Abstract
Kinetic analyses of bacterial growth, carbohydrate consumption, and metabolite production of five butyrate-producing clostridial cluster XIVa colon bacteria grown on acetate plus fructose, oligofructose, inulin, or lactate were performed. A gas chromatography method was set up to assess H2 and CO2 production online and to ensure complete coverage of all metabolites produced. Method accuracy was confirmed through the calculation of electron and carbon recoveries. Fermentations with Anaerostipes caccae DSM 14662T, Roseburia faecis DSM 16840T, Roseburia hominis DSM 16839T, and Roseburia intestinalis DSM 14610T revealed similar patterns of metabolite production with butyrate, CO2, and H2 as the main metabolites. R. faecis DSM 16840T and R. intestinalis DSM 14610T were able to degrade oligofructose, displaying a nonpreferential breakdown mechanism. Lactate consumption was only observed with A. caccae DSM 14662T. Roseburia inulinivorans DSM 16841T was the only strain included in the present study that was able to grow on fructose, oligofructose, and inulin. The metabolites produced were lactate, butyrate, and CO2, without H2 production, indicating an energy metabolism distinct from that of other Roseburia species. Oligofructose degradation was nonpreferential. In a coculture of R. inulinivorans DSM 16841T with the highly competitive strain Bifidobacterium longum subsp. longum LMG 11047 on inulin, hardly any production of butyrate and CO2 was detected, indicating a lack of competitiveness of the butyrate producer. Complete recovery of metabolites during fermentations of clostridial cluster XIVa butyrate-producing colon bacteria allowed stoichiometric balancing of the metabolic pathway for butyrate production, including H2 formation.
The implementation of 16S rRNA gene-based analytical techniques in the ongoing exploration of the microbial diversity of the human colon ecosystem has both broadened and sharpened the prevailing image of its population (17, 24, 32). While a rather conservative perception of the composition of the colon microbiota has dominated gut research for several decades (36), recent studies have revealed the importance of previously largely neglected bacterial groups and have reduced historically numerically overestimated subpopulations to their actual (marginal) size (8, 22, 52). The human colon has been shown to be a remarkably selective environment, which is reflected by a rather shallow microbial diversity (32). Species belonging to the bacterial divisions Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria make up more than 98% of the bacterial population of the human colon (2, 17, 24). However, this superficial uniformity only covers an overwhelming diversity at the lower taxonomic levels; the human colon has been estimated to harbor between 500 and 1,000 species, representing over 7,000 strains, with up to 80% of them considered uncultivable using presently available methodologies (14, 28, 53).
Assessing identity and abundance of the major microbial groups composing the colon microbiota is a first and indispensable step toward a better understanding of the ecosystem of the large intestine (48). However, defining a complex ecosystem such as the human colon requires more than the construction of a catalog of its members (32). A major challenge of gastrointestinal microbiology lies in linking phylogenetic subgroups with particular ecological habitats and niches (7, 8, 23). The latter requires further development of highly discriminating 16S rRNA gene-targeted probes to monitor spatial bacterial distribution, combined with renewed efforts toward species isolation through the application of innovative cultivation methods and media, and extensive metabolic characterization of representative strains (19, 35, 48).
Recently, a global ecological approach, combining efforts in probe development (1, 27), species isolation (3), and metabolic characterization (4, 11, 15, 20), has led to the identification of a functional group of microorganisms, composed of species belonging to the clostridial clusters IV and XIVa, that are responsible for colon butyrate production. As butyrate is regarded as a key metabolite for the maintenance of colon health, this functional subunit of the colon microbiota could have a major influence on human well-being and might be considered as a target for prebiotic dietary interventions (25, 35, 45). Some recently described lactate- and/or acetate-converting colon butyrate producers have been reported to be able to degrade prebiotic inulin-type fructans, although the kinetics of their respective breakdown mechanisms have hardly been investigated (10, 20). The enhancement of colon butyrate production observed after consumption of oligofructose or inulin (6, 31, 40)—the so-called butyrogenic effect—as well as the limited stimulatory effect of these prebiotics on the clostridial cluster IV and XIVa colon populations (16, 30) have been attributed to cross-feeding with bifidobacteria, which are still considered the primary fructan degraders (5, 38). Anaerostipes caccae as well as Roseburia spp. have been shown to be able to (co)metabolize end products of bifidobacterial fructan fermentation (lactate and/or acetate) or to grow on short oligosaccharides and monosaccharides released by Bifidobacterium spp. during fructan degradation (4, 20).
Recently, many clostridial cluster IV and XIVa butyrate producers characterized in detail have been shown to produce gases, mainly CO2 and H2 (12, 15, 20, 46). Consequently, they might be responsible for an enhancement of gas production as a result of fructan fermentation, through either cross-feeding or direct degradation of inulin-type fructans (15, 16). Indeed, inulin-type fructan consumption has been reported to cause some gastrointestinal discomfort related to gas production—essentially, flatulence and bloating (43)—while bifidobacteria, the main beneficiaries of dietary fructan intake, do not produce gases (19, 49). Although CO2 and H2 production by colon butyrate producers could have implications for human intestinal well-being, (in vitro) production has not been satisfactorily monitored up to now, probably due to limited availability of a performant apparatus for (online) gas analysis (15, 20). Moreover, the currently proposed pathway for colon butyrate production does not provide a conclusive quantitative link between bacterial (co)substrate metabolism and H2 formation (11).
This study investigated the kinetics of inulin-type fructan degradation by representatives of the genera Anaerostipes and Roseburia. A method based on online gas chromatography (GC) was developed to assess gas production qualitatively and quantitatively in a continuously sparged fermentation vessel for complete coverage of metabolite production. The competitiveness of inulin-degrading butyrate producers was investigated through coculture fermentations with Bifidobacterium longum subsp. longum LMG 11047, a strain representing a highly competitive cluster of bifidobacteria that share both high fructose consumption and oligofructose degradation rates and are able to perform partial breakdown of inulin (18, 20). A stoichiometrically balanced pathway for butyrate production, including H2 production, is proposed.
MATERIALS AND METHODS
Microorganisms and media.
