose beads (1). In addition, the GOS yield remained at high levels even after seven rounds of batch synthesis (40.80%), with 92% of the initial yield (43.64%). This is considerably higher than the 24.6% GOS yield from wild-type immobilized enzyme that had 85% of the initial yield (28.7%) (9). In previous reports, the yield of GOS after seven batches was less than 90% of the initial yield (1, 20). This suggested that the BGase-anchored yeast system was very effective and provided stable GOS synthesis.
Our results can be understood based on the working principle of BGase-anchored yeast shown in Fig. 1. The reaction process was regulated similarly to enzymatic feedback regulation. The BGase on the surface of the yeast used lactose to form GOS, as well as glucose and a small quantity of galactose. The glucose was consumed by the yeast, which contributed to its proliferation, while the galactose in the medium increased the expression of BGase. Thus, the BGase-anchored yeast both proliferated and had enhanced BGase expression while it utilized successive batches of lactose to synthesize GOS. After each batch synthesis, BGase-anchored yeast had much higher BGase activity and was recycled for GOS synthesis. This strategy simultaneously achieved immobilization of enzyme, cultivation of yeast, expression of BGase on the cell surface, and removal of the glucose that would inhibit GOS synthesis. There are several advantages for GOS synthesis through BGase-anchored yeast: (i) BGase-anchored yeast as an immobilized enzyme possessed reusability, safety, simplicity, efficiency, and high productivity; (ii) BGase was sufficiently replenished on the surface of the yeast during GOS synthesis, which alleviated the problem of low recovery of enzyme activity that occurs in conventional immobilization, as well as the gradual loss of recycled enzymes; (iii) the removal of glucose during yeast growth not only facilitated GOS synthesis, but also simplified the purification of GOS; (iv) the BGase-anchored yeast maintained high productivity after a single batch reaction and could be recycled for multiple batches of GOS synthesis.
In conclusion, a novel BGase from P. expansum F3 was successfully expressed on the cell surface of S. cerevisiae. This surface-engineered yeast functioned as an immobilized biocatalyst and provided a new platform for GOS synthesis. Using this method, GOS synthesis was effective, stable, and automatically regulated. The yield of GOS remained at high levels (over 40% in each batch). This could decrease the industrial production costs of GOS. In addition, the value of the approach presented here extends far beyond GOS synthesis using the BGase from P. expansum. It is likely other BGases could be anchored on the surface of yeast using cell surface display technology. Moreover, the engineered yeast or other hosts with immobilized BGase could also be used to synthesize galactose-containing chemicals, which play important roles in many biological processes.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (no. 2006AA10Z338).
We thank Roberta Greenwood for help in editing the manuscript.
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Albayrak, N., and S. T. Yang. 2002. Production of galactooligosaccharides from lactose by Aspergillus oryzae beta-galactosidase immobilized on cotton cloth. Biotechnol. Bioeng. 77:8-19. [DOI] [PubMed] [Google Scholar]
- 2.del Val, M. I., and C. Otero. 2003. Biphasic aqueous media containing polyethylene glycol for the enzymatic synthesis of oligosaccharides from lactose. Enzyme Microb. Technol. 33:118-126. [Google Scholar]
- 3.Giacomini, C., G. Irazoqui, P. Gonzalez, F. Batista-Viera, and B. M. Brena. 2002. Enzymatic synthesis of galactosyl-xylose by Aspergillus oryzae β-galactosidase. J. Mol. Catal. B 19:159-165. [Google Scholar]