The butyrate-producing colon bacteria Anaerostipes caccae DSM 14662T, Roseburia faecis DSM 16840T, Roseburia hominis DSM 16839T, Roseburia intestinalis DSM 14610T, and Roseburia inulinivorans DSM 16841T were obtained from the Deutsche Sammlung von Mikro-Organismen und Zellkulturen (Göttingen, Germany); the highly competitive B. longum subsp. longum LMG 11047 strain was purchased from the Belgian Co-Ordinated Collections of Micro-Organisms/Laboratory for Microbiology Ghent (Ghent, Belgium). For inoculum preparation and storage purposes, all strains were grown anaerobically in reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, United Kingdom). Strains were stored at −80°C in RCM supplemented with 25% (vol/vol) glycerol as a cryoprotectant.
Fermentation experiments were performed in a medium for colon bacteria (MCB [51]) supplemented with 50 mM acetate (6.8 g liter−1 of CH3COO− Na+·3H2O; modified MCB [mMCB]). The pH of the medium was adjusted to 6.3 before autoclaving at 210 kPa and 121°C for 20 min. After separate sterilization, fructose (VWR International GmbH, Darmstadt, Germany), oligofructose (OraftiP95; BENEO-Orafti NV, Tienen, Belgium), inulin (OraftiHP; BENEO-Orafti), or lactate (VWR International) was added aseptically to mMCB as an energy source. Fructose, oligofructose, and inulin were added at concentrations of 50 mM fructose equivalents (FE); lactate was used at a concentration of 100 mM. Fructose and lactate were autoclaved under the same conditions as the mMCB; oligofructose and inulin were sterilized through membrane filtration using Minisart filters (pore size, 0.2 μm; Sartorius AG, Göttingen, Germany). OraftiP95 and OraftiHP are commercial powders derived from chicory roots. OraftiP95 is obtained through enzymatic hydrolysis of chicory inulin. It consists mainly of oligofructose (≥93.2%, wt/wt) but contains also some minor amounts of glucose, fructose, and sucrose (<6.8%, wt/wt). The degree of polymerization (DP) of the oligofructose fractions varies between 2 and 8, with an average of 4. OraftiHP contains inulin (≥99.5%, wt/wt), with a DP ranging from 12 to 65 and minor amounts of glucose, fructose, and sucrose (<0.5%, wt/wt). The average DP of the inulin chains exceeds 23, due to removal of the smaller molecules during processing.
Solid RCM was prepared by adding 1.5% (wt/vol) agar (Oxoid) to RCM.
Fermentation experiments.
Mono- and coculture fermentations were carried out in 2-liter Biostat B-DCU fermentors (Sartorius) containing 1.5 liters of mMCB supplemented with the energy source under study. Inocula were prepared as follows: strains were transferred from −80°C to RCM and incubated anaerobically at 37°C for 12 h in a modular atmosphere-controlled system (MG Anaerobic Work Station; Don Whitley Scientific Ltd., West Yorkshire, United Kingdom) that was continuously sparged with a mixture of 80% N2, 10% CO2, and 10% H2 (Air Liquide, Paris, France). Subsequently, the strains were propagated twice in mMCB with fructose as the sole energy source and finally added to the fermentor. During inoculum buildup, the transferred volume was always 5% (vol/vol). Anaerobic conditions during fermentation experiments were assured by continuously sparging the medium with N2 (Air Liquide) at a flow rate of 1.25 ml s−1. The fermentation temperature was kept constant at 37°C. A constant pH of 6.3 was imposed and controlled automatically, using 1.5 M solutions of NaOH and H3PO4. To keep the medium homogeneous, a gentle stirring of 100 rpm was applied. Temperature, pH, and agitation speed were controlled online (MFCS/win 2.1; Sartorius). Fermentations were followed for 48 h; samples for further analysis were taken at regular time intervals. All fermentations were performed in duplicate. The results and figures presented onward are representative for both fermentations.
Analyses of bacterial growth, carbohydrate consumption, and metabolite production. (i) Bacterial growth.
Growth was followed throughout all fermentations by measuring the optical density at 600 nm. During coculture fermentations with B. longum subsp. longum LMG 11047, bifidobacterial growth was quantified through plating on RCM agar. Plates were incubated for 24 h under anaerobic conditions as indicated above. Roseburia spp. and A. caccae grow poorly on solid culture media (20).
(ii) Carbohydrate, organic acid, and ethanol determinations.
Residual concentrations of glucose, fructose, oligofructose, and inulin (the latter two expressed in mM FE), as well as concentrations of acetate, butyrate, ethanol, formate, and lactate were determined through high-performance liquid chromatography as described previously (19). All samples were analyzed in triplicate. Concentrations not exceeding 10 mM were considered trace concentrations and are not represented in figures but only mentioned in the text and tables.
(iii) Breakdown of oligofructose and inulin.
Breakdown of the different fractions of oligofructose was analyzed in detail by gas chromatography as described previously (19, 29). Qualitative analysis of inulin breakdown (OraftiHP) was performed using high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD) as described previously (19). Samples were analyzed in duplicate.
(iv) Analysis of gas production.
Concentrations of H2 and CO2 in the fermentor gas effluents were determined online through GC (CompactGC; Interscience, Breda, The Netherlands). The apparatus was equipped with two analytical channels, each composed of a carrier gas module, an injection valve, a column oven, and a thermal conductivity detector. Transfer of effluent gases from the fermentor outlet to the CompactGC was performed using a sample pump coupled to a four-position selection valve, allowing sampling of three fermentation vessels and injection of calibration gas mixtures (Saphir calibration gases; Air Liquide). Reference gas flows for both thermal conductivity detectors were controlled by a digital gas module. CompactGC operating conditions were similar for both channels: valve temperature, 60°C; injection volume, 20 μl; carrier gas module mode, constant pressure (70 kPa); split flow, 5 ml min−1; reference flow, 1 ml min−1; column temperature, 60°C; detector temperature, 110°C. Injections were performed at atmospheric pressure. The CompactGC was controlled using the CGC Editor 1.53 software package (Interscience).
A first analytical channel, used for the determination of H2 concentrations in the effluent gases, consisted of a 2-m PoraBOND Q guard column (Varian Inc., Palo Alto, CA) coupled to a 10-m Molsieve 5A column (Varian). N2 (Air Liquide) was used as a carrier gas. A 54.5-s lasting backflush was imposed through the guard column 6.5 s after injection, preventing contaminants including H2O from reaching the Molsieve 5A column. CO2 concentrations in the effluent gases were quantified using a second analytical channel. It consisted of a 2-m PoraBOND Q guard column followed by a 10-m main column of the same type (Varian). Helium was used as a carrier gas. A 55.5-s lasting backflush was sent through the guard column 5.5 s after injection. The EZChrom Elite 3.2 software package (Agilent Technologies, Palo Alto, CA) was used for peak integration and subsequent component identification and quantification.
Gas flow through the fermentor vessels was quantified using a Flow Tracker 1000 (Agilent). H2 as well as CO2 concentrations in the gas effluents of each fermentor were determined every 30 min. Integration of the data acquired over 48 h of fermentation allowed calculation of the total gas production. To facilitate stoichiometric interpretation, data concerning gas production are represented as dissolved metabolites (in mM), corresponding with the amounts of CO2 or H2 produced from 1 liter of fermentation medium.
Carbon and electron recoveries.
Carbon recoveries (CRs; expressed in percentages) were calculated by dividing the total amount of carbon recovered in the sugar metabolites by the total amount of carbon present in the added energy source (20). Electron recoveries (ERs; expressed in percentages) for growth in mMCB supplemented with fructose or lactate were calculated by dividing the total amount of electrons released upon formal oxidation of the sugar metabolites to CO2 by the total amount of electrons released upon formal oxidation of the energy source consumed (15). The complex nature of oligofructose and inulin (mixes of different-chain-length polymers with equally different numbers of glycosidic bonds) did not allow ER calculations in these cases.
RESULTS
Inulin-type fructan degradation fingerprint of a monoculture of Bifidobacterium longum subsp. longum LMG 11047.
Monocultures of B. longum subsp. longum LMG 11047 in MCB supplemented with 50 mM FE of fructose, oligofructose, or inulin were described in a previous paper (19). An inulin-type fructan degradation fingerprint, encompassing growth, carbohydrate consumption, and metabolite production profiles for each substrate, detailed quantitative analysis of oligofructose degradation, and an HPAEC-PAD chromatogram providing qualitative data concerning inulin degradation, has been constructed (19). The main metabolites of B. longum subsp. longum LMG 11047 monoculture fermentations were acetate, lactate, formate, and ethanol. No gas production was observed. CRs varied between 96.4 and 116.2%. Detailed quantitative analysis of oligofructose breakdown showed simultaneous degradation of all different chain length fractions.
Inulin-type fructan degradation fingerprint of a monoculture of Roseburia inulinivorans DSM 16841T.
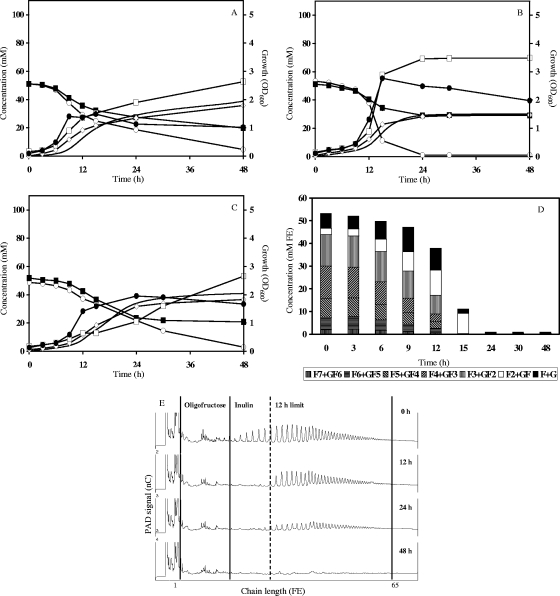
Of all clostridial cluster XIVa strains tested, R. inulinivorans DSM 16841T was the only strain that grew in mMCB supplemented with 50 mM FE of either fructose, oligofructose, or inulin (Fig. 1A, B, and C). CRs of these three monoculture fermentations were 97.7, 96.3, and 99.7%, respectively. ER for growth in mMCB supplemented with fructose was 96.8%. The metabolites produced included lactate, CO2, and butyrate; acetate was partially consumed during all fermentations. No H2 production was found. Growth on oligofructose was substantially faster than that on fructose and inulin. In all fermentations, a butyrate/CO2 production molar ratio of approximately 1:1 was observed. A metabolic shift toward relatively less acetate consumption and more lactate production—at the expense of butyrate and CO2 production—was observed with growth on oligofructose compared to growth on fructose or inulin. Detailed quantitative analysis of oligofructose breakdown by R. inulinivorans DSM 16841T revealed simultaneous degradation of all different chain length fractions, accompanied by a momentary accumulation of free fructose and inulobiose in the fermentation medium (Fig. 1D). Inulin degradation was characterized by initial breakdown of short fractions during the first 12 h of fermentation, followed by slow degradation of the long fractions upon further fermentation (Fig. 1 E).
FIG. 1.
Inulin-type fructan degradation fingerprint of Roseburia inulinivorans DSM 16841T. Growth, carbohydrate consumption, and metabolite production in mMCB supplemented with 50 mM FE of fructose (A), oligofructose (OraftiP95) (B), or inulin (OraftiHP) (C) are shown. ○, carbohydrate (FE); ▪, acetate; ⋄; butyrate; □, lactate; _, CO2; •, growth. (D) Oligofructose degradation. F, fructose; G, glucose. (E) Qualitative inulin degradation. An HPAEC-PAD chromatogram is shown.
Inulin-type fructan degradation fingerprints of monocultures of Anaerostipes caccae DSM 14662T, Roseburia faecis DSM 16840T, Roseburia hominis DSM 16839T, and Roseburia intestinalis DSM 14610T.