- 4.Haider, T., and Q. Husain. 2007. Calcium alginate entrapped preparations of Aspergillus oryzae β galactosidase: its stability and applications in the hydrolysis of lactose. Int. J. Biol. Macromol. 41:72-80. [DOI] [PubMed] [Google Scholar]
- 5.Henrissat, B., and A. Bairoch. 1996. Updating the sequence based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki, K., M. Nakajima, and S. Nakao. 1996. Galacto-oligosaccharide production from lactose by enzymic batch reaction using β-galactosidase. Process Biochem. 31:69-76. [Google Scholar]
- 7.Kaya, M., J. Ito, A. Kotaka, K. Matsumura, H. Bando, H. Sahara, C. Ogino, S. Shibasaki, K. Kuroda, M. Ueda, A. Kondo, and Y. Hata. 2008. Isoflavone aglycones production from isoflavone glycosides by display of β-glucosidase from Aspergillus oryzae on yeast cell surface. Appl. Microbiol. Biotechnol. 79:51-60. [DOI] [PubMed] [Google Scholar]
- 8.Lee, S. Y., J. H. Choi, and Z. Xu. 2003. Microbial cell-surface display. Trends Biotechnol. 21:45-52. [DOI] [PubMed] [Google Scholar]
- 9.Li, Z., M. Xiao, L. Lu, and Y. Li. 2008. Production of non-monosaccharide and high-purity galactooligosaccharides by immobilized enzyme catalysis and fermentation with immobilized yeast cells. Process Biochem. 43:896-899. [Google Scholar]
- 10.Mahoney, R. R. 1998. Galactosyl-oligosaccharide formation during lactose hydrolysis: a review. Food Chem. 63:147-154. [Google Scholar]
- 11.Murai, T., M. Ueda, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Imanaka, and A. Tanaka. 1999. Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface. Appl. Microbiol. Biotechnol. 51:65-70. [DOI] [PubMed] [Google Scholar]
- 12.Neri, D. F. M., V. M. Balcão, R. S. Costa, I. C. A. P. Rocha, E. M. F. C. Ferreira, D. P. M. Torres, L. R. M. Rodrigues, L. B. Carvalho, Jr., and J. A. Teixeira. 2008. Galacto-oligosaccharides production during lactose hydrolysis by free Aspergillus oryzae β-galactosidase and immobilized on magnetic polysiloxane-polyvinyl alcohol. Food Chem. doi: 10.1016/j.200811.068. [DOI]
- 13.Onishi, N., A. Yamashiro, and K. Yokozeki. 1995. Production of galactooligosaccharide from lactose by Sterigmatomyces elviae CBS8119. Appl. Environ. Microbiol. 61:4022-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojas, A. L., R. A. P. Nagem, and K. N. Neustroev. 2004. Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. J. Mol. Biol. 343:1281-1292. [DOI] [PubMed] [Google Scholar]
- 15.Sako, T., K. Matsumoto, and R. Tanaka. 1999. Recent progress on research and applications of non-digestible galactooligosaccharides. Int. Dairy J. 9:69-80. [Google Scholar]
- 16.Sears, P., and C. H. Wong. 2001. Toward automated synthesis of oligosaccharides and glycoproteins. Science 291:2344-2350. [DOI] [PubMed] [Google Scholar]
- 17.Shibasaki, S., M. Ueda, T. Iizuka, M. Hirayama, Y. Ikeda, N. Kamasawa, M. Osumi, and A. Tanaka. 2001. Quantitative evaluation of the enhanced green fluorescent protein displayed on the cell surface of Saccharomyces cerevisiae by fluorometric and confocal laser scanning microscopic analyses. Appl. Microbiol. Biotechnol. 55:471-475. [DOI] [PubMed] [Google Scholar]
- 18.Tuohy, K. M., G. C. M. Rouzaud, W. M. Brück, and G. R. Gibson. 2005. Modulation of the human gut flora towards improved health using prebiotics—assessment of efficacy. Curr. Pharm. Des. 11:75-90. [DOI] [PubMed] [Google Scholar]
- 19.Wang, Z., Q. Qi, and P. G. Wang. 2006. Engineering of cyclodextrin glucanotransferase on the cell surface of Saccharomyces cerevisiae for improved cyclodextrin production. Appl. Environ. Microbiol. 72:1873-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, P., H. Yu, Z. Sun, Y. Ni, W. Zhang, Y. Fan, and Y. Xu. 2006. Production of galactooligosaccharides by immobilized recombinant beta-galactosidase from Aspergillus candidus. Biotechnol. J. 12:1464-1470. [DOI] [PubMed] [Google Scholar]