Graphical representations of the inulin-type fructan degradation fingerprints of monocultures of A. caccae DSM 14662T, R. faecis DSM 16840T, R. hominis DSM 16839T, and R. intestinalis DSM 14610T are provided in the supplemental material (Fig. S1 to S4); fermentation characteristics of monocultures showing substantial growth are summarized in Table 1. All strains were able to grow in mMCB supplemented with 50 mM of fructose. The main metabolites were CO2, H2, and butyrate; traces of lactate and formate were only found during fermentations with R. hominis DSM 16839T. Acetate was partially consumed during fermentations of all strains on fructose except for those with R. faecis DSM 16840T. A fructose/CO2 consumption/production molar ratio of approximately 1:2 was observed for all strains.
TABLE 1.
Growth, carbohydrate consumption, and metabolite production of Anaerostipes caccae DSM 14662T, Roseburia faecis DSM 16840T, Roseburia hominis DSM 16839T, Roseburia intestinalis DSM 14610T, and Roseburia inulinivorans DSM 16841T in mMCB supplemented with 50 mM FE fructose, oligofructose, or inulin or 100 mM lactate
| Strain | Mean ± SD consumption (mM) of substrate (after 48 h)
|
Mean ± SD production (mM) of metabolite (after 48 h)
|
Carbon recovery (%) | Electron recovery (%) | Substrate depletion time (h) | Max. cell populationf (time [h] max. was reached) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Energy source | Acetate | Butyrate | Carbon dioxide | Hydrogen gas | Lactate | |||||
| A. caccae DSM 14662T | 50.2 ± 0.1a | 28.7 ± 1.0 | 58.6 ± 0.4 | 104.0 ± 0.1 | 67.4 ± 0.1 | 0 | 94.3 | 91.0 | 24-48 | 2.7 (24) |
| A. caccae DSM 14662T | 86.1 ± 1.4b | 39.9 ± 1.8 | 63.2 ± 0.4 | 85.3 ± 0.1 | 42.8 ± 0.1 | 0 | 100 | 99.8 | >48 | 1.8 (9) |
| R. faecis DSM 16840T | 38.8 ± 0.2a | 0 | 30.1 ± 0.5 | 84.6 ± 0.1 | 90.1 ± 0.1 | 0 | 88.1 | 84.0 | >48 | 1.1 (6) |
| R. faecis DSM 16840T | 12.8 ± 0.2c | 6.5 ± 1.0 | 15.0 ± 0.4 | 23.4 ± 0.1 | 13.6 ± 0.1 | 0 | 92.6 | >48 | 1.1 (3) | |
| R. hominis DSM 16839T | 50.9 ± 0.2a | 20.8 ± 0.9 | 55.7 ± 0.1 | 95.2 ± 0.1 | 65.1 ± 0.1 | 7.0 ± 0.2 | 99.4d | 96.5d | 15-24 | 4.4 (15) |
| R. intestinalis DSM 14610T | 51.8 ± 1.3a | 36.8 ± 1.1 | 74.5 ± 0.9 | 103.5 ± 0.1 | 57.7 ± 0.1 | 0 | 104.4 | 104.4 | 15-24 | 3.1 (15) |
| R. intestinalis DSM 14610T | 56.0 ± 0.2c | 33.0 ± 0.4 | 67.5 ± 0.5 | 101.7 ± 0.1 | 62.9 ± 0.1 | 0 | 91.7 | >48 | 2.2 (48) | |
| R. inulinivorans DSM 16841T | 46.4 ± 0.4a | 31.1 ± 0.7 | 35.7 ± 0.2 | 38.9 ± 0.1 | 0 | 50.4 ± 0.3 | 97.7 | 96.8 | >48 | 1.5 (15) |
| R. inulinivorans DSM 16841T | 52.3 ± 0.2c | 22.0 ± 0.6 | 28.6 ± 0.7 | 30.1 ± 0.1 | 0 | 66.7 ± 0.6 | 96.3 | 15-24 | 2.8 (15) | |
| R. inulinivorans DSM 16841T | 45.8 ± 0.6e | 31.0 ± 1.1 | 35.5 ± 0.8 | 40.9 ± 0.1 | 0 | 51.0 ± 0.2 | 99.7 | >48 | 2.0 (24) | |
The energy source was fructose.
The energy source was lactate.
The energy source was oligofructose (FE).
Traces of formate (6.0 ± 0.3 mM) were included in the calculation.
The energy source was inulin (FE).
Determined by monitoring the optical density at 600 nm.
Only R. intestinalis DSM 14610T and, to a lesser extent, R. faecis DSM 16840T proved able to degrade oligofructose (Table 1). In both cases, substrate degradation was slower than with growth in mMCB supplemented with fructose. The main metabolites were CO2, H2, and butyrate; no lactate production was observed. Oligofructose degradation profiles revealed a nonpreferential breakdown mechanism, characterized by simultaneous degradation of all fractions (DP3 and DP4) and a minor accumulation of short fractions of oligofructose (mainly inulobiose) and fructose monomers in the fermentation medium (see Fig. S2 and S4 in the supplemental material). Both strains consumed acetate, although this was limited to trace amounts in the case of R. faecis DSM 16840T. An approximate oligofructose (FE)/CO2 consumption/production molar ratio of 1:2 was maintained. No explicit metabolic shift was observed in fermentations with R. intestinalis DSM 14610T on oligofructose compared to growth on fructose; acetate consumption by R. faecis DSM 16840T resulted in relatively more butyrate and less H2 production levels. Minor growth of A. caccae DSM 14662T and R. hominis DSM 16839T in mMCB supplemented with 50 mM FE of oligofructose could be attributed to the presence of contaminating monosaccharides in the commercial preparation administered (see Fig. S1 and S3 in the supplemental material). Also, in mMCB supplemented with 50 mM FE inulin, A. caccae DSM 14662T, R. faecis DSM 16840T, R. hominis DSM 16839T, and R. intestinalis DSM 14610T fermentations revealed only minor growth, which was caused by the presence of monosaccharides in the commercial substrate (see Fig. S1 to S4 in the supplemental material).
A. caccae DSM 14662T was able to grow in mMCB supplemented with 100 mM lactate (Table 1). Metabolite production levels represented an approximate lactate/acetate/butyrate/CO2/H2 consumption/production molar ratio of 4:2:3:4:2. No lactate consumption by Roseburia spp. was observed.
Butyrate production pathway in Anaerostipes caccae and Roseburia species.
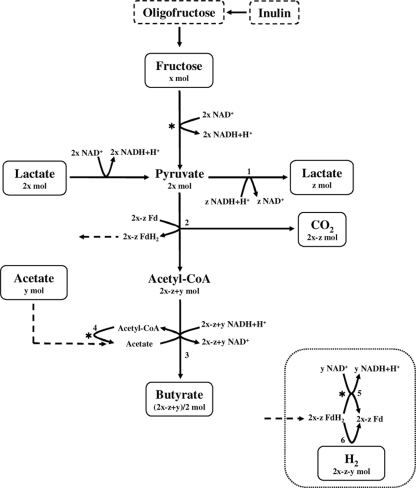
Analysis of substrate consumption and metabolite production profiles of growth of A. caccae DSM 14662T, R. faecis DSM 16840T, R. hominis DSM 16839T, and R. intestinalis DSM 14610T in mMCB supplemented with fructose or oligofructose (or lactate, if 2 mol of lactate is considered equal to 1 mol of FE) revealed acetate/fructose (FE) consumption rates varying between 0 (R. faecis DSM 16840T) and 0.6 (A. caccae DSM 14662T). Acetate consumption was negatively correlated with H2 production. Based on these observations, a modified metabolic pathway for these strains was proposed (Fig. 2). As metabolite production profiles of R. inulinivorans DSM 16841T revealed considerable lactate production and the absence of H2 formation, an alternative route for NADH+H+ disposal using lactate dehydrogenase was included (Fig. 2). The metabolic pathways proposed allowed theoretical calculations of butyrate, H2, and CO2 production levels based on fructose/oligofructose/inulin (in FE), lactate, and acetate consumption and lactate production measurements (Table 2).
FIG. 2.
Scheme for butyrate production in clostridial cluster XIVa colon bacteria. 1, lactate dehydrogenase; 2, pyruvate:ferredoxin oxidoreductase; 3, butyryl-CoA:acetate CoA transferase; 4, acetate kinase; 5, membrane-associated NADH:ferredoxin oxidoreductase; 6, hydrogenase; *, generation of ATP; x, mM FE fructose/oligofructose/inulin consumed; 2x, mM lactate consumed; y, mM acetate consumed; z, mM lactate produced.
TABLE 2.
Comparisons between calculated and measured butyrate, CO2, and H2 productiona
| Strain (fermentation substrate) | Production level (mM)
|
|||||
|---|---|---|---|---|---|---|
| Butyrate
|
CO2
|
H2
|
||||
| Calculated | Measured | Calculated | Measured | Calculated | Measured | |
| A. caccae DSM 14662T (fructose) | 64.6 | 58.6 | 100.5 | 104.0 | 71.8 | 67.4 |
| A. caccae DSM 14662T (lactate) | 63.0 | 63.2 | 86.1 | 85.3 | 46.2 | 42.8 |
| R. faecis DSM 16840T (fructose) | 38.8 | 30.1 | 77.7 | 84.6 | 77.7 | 90.1 |
| R. faecis DSM 16840T (oligofructose) | 16.0 | 15.0 | 25.6 | 23.4 | 19.2 | 13.6 |
| R. hominis DSM 16839T (fructose) | 57.8 | 55.7 | 94.8 | 95.2 | 74.0 | 65.1 |
| R. intestinalis DSM 14610T (fructose) | 70.2 | 74.5 | 103.7 | 103.5 | 66.9 | 57.7 |
| R. intestinalis DSM 14610T (oligofructose) | 73.0 | 67.5 | 113.1 | 101.7 | 80.1 | 62.9 |
| R. inulinivorans DSM 16841T (fructose) | 36.8 | 35.7 | 42.4 | 38.9 | 11.3b | 0.0 |
| R. inulinivorans DSM 16841T (oligofructose) | 30.0 | 28.6 | 38.0 | 30.1 | 16.0b | 0.0 |
| R. inulinivorans DSM 16841T (inulin) | 35.8 | 35.5 | 40.6 | 40.9 | 9.6b | 0.0 |
Based on fructose/oligofructose/inulin, lactate, and acetate consumption and lactate production by clostridial cluster XIVa butyrate producers in mMCB supplemented with 50 mM FE fructose, oligofructose, or inulin or 100 mM lactate (Fig. 1; Table 1). Calculations are based on the metabolic pathway proposed in Fig. 2.
Theoretical (Fig. 2) and measured H2 production levels were 0; deviant calculated values are to be attributed to measuring errors for input data (fructose/oligofructose/inulin and acetate consumption or lactate production).
Coculture fermentation with Bifidobacterium longum subsp. longum LMG 11047 and Roseburia inulinivorans DSM 16841T in mMCB supplemented with inulin.
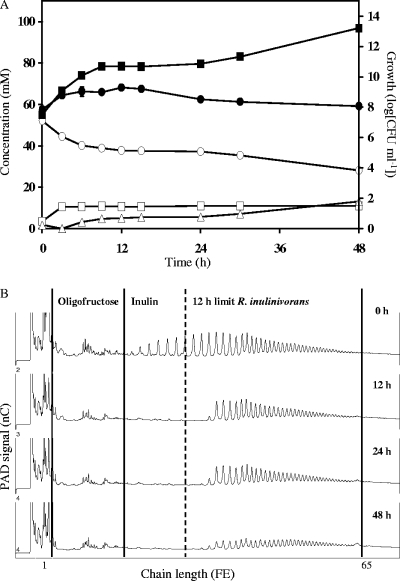
In a coculture of B. longum subsp. longum LMG 11047 with R. inulinivorans DSM 16841T, 24.0 ± 0.8 mM (mean ± standard deviation) FE of inulin was consumed within 48 h of fermentation (Fig. 3A). The main metabolites included acetate (41.8 ± 1.2 mM) and formate (11.3 ± 1.2 mM). Traces of lactate (8.7 ± 0.2 mM), ethanol (7.2 ± 0.8 mM), butyrate (4.0 ± 0.4 mM), and CO2 (3.1 ± 0.1 mM) were found. No H2 production was observed. The CR was 107.1%.
FIG. 3.
Growth, carbohydrate consumption, and metabolite production by a coculture of Bifidobacterium longum subsp. longum LMG 11047 with Roseburia inulinivorans DSM 16841T in mMCB supplemented with 50 mM FE of inulin (OraftiHP). (A) ○, inulin (FE); ▪, acetate; □, lactate; ▵, formate; •, growth of Bifidobacterium. (B) Qualitative inulin degradation. An HPAEC-PAD chromatogram is shown.
Qualitative analysis of inulin degradation revealed fast degradation of short-chain-length fractions of inulin during the first 12 h of fermentation, followed by slow degradation of long fractions (Fig. 3B). No accumulation of oligofructose or short fractions of inulin was observed. Qualitative comparison of HPAEC-PAD chromatograms for inulin degradation in the coculture with those of inulin degradation in the monoculture of R. inulinivorans DSM 16841T (Fig. 1E) revealed initial breakdown of longer-chain-length fractions in the coculture fermentation versus the monoculture.
DISCUSSION
Clostridial cluster XIVa Firmicutes are one of the most abundant bacterial groups in the human colon, making up around 25% of the colon microbiota (22). The cluster is composed of a broad group of bacteria, including species of Anaerostipes, Clostridium, Coprococcus, Eubacterium, Roseburia, and Ruminococcus (14). Many of its members, representing more than 7% of the fecal microbiota (1), have been reported to be major producers of butyrate in the colon (3). These acetate consumers include saccharolytic species belonging to the genus Roseburia (10, 12), as well as lactate converters such as A. caccae (46). Until now, studies including (partial) metabolic characterization of Roseburia spp. and A. caccae did not (10, 20) or did not satisfactorily (11, 12, 15, 46) assess gas production by these species quantitatively, although CO2 and H2 production are considered key elements of their energy metabolism. During the present study, a GC method was implemented to quantify both CO2 and H2 production through the online determination of their respective concentrations in the effluent gases of a fermentor system continuously sparged with N2. Although GC analyses are per definition discontinuous and offline, requiring subsequent injection, separation, and detection of components in a defined sample volume, the short analysis time (<120 s) of the CompactGC-based method presented here assured its semicontinuous (online) character. Regular sampling of effluent gases combined with gas flow quantification allowed accurate calculation of H2 and CO2 concentrations, with CRs and ERs for monoculture fermentations ranging between 88 and 105% and between 84 and 105%, respectively.
Complete quantitative recovery of gaseous and aqueous fermentation products through HPLC and CompactGC during the present study granted a deeper insight into the metabolic routes leading to clostridial cluster XIVa butyrate production. In general, the microbial pathway for butyrate production involves the condensation of two molecules of acetyl coenzyme A (CoA) and their subsequent reduction to butyryl-CoA (9, 11). For the final step of the pathway, the actual butyrate formation, two alternative metabolic routes have been described. Butyrate can be produced using a butyrate kinase, as has been demonstrated in some strains of Butyrivibrio fibrisolvens (9). Alternatively, a butyryl-CoA:acetate CoA transferase can move the CoA moiety to external acetate, leading to the production of butyrate and acetyl-CoA, as is the case for Roseburia spp. and A. caccae (34). Given the abundance of acetate in the human colon, it is not surprising that butyryl-CoA:acetate CoA transferase activity has been proven common among butyrate-producing strains, in contrast to butyrate kinase activity (13, 34, 40). Acetate is produced by nearly all heterotrophic colon anaerobes (8, 22, 36), including bifidobacteria (19, 49), and has been shown to be a key intermediate in cross-feeding interactions involving Bifidobacterium spp. and butyrate producers during growth on inulin-type fructans (4, 20). Metabolic studies of butyrate producers using the butyryl-CoA:acetate CoA transferase pathway have revealed a high degree of variation among such bacteria concerning their respective needs for external acetate as a cosubstrate. Growth of A. caccae DSM 14662T, a net acetate converter (46), in an acetate-free medium has been reported (20), while R. intestinalis DSM 14610T shows an absolute requirement for the presence of acetate as a cosubstrate (11, 20). Implementation of online GC during the present study revealed a negative correlation between acetate consumption and H2 production, leading to the formulation of a stoichiometrically balanced pathway for clostridial cluster XIVa butyrate production. The latter includes H2 production (11) and allows major flexibility regarding internal acetate recycling, probably using an acetate kinase (15). However, the pathway proposed in the present study should not be considered absolute, as the presence/absence of the production of certain metabolites during fermentation might indicate adaptation to particular growth conditions rather than the presence/absence of an operational enzyme system.
Clostridial cluster XIVa H2 production is thought to be mediated through the combined action of a pyruvate:ferredoxin oxidoreductase and a hydrogenase (26). Production of H2 using pyruvate:ferredoxin oxidoreductase is exergonic and generally unaffected by the partial pressure of H2 (37). Indeed, growth suppression associated with high partial H2 pressure was only observed for R. faecis DSM 16840T during growth in mMCB supplemented with fructose. This strain was shown unable to cometabolize acetate, leading to elevated H2 production levels, probably affecting final hydrogenase efficiency. Reverse activity of NADH:ferredoxin oxidoreductase, probably favored by high partial H2 pressures (37), has been reported previously in butyrate-producing bacteria (33, 47). Ferredoxin oxidation by a membrane-associated NADH:ferredoxin oxidoreductase would not only lead to NADH+H+ formation but is also thought to create a proton motive force, which allows extra ATP production (26).
Producing considerable amounts of lactate and no H2, R. inulinivorans DSM 16841T occupies a singular metabolic position among Roseburia species. As it probably lacks a (functional) hydrogenase, this strain has two routes to regenerate NAD+ out of the NADH+H+ produced during initial glycolysis (41). One route involves butyrate production using the butyryl-CoA:acetate CoA transferase pathway, with an obligatory equimolar consumption/production of acetate, butyrate, and CO2 (11, 15, 37). The other pathway generates lactate out of pyruvate using lactate dehydrogenase (37). The first route, which generates even more NADH+H+ through reverse activity of NADH:ferredoxin oxidoreductase during its initial reactions, is preferred over the second one in the case of slow growth, as shown for fermentations on inulin and fructose. The latter is probably linked with the generation of additional ATP through the generation of the proton motive force discussed above (26). During growth on oligofructose, lactate production was considerably higher.
Although butyrate production is considered a key element for the maintenance of human colon health (25, 45), the susceptibility of butyrate-producing strains toward prebiotic stimulation has hardly been investigated (42). However, a link between bacterial inulin-type fructan degradation fingerprints (20, 39, 50, 51), breakdown and/or uptake mechanisms (19), and in vitro competitiveness of colon bacteria (18) has been established. Extracellular fructan degradation, as is the case for Lactobacillus paracasei subsp. paracasei 8700:2 (39) and Bacteroides thetaiotaomicron LMG 11262 (18), has been shown to be competitively disadvantageous compared to its intracellular or cell-associated counterparts usually encountered in bifidobacteria (19, 49-51). Oligofructose degradation by R. faecis DSM 16840T, R. intestinalis DSM 14610T, and R. inulinivorans DSM 16841T revealed simultaneous degradation of all chain length fractions, a breakdown pattern that is associated with extracellular fructan degradation (39, 51). As such a degradation mechanism is accompanied by elaboration of free fructose into the extracellular environment, providing opportunistic competitors that are not able to degrade inulin-type fructans themselves with a valuable source of energy (21, 23), a decrease in competitiveness is unavoidable. Indeed, during coculture fermentations with R. inulinivorans DSM 16841T and B. longum subsp. longum LMG 11047, hardly any end products exclusively attributable to the roseburial metabolism (butyrate and CO2) were found. HPAEC-PAD chromatograms of fructan degradation during the first 12 h of fermentation showed a striking resemblance with those observed during B. longum subsp. longum LMG 11047 monoculture fermentations (19). In a monoculture, R. inulinivorans DSM 16841T was able to perform complete inulin degradation, an ability previously reported for Bacteroides thetaiotaomicron LMG 11262 (18) and L. paracasei subsp. paracasei 8700:2 (39), both extracellular fructan degraders. Most Bifidobacterium spp. (including B. longum subsp. longum LMG 11047) are—due their intracellular or cell wall-associated fructan degradation mechanisms—limited to degradation of short inulin fractions at the most (19, 44). However, HPAEC-PAD analysis of inulin breakdown by R. inulinivorans DSM 16841T revealed a preference for even shorter fractions than previously reported for B. longum subsp. longum LMG 11047 (18, 19). The ability of the latter strain to quickly metabolize longer-chain-length fractions of inulin than R. inulinivorans DSM 16841T appears decisive for competitiveness. All together, these findings seem to sustain the hypothesis that in vivo stimulation of butyrate-producing clostridial cluster XIVa colon bacteria (16, 30) is rather to be attributed to cross-feeding interactions than to primary fructan breakdown by these microorganisms (4, 20), stressing the importance of the presence of primary inulin-type fructan degraders, such as some bifidobacteria (19, 44), in the colon.
During the present study, an online GC-based method was implemented to assess gas production during growth of butyrate-producing colon bacteria both qualitatively and quantitatively in a continuously sparged fermentor system. Carbon and electron balances demonstrated the accuracy of the technique. The kinetics of both aqueous and gaseous metabolite production by butyrate-producing clostridial cluster XIVa colon bacteria during growth on fructose and inulin-type fructans revealed a deeper insight into their energy metabolism and substrate consumption patterns. The latter is indispensable for improving current understandings of the functional role of butyrate-producing colon bacteria in a complex ecosystem such as the human colon.
Supplementary Material
Acknowledgments
This research was funded by the Research Council of the Vrije Universiteit Brussel, the Fund for Scientific Research—Flanders (FWO-AL418), the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen; GBOU project IWT-010054), Yakult Belgium NV, and Institute Danone. Gwen Falony was the recipient of a Ph.D. grant from the IWT-Vlaanderen. Vicky De Preter and Frédéric Leroy were supported by a postdoctoral fellowship from the FWO.
We thank Gino Vrancken (Vrije Universiteit Brussel) and the analytical laboratory team of BENEO-Orafti NV for their support.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aminov, R. I., A. W. Walker, S. H. Duncan, H. J. M. Harmsen, G. W. Welling, and H. J. Flint. 2006. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl. Environ. Microbiol. 72:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belenguer, A., S. H. Duncan, G. Calder, G. Holtrop, P. Louis, G. E. Lobley, and H. J. Flint. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosscher, D., J. Van Loo, and A. Franck. 2006. Inulin and oligofructose as prebiotics in the prevention of intestinal infections and diseases. Nutr. Res. Rev. 19:216-226. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, J. M., G. C. Fahey, and B. W. Wolf. 1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 127:130-136. [DOI] [PubMed] [Google Scholar]
- 7.Chassard, C., K. P. Scott, P. Marquet, J. C. Martin, C. Del'homme, M. Dapoigny, H. J. Flint, and A. Bernalier-Donadille. 2008. Assessment of metabolic diversity within the intestinal microbiota from healthy humans using combined molecular and cultural approaches. FEMS Microbiol. Ecol. 66:496-504. [DOI] [PubMed] [Google Scholar]
- 8.Dethlefsen, L., P. B. Eckburg, E. M. Bik, and D. A. Relman. 2006. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21:517-523. [DOI] [PubMed] [Google Scholar]
- 9.Diez-Gonzalez, F., D. R. Bond, E. Jennings, and J. B. Russell. 1999. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 171:324-330. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, S. H., R. I. Aminov, K. P. Scott, P. Louis, T. B. Stanton, and H. J. Flint. 2006. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov., and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int. J. Syst. Evol. Microbiol. 56:2437-2441. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, S. H., A. Barcenilla, C. S. Stewart, S. E. Pryde, and H. J. Flint. 2002. Acetate utilization and butyryl-coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52:1615-1620. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, S. H., G. Holtrop, G. E. Lobley, A. G. Calder, C. S. Stewart, and H. J. Flint. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91:915-923. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, S. H., P. Louis, and H. J. Flint. 2007. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 44:343-350. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70:5810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falony, G., T. Calmeyn, F. Leroy, and L. De Vuyst. 2009. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl. Environ. Microbiol. 75:2312-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falony, G., K. Lazidou, A. Verschaeren, S. Weckx, D. Maes, and L. De Vuyst. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl. Environ. Microbiol. 75:454-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falony, G., A. Vlachou, K. Verbrugghe, and L. De Vuyst. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72:7835-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flint, H. J. 2004. Polysaccharide breakdown by anaerobic microorganisms inhabiting the mammalian gut. Adv. Appl. Microbiol. 56:89-120. [DOI] [PubMed] [Google Scholar]
- 22.Flint, H. J. 2006. The significance of prokaryote diversity in the human gastrointestinal tract, p. 65-90. In N. A. Logan, H. M. Lappin-Scott, and P. C. F. Oyston (ed.), Prokaryotic diversity: mechanisms and significance. SGM Symposium, vol. 66. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 23.Flint, H. J., E. A. Bayer, M. T. Rincon, R. Lamed, and B. A. White. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121-131. [DOI] [PubMed] [Google Scholar]
- 24.Frank, D. N., A. L. S. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamer, H. M., D. Jonkers, K. Venema, S. Vanhoutvin, F. J. Troost, and R. J. Brummer. 2008. The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27:104-119. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann, G., E. Jayamani, G. Mai, and W. Buckel. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 29.Joye, D., and H. Hoebregs. 2000. Determination of oligofructose, a soluble dietary fiber, by high-temperature capillary gas chromatography. J. AOAC Int. 83:1020-1025. [PubMed] [Google Scholar]
- 30.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa-associated microflora of the human large bowel. Gut 53:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Blay, G., C. Michel, H. M. Blottiere, and C. Cherbut. 1999. Prolonged intake of fructooligosaccharides induces a short-term elevation of lactic acid-producing bacteria and a persistent increase in cecal butyrate in rats. J. Nutr. 129:2231-2235. [DOI] [PubMed] [Google Scholar]
- 32.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 33.Li, F., J. Hinderberger, H. Seedorf, J. Zhang, W. Buckel, and R. K. Thauer. 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis, P., K. P. Scott, S. H. Duncan, and H. J. Flint. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102:1197-1208. [DOI] [PubMed] [Google Scholar]
- 36.Macfarlane, G. T., and J. H. Cummings. 1991. The colonic flora, fermentation and large bowel digestive function, p. 51-92. In S. F. Phillips, J. H. Pemberton, and R. G. Shorter (ed.), The large intestine: physiology, pathophysiology and disease. Raven Press Ltd., New York, NY.
- 37.Macfarlane, S., and G. T. Macfarlane. 2003. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62:67-72. [DOI] [PubMed] [Google Scholar]
- 38.Macfarlane, S., G. T. Macfarlane, and J. H. Cummings. 2006. Prebiotics in the gastrointestinal tract. Aliment. Pharmacol. Ther. 24:701-714. [DOI] [PubMed] [Google Scholar]
- 39.Makras, L., G. Van Acker, and L. De Vuyst. 2005. Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans exhibiting different degrees of polymerization. Appl. Environ. Microbiol. 71:6531-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison, D. J., W. G. Mackay, C. A. Edwards, T. Preston, B. Dodson, and L. T. Weaver. 2006. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br. J. Nutr. 96:570-577. [PubMed] [Google Scholar]
- 41.Nelson, D. L., and M. M. Cox. 2005. Lehninger's principles of biochemistry. W. H. Freeman and Company, New York, NY.
- 42.Ramirez-Farias, C., K. Slezak, Z. Fuller, A. Duncan, G. Holtrop, and P. Louis. 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 101:541-550. [DOI] [PubMed] [Google Scholar]
- 43.Roberfroid, M. B. 2005. The digestive functions: inulin and oligofructose as dietary fiber, p. 103-131. In M. B. Roberfroid and I. Wolinsky (ed.), Inulin-type fructans: functional food ingredients. CRC Press, Boca Raton, FL.
- 44.Rossi, M., C. Corradini, A. Amaretti, M. Nicolini, A. Pompei, S. Zanoni, and D. Matteuzzi. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 71:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheppach, W., and F. Weiler. 2004. The butyrate story: old wine in new bottles? Curr. Opin. Clin. Nutr. Metab. Care 7:563-567. [DOI] [PubMed] [Google Scholar]
- 46.Schwiertz, A., G. L. Hold, S. H. Duncan, B. Gruhl, M. D. Collins, P. A. Lawson, H. J. Flint, and M. Blaut. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilizing, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25:46-51. [DOI] [PubMed] [Google Scholar]
- 47.Seedorf, H., W. F. Fricke, B. Veith, H. Bruggemann, H. Liesegang, A. Strittimatter, M. Miethke, W. Buckel, J. Hinderberger, F. Li, C. Hagemeier, R. K. Thauer, and G. Gottschalk. 2008. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. USA 105:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van der Meulen, R., T. Adriany, K. Verbrugghe, and L. De Vuyst. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl. Environ. Microbiol. 72:5204-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Meulen, R., L. Avonts, and L. De Vuyst. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Meulen, R., L. Makras, K. Verbrugghe, T. Adriany, and L. De Vuyst. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl. Environ. Microbiol. 72:1006-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter, J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74:4985-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, J., M. A. Mahowald, R. E. Ley, C. A. Lozupone, M. Hamady, E. C. Martens, B. Henrissat, P. M. Coutinho, P. Minx, P. Latreille, H. Cordum, A. Van Brunt, K. Kim, R. S. Fulton, L. A. Fulton, S. W. Clifton, R. K. Wilson, R. D. Knight, and J. I. Gordon. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